Abstract
Proteins homologous to the protein NPS (neck passage structure) are widespread among lactococcal phages. We investigated the hypothesis that NPS is involved in the infection of phage TP901-1 by analysis of an NPS− mutant. NPS was determined to form a collar-whisker complex but was shown to be nonessential for infection, phage assembly, and stability.
Bacteriophages of lactic acid bacteria (LAB) are an economic problem in the dairy industry, as phage infections can lead to slow or failed milk fermentation. Temperate Lactococcus lactis phage TP901-1 belongs to the P335 species of the Siphoviridae family, which is characterized by a small isometric head and a long noncontractile tail (1, 3, 18). The phage contains a collar and whiskers, which previous antibody-gold labeling experiments suggested were formed by the protein NPS (neck passage structure) (20), but the function of these structures is unknown. Recently, the collar-whisker labeling was confirmed for the similar lactococcal phage Tuc2009 (26). Collars and/or whiskers have often been observed on LAB phages (16, 17, 19, 21, 23), but these structures have not been further investigated. The corresponding complex of Escherichia coli phage T4 is formed from protein Gpwac, which forms both the fiber-shaped whiskers and the ring structure attached to the phage neck (8, 9). The function of the T4 whiskers is to assist long tail fiber assembly and retraction (7, 30). However, only a single, relatively short tail fiber is found in TP901-1 (32), and it is therefore unlikely that the TP901-1 whiskers have a function equivalent to that of the whiskers of T4. Proteins homologous to NPS are encoded by more than 20 sequenced LAB phage genomes, and many of these proteins have been proposed to constitute host-interacting proteins (5, 11, 12, 29). In order to establish whether NPS does constitute the collar-whisker complex of TP901-1 and to explore the function of these structures, we have mutated nps of TP901-1 and analyzed the resulting mutant with respect to morphology, protein profile, infection efficiency, and stability.
A TP901-1 NPS− prophage mutant was constructed by altering codons 76 and 77 of the nps gene (3) from ATA TCC AAG to ATC TAG AAG, hence introducing an in-frame amber mutation and an XbaI site. This was accomplished using the pGhost8 vector as previously described (25, 32). NPS− phage were subsequently induced with mitomycin C, purified by isopycnic gradient centrifugation, and compared to the wild type (wt) phage and the previously described tailless TP901-1 Dit− mutant (32).
The morphology of the collar and whiskers was investigated by transmission electron microscopy (TEM) as described previously (32). The diameter of the collar of the wt phage was determined to be 16 ± 2 nm (n = 17), and at least two whiskers were found attached to the collar. The thin whiskers had a total length of 33 ± 4 nm (n = 44) and were found to carry small globular ends (Fig. 1A). The latter is a novel feature observed for whiskers of phages. The NPS− mutant was found to lack both collar and whiskers, while all other structures of this mutant were morphologically indistinguishable from those of the wt phage (Fig. 1B). Collar and whiskers were, however, found at the DNA-filled heads of the tailless Dit− mutant (Fig. 1C), showing that the collar-whisker complex actually is assembled on the head structure. This is in contrast to the whiskers of E. coli phage T4, which are assembled on the virion after the joining of head and tail (9).
FIG. 1.
Transmission electron micrographs of TP901-1 phages negatively stained with uranyl acetate. A, wt TP901-1. Arrows indicate collar and whiskers. B, NPS− mutant. Arrows indicate missing collar and whiskers. C, tailless Dit− mutant. Arrows indicate collar and whiskers. Bar, approximately 50 nm.
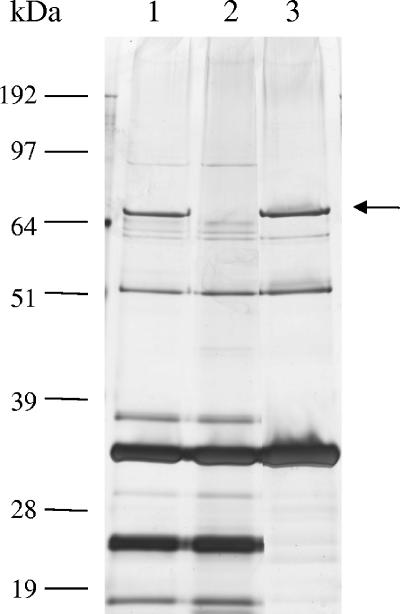
The protein contents of the wt, NPS−, and Dit− phages were examined in 10% Novex bis-Tris polyacrylamide gels (Invitrogen) under reducing conditions (Fig. 2). This showed the NPS− mutant to be lacking a single band with the 72-kDa predicted size for the NPS protein. In view of the results of the morphological analysis, it was therefore concluded that the NPS protein forms the collar-whisker complex of TP901-1. However, it cannot be ruled out that additional minor structural proteins also may be present in the complex. The protein profiles furthermore confirmed that NPS is not required for assembly of the other structural components, as no additional protein bands were missing from the NPS− mutant, which supports a similar observation made for a comparable mutant of lactococcal phage bIL41 (10). In agreement with the TEM analysis, the 72-kDa NPS protein band was found to be present in the Dit− mutant, which therefore confirmed that NPS must be regarded as a head-associated protein. This result was unexpected, since nps is located downstream of the tail genes and 11 kbp away from the major head protein gene (3). Moreover, the NPS-like protein of lactococcal phage bIL170 was recently found to be lacking in a head-enriched fraction of bIL170 (10). However, the bIL170 heads examined were separated from tails by heat treatment in acidic, EDTA-containing buffer (10), and we therefore speculate that the NPS-like protein was lost during this stringent procedure.
FIG. 2.
Silver stained sodium dodecyl sulfate-polyacrylamide gel with protein profiles of TP901-1 phages. Lane 1, wt TP901-1; lane 2, NPS− mutant; lane 3, tailless Dit− mutant. Molecular masses are indicated. The arrow indicates the 72-kDa NPS protein.
The exact number of whiskers could not be determined by examining negatively stained phages with TEM, because this procedure can visualize structures only in one plane. However, we speculate that TP901-1 contains three whiskers, each formed by NPS trimers. This hypothesis is based on the facts that (i) no more than two whiskers were observed on wt TP901-1 and the tailless Dit− mutant, (ii) the general symmetry of the portal vertex is 12-fold (4, 24), (iii) the NPS copy number is roughly estimated from the protein profile to be nine (results not shown), and (iv) the T4 whiskers and phage fibers in general appear to be trimeric (27).
In order to explore the possible host-interacting function of the NPS protein, several aspects of the infection process were examined and compared for the NPS− mutant and the wt phage. The infection efficiency of induced phage lysates was tested in plaque assays on host strain L. lactis 3107 essentially as described by Lillehaug (22). These experiments revealed NPS− mutant and wt phages to produce identical plaques (1 mm and turbid) at approximately the same titer (4 × 109 PFU/ml [NPS−] and 2 × 109 PFU/ml [wt]). The infectious phage of the NPS− lysate were subsequently verified as actual nps mutants by amplifying and sequencing the mutated regions from 10 plaque-forming isolates. All sequenced phages contained the amber mutation (results not shown), hence demonstrating that NPS is indeed not required for TP901-1 infection of L. lactis 3107. To determine whether the NPS protein played a role in host range extension, as previously hypothesized for a homologous protein (11), the infectivity of the NPS− lysate was tested in plaque assays on the two known TP901-1 indicator strains L. lactis 3107 and Wg2 (6). However, no differences in host range or infectivity were observed, as identical titers were obtained on both strains (data not shown). To determine if the collar-whisker complex assists the adsorption process, analogously to the long tail fibers of E. coli phage T5 (14, 15), the rate of TP901-1 adsorption to L. lactis 3107 was monitored essentially as described by Garvey et al. (13). Several independent experiments at different multiplicities of infection (MOIs) did not show a difference in the adsorption kinetics between mutant and wt phages (Table 1 shows the results at an MOI of 0.1). Based on these experiments, it was concluded that the NPS protein does not have a role in the infection process of TP901-1.
TABLE 1.
Rate of TP901-1 adsorption to L. lactis 3107
| TP901-1 | Adsorption (%) at indicated mina
|
||
|---|---|---|---|
| 1 | 2.5 | 10 | |
| wt | 77 | 94 | 98 |
| NPS− mutant | 84 | 96 | 99 |
Percentage of adsorbed phages at different time points after mixing of phages and bacteria at an MOI of 0.1.
In order to determine if the NPS protein had an effect on the stability of the TP901-1 virion, i.e., the capacity to keep an intact and infectious virion, we compared the long-term stability and robustness of NPS− mutant and wt phages. Purified phages (2.1 × 1011 PFU/ml [NPS−] and 5.2 × 1011 PFU/ml [wt]) were incubated at 4°C, and the amounts of intact and infectious phages were determined by TEM and plaque assays at 1- to 2-month intervals over a 1-year period. The titers of both phages were found to decrease at approximately the same rate, and the overall reductions were found to be 58% (8.8 × 1010 PFU/ml [NPS−]) and 90% (5.2 × 1010 PFU/ml [wt]) after 1 year of storage at 4°C. The effect of harsh handling was examined by measuring PFU/ml before and after pressure filtration through a 0.45-μm filter or intensive shaking (FastPrep 120; BIO101). Surprisingly, and in contrast to the general practice in phage handling, neither NPS− mutant nor wt phage were significantly affected by these rough treatments (results not shown). Taken together, these experiments strongly indicate that the NPS protein does not have a stabilizing function in phage TP901-1.
Proteins homologous to NPS are widespread among phages infecting L. lactis and Streptococcus thermophilus, and several of these proteins are putative tail fibers and/or determinants of host specificity. However, this study shows that proteins having fiber structure are not synonymous with host interaction proteins, as the fiber-shaped whiskers of TP901-1 were shown not to be involved in the infection process. We propose that proteins showing high sequence similarity to NPS, e.g., proteins encoded by the lactococcal phages r1t, ϕLC3, Tuc2009, bIL170, and bIL41 (2, 10, 11, 28, 29, 31), form a collar-whisker complex similar to that of TP901-1. Phage proteins with less pronounced similarity to NPS (e.g., ORF18 of streptococcal phage DT2 and ORF53 of lactococcal bIL309) share two conserved motifs, which have been described by Crutz-Le Coq et al. (11), and we hypothesize that these motifs form a structural conformation associated with phage fibers. The function of the TP901-1 collar-whisker complex awaits further studies, but our study shows the collar-whisker forming protein NPS to be unnecessary for TP901-1 assembly, infection, and virion stability. However, the prevalence of genes highly similar to nps among lactococcal phages indicates that these proteins have a somehow beneficial function.
Acknowledgments
This work was supported by The Royal Veterinary and Agricultural University of Denmark.
We thank Bernd Fahrenholz (FRCNF Kiel) for assistance with the TEM.
REFERENCES
- 1.Ackermann, H. W. 1998. Tailed bacteriophages: the order Caudovirales. Adv. Virus Res. 51:135-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blatny, J. M., L. Godager, M. Lunde, and I. F. Nes. 2004. Complete genome sequence of the Lactococcus lactis temperate phage φLC3: comparative analysis of φLC3 and its relatives in lactococci and streptococci. Virology 318:231-244. [DOI] [PubMed] [Google Scholar]
- 3.Brøndsted, L., S. Østergaard, M. Pedersen, K. Hammer, and F. K. Vogensen. 2001. Analysis of the complete DNA sequence of the temperate bacteriophage TP901-1: evolution, structure, and genome organization of lactococcal bacteriophages. Virology 283:93-109. [DOI] [PubMed] [Google Scholar]
- 4.Casjens, S., and R. Hendrix. 1988. Control mechanisms in dsDNA bacteriophage assembly, p. 15-91. In R. Calendar (ed.), The bacteriophages, vol. 1. Plenum Press, New York, N.Y. [Google Scholar]
- 5.Chopin, A., A. Bolotin, A. Sorokin, S. D. Ehrlich, and M.-C. Chopin. 2001. Analysis of six prophages in Lactococcus lactis IL1403: different genetic structure of temperate and virulent phage populations. Nucleic Acids Res. 29:644-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Christiansen, B., M. G. Johnsen, E. Stenby, F. K. Vogensen, and K. Hammer. 1994. Characterization of the lactococcal temperate phage TP901-1 and its site-specific integration. J. Bacteriol. 176:1069-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conley, M. P., and W. B. Wood. 1975. Bacteriophage T4 whiskers: a rudimentary environment-sensing device. Proc. Natl. Acad. Sci. USA 72:3701-3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coombs, D. H., and F. Arisaka. 1994. T4 tail structure and function, p. 259-281. In J. D. Karam (ed.), Molecular biology of T4. American Society for Microbiology, Washington, D.C.
- 9.Coombs, D. H., and F. A. Eiserling. 1977. Studies on the structure, protein composition and assembly of the neck of bacteriophage T4. J. Mol. Biol. 116:375-405. [DOI] [PubMed] [Google Scholar]
- 10.Crutz-Le Coq, A.-M., F. Cantele, S. Lanzavecchia, and S. Marco. 2006. Insights into structural proteins of 936-type virulent lactococcal bacteriophages. Arch. Virol. 151:1039-1053. [DOI] [PubMed] [Google Scholar]
- 11.Crutz-Le Coq, A.-M., B. Cesselin, J. Commissaire, and J. Anba. 2002. Sequence analysis of the lactococcal bacteriophage bIL170: insights into structural proteins and HNH endonucleases in dairy phages. Microbiology 148:985-1001. [DOI] [PubMed] [Google Scholar]
- 12.Desiere, F., W. M. McShan, D. van Sinderen, J. J. Ferretti, and H. Brüssow. 2001. Comparative genomics reveals close genetic relationships between phages from dairy bacteria and pathogenic streptococci: evolutionary implications for prophage-host interactions. Virology 288:325-341. [DOI] [PubMed] [Google Scholar]
- 13.Garvey, P., C. Hill, and G. F. Fitzgerald. 1996. The lactococcal plasmid pNP40 encodes a third bacteriophage resistance mechanism, one which affects phage DNA penetration. Appl. Environ. Microbiol. 62:676-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heller, K., and V. Braun. 1982. Polymannose O-antigens of Escherichia coli, the binding sites for the reversible adsorption of bacteriophage T5+ via the L-shaped tail fibers. J. Virol. 41:222-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heller, K., and V. Braun. 1979. Accelerated adsorption of bacteriophage T5 to Escherichia coli F, resulting from reversible tail fiber-lipopolysaccharide binding. J. Bacteriol. 139:32-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jarvis, A. W. 1989. Bacteriophages of lactic acid bacteria. J. Dairy Sci. 72:3406-3428. [Google Scholar]
- 17.Jarvis, A. W. 1984. Differentiation of lactic streptococcal phages into phage species by DNA-DNA homology. Appl. Environ. Microbiol. 47:343-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarvis, A. W., G. F. Fitzgerald, M. Mata, A. Mercenier, H. Neve, I. B. Powell, C. Ronda, M. Saxelin, and M. Teuber. 1991. Species and type phages of lactococcal bacteriophages. Intervirology 32:2-9. [DOI] [PubMed] [Google Scholar]
- 19.Jarvis, A. W., and J. Meyer. 1986. Electron microscopic heteroduplex study and restriction endonuclease cleavage analysis of the DNA genomes of three lactic streptococcal bacteriophages. Appl. Environ. Microbiol. 51:566-571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnsen, M. G., H. Neve, F. K. Vogensen, and K. Hammer. 1995. Virion positions and relationships of lactococcal temperate bacteriophage TP901-1 proteins. Virology 212:595-606. [DOI] [PubMed] [Google Scholar]
- 21.Lembke, J., U. Krusch, A. Lompe, and M. Teuber. 1980. Isolation and ultrastructure of bacteriophages of group N (lactic) streptococci. Zentbl. Bakteriol. Mikrobiol. Hyg. Abt. 1 Orig. C 1:79-91. [Google Scholar]
- 22.Lillehaug, D. 1997. An improved plaque assay for poor plaque-producing temperate lactococcal bacteriophages. J. Appl. Microbiol. 83:85-90. [DOI] [PubMed] [Google Scholar]
- 23.Loof, M., and M. Teuber. 1986. Heteroduplex analysis of the genomes of Steptococcus lactis “subsp. diacetylactis” bacteriophages of the P008-type isolated from German cheese factories. Syst. Appl. Microbiol. 8:226-229. [DOI] [PubMed] [Google Scholar]
- 24.Lurz, R., E. V. Orlova, D. Günther, P. Dube, A. Dröge, F. Weise, M. van Heel, and P. Tavares. 2001. Structural organisation of the head-to-tail interface of a bacterial virus. J. Mol. Biol. 310:1027-1037. [DOI] [PubMed] [Google Scholar]
- 25.Maguin, E., H. Prévost, S. D. Ehrlich, and A. Gruss. 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J. Bacteriol. 178:931-935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mc Grath, S., H. Neve, J. F. Seegers, R. Eijlander, C. S. Vegge, L. Brøndsted, K. J. Heller, G. F. Fitzgerald, F. K. Vogensen, and D. van Sinderen. 2006. Anatomy of a lactococcal phage tail. J. Bacteriol. 188:3972-3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitraki, A., S. Miller, and M. J. van Raaij. 2002. Conformation and folding of novel beta-structural elements in viral fiber proteins: the triple beta-spiral and triple beta-helix. J. Struct. Biol. 137:236-247. [DOI] [PubMed] [Google Scholar]
- 28.Parreira, R., R. Valyasevi, A. L. Lerayer, S. D. Ehrlich, and M.-C. Chopin. 1996. Gene organization and transcription of a late-expressed region of a Lactococcus lactis phage. J. Bacteriol. 178:6158-6165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seegers, J. F., S. Mc Grath, M. O'Connell-Motherway, E. K. Arendt, M. van de Guchte, M. Creaven, G. F. Fitzgerald, and D. van Sinderen. 2004. Molecular and transcriptional analysis of the temperate lactococcal bacteriophage Tuc2009. Virology 329:40-52. [DOI] [PubMed] [Google Scholar]
- 30.Terzaghi, B. E., E. Terzaghi, and D. Coombs. 1979. The role of the collar/whisker complex in bacteriophage T4D tail fiber attachment. J. Mol. Biol. 127:1-14. [DOI] [PubMed] [Google Scholar]
- 31.van Sinderen, D., H. Karsens, J. Kok, P. Terpstra, M. H. Ruiters, G. Venema, and A. Nauta. 1996. Sequence analysis and molecular characterization of the temperate lactococcal bacteriophage r1t. Mol. Microbiol. 19:1343-1355. [DOI] [PubMed] [Google Scholar]
- 32.Vegge, C. S., L. Brøndsted, H. Neve, S. Mc Grath, D. van Sinderen, and F. K. Vogensen. 2005. Structural characterization and assembly of the distal tail structure of the temperate lactococcal bacteriophage TP901-1. J. Bacteriol. 187:4187-4197. [DOI] [PMC free article] [PubMed] [Google Scholar]