Abstract
Furanone metabolites called AI-2 (autoinducer 2), used by some bacterial species for signaling and cell density-regulated changes in gene expression, are made while regenerating S-adenosyl methionine (SAM) after its use as a methyl donor. The luxS-encoded enzyme, in particular, participates in this activated methyl cycle by generating both a pentanedione, which is transformed chemically into these AI-2 compounds, and homocysteine, a precursor of methionine and SAM. Helicobacter pylori seems to contain the genes for this activated methyl cycle, including luxS, but not genes for AI-2 uptake and transcriptional regulation. Here we report that deletion of luxS in H. pylori reference strain SS1 diminished its competitive ability in mice and motility in soft agar, whereas no such effect was seen with an equivalent ΔluxS derivative of the unrelated strain X47. These different outcomes are consistent with H. pylori's considerable genetic diversity and are reminiscent of phenotypes seen after deletion of another nonessential metabolic gene, that encoding polyphosphate kinase 1. We suggest that synthesis of AI-2 by H. pylori may be an inadvertent consequence of metabolite flux in its activated methyl cycle and that impairment of this cycle and/or pathways affected by it, rather than loss of quorum sensing, is deleterious for some H. pylori strains. Also tenable is a model in which AI-2 affects other microbes in H. pylori's gastric ecosystem and thereby modulates the gastric environment in ways to which certain H. pylori strains are particularly sensitive.
There has been great interest in the ability of some bacterial taxa to signal their presence and abundance by secreting specific metabolites; to use these metabolites to monitor the density of members of their own species and of other species; and to respond with changes in patterns of gene expression, cellular phenotypes, and interactions with other microbes or host tissues—a set of behaviors termed “quorum sensing” (6, 17, 25, 27). Quorum sensing within individual species is often achieved using acyl-homoserine lactones (collectively called autoinducer 1 or AI-1), with much of the species specificity of AI-1 action stemming from differences in acyl chain length.
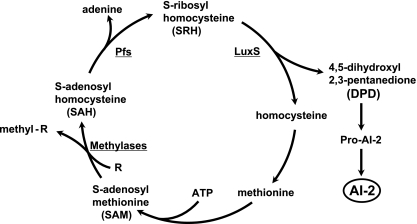
Chemically distinct furanone metabolites called AI-2 are also used by some species as signals for sensing cell density, both of unrelated taxa and of the same species. These furanones are by-products of a cyclic pathway that uses S-adenosyl methionine (SAM) as a methyl donor and then regenerates it (Fig. 1). In this pathway, methyl group transfer from SAM generates S-adenosyl homocysteine (SAH), a potentially toxic methyltransferase inhibitor. SAH is then deadenylated by a nucleosidase (Pfs) to generate S-ribosyl homocysteine (SRH). In the final enzymatic step in AI-2 synthesis, SRH is cleaved by the LuxS enzyme to generate 4,5-dihydroyxl-2,3-pentanedione, which undergoes chemical rearrangement and in some cases addition of boron or other substituents to generate a variety of furanones, some of which have AI-2 signaling activity (6, 25). The other cleavage product, homocysteine, serves as a precursor for methionine and then SAM synthesis. Despite use of AI-2 for quorum sensing by some taxa, AI-2s synthesized in other taxa could be simple by-products of the activated methyl cycle and of no regulatory significance (20, 25). It is not known if LuxS-mediated SRH consumption contributes importantly to depletion of the potentially toxic SAH intermediate and thus to fitness, or if Pfs-catalyzed conversion of SAH to SRH is sufficient, in any species in which AI-2 synthesis has been studied.
FIG. 1.
AI-2 synthesis as a by-product of metabolic flux in the activated methyl cycle. For simplicity, only those metabolites and enzymes most relevant to the present studies are presented. More-detailed descriptions of this cycle, including structures of intermediates, are presented elsewhere (20, 25).
Helicobacter pylori, the genetically diverse pathogen implicated in peptic ulcer disease and gastric cancer (4, 15), contains a luxS gene and exhibits luxS-dependent AI-2 synthesis but seems to lack close homologs of genes known to be involved in AI-2 uptake or AI-2-responsive transcriptional regulation (2, 20, 25). Furthermore, no effect of luxS gene inactivation on overall growth in culture, VacA toxin synthesis, protein profiles in two-dimensional gels, or motility in liquid cultures was detected for the H. pylori strains tested (10, 12). Additional studies indicated that a functional luxS gene diminishes the capacity of H. pylori to form a biofilm at air-liquid-glass interfaces (9) and that a functional luxS gene contributes to a cell density-dependent induction of expression of flaA-lacZ and flaA-cat reporter constructs (14). However, flaA expression was less stimulated by conditioned medium than was typical of responses to AI-2 in well-established models. These studies were carried out using pure cultures of H. pylori, however, whereas H. pylori's natural habitat consists primarily of gastric epithelial cell surfaces and overlying mucin, a niche that also can contain numerous other microbial species (3). Thus, if AI-2 signaling were important for H. pylori at all, this might be most evident in vivo, where AI-2 might affect coexisting microbes in ways that, in turn, impact on receptivity of the gastric mucosal environment to H. pylori.
Here we studied effects of luxS inactivation on colonization using two unrelated mouse-colonizing H. pylori strains, SS1 and X47. These two strains were chosen because they differ in their preferred sites of gastric colonization (antrum versus corpus, respectively [1]) and in their need for another gene of central metabolism, that encoding polyphosphate kinase 1 (PPK1) (responsible for inorganic polyphosphate synthesis [23]).
MATERIALS AND METHODS
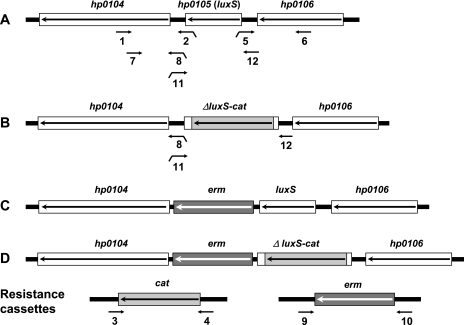
H. pylori strains SS1 and X47-2AL (for simplicity, here called X47) and derivatives diagrammed in Fig. 2 were used in these experiments. Standard growth conditions were used, including culture on brain heart infusion or brucella agar (supplemented with 7% blood or 7% serum) in microaerobic atmospheres (5% O2, 10% CO2, or simply 5% CO2, at Washington and Vanderbilt universities, respectively) (1, 14, 22, 23). These media were supplemented with 0.4% Isovitalex and the antibiotics amphotericin B (8 μg/ml), trimethoprim (5 μg/ml), and vancomycin (6 μg/ml). Nalidixic acid (10 μg/ml), polymyxin B (10 μg/ml), and bacitracin (200 μg/ml) were added to this medium when culturing H. pylori from mouse stomachs (referred to as H. pylori selective agar), thereby ensuring that all colonies recovered are H. pylori (1). Motility was assayed by inoculating patches of H. pylori growth into plates containing brucella serum medium solidified with 0.35% agar, as described previously (22). Deletion of most of the luxS gene and its replacement with a nonpolar cat (resistance) cassette, as well as insertion of a nonpolar erm resistance cassette just downstream of both ΔluxS-cat and luxS wild-type alleles (Fig. 2), was done by PCR without cloning (7, 22), using primers listed in Table 1. The structures of transformants made with PCR products diagrammed in Fig. 2 were tested by PCR (as in reference 23); all transformants had the expected allele replacements. Inoculation of 8- to 10-week-old BALB/cJ and C57BL/6J mice, their sacrifice 2 weeks later, culturing of H. pylori from them, and testing for bacterial genotype were carried out according to Washington Animal Studies Committee-approved protocols, as described previously (1, 23).
FIG. 2.
Structures of luxS-cat deletions and erm-marked insertions used in these studies. Alleles were constructed by assembly of separate PCR products using primers 1 through 12 diagrammed here, with sequences given in Table 1. Downward tails indicate extensions at 5′ ends of primers that overlap with and are complementary to other specific primers used here and allow assembly of alleles from individual PCR products, as diagrammed in reference 22. The extensions on primers 2 and 5 overlap with primers 3 and 4, respectively, and extensions on primers 8 and 11 overlap with primers 9 and 10, respectively.
TABLE 1.
PCR primersa
| Primer function, no., and name | Sequence | Location of 5′ end |
|---|---|---|
| Generation of ΔluxS deletion allele marked with cat | ||
| 1. HP105F2 | 5′-GCTATTGCCTTGCAACAAATCCCCGC | 242 bp after 5′ end of hp0104 |
| 2. HP105R3 | 5′-CCCAGTTTGTCGCACTGATAATTAGACAAACGCGCTGAGTGGTC | Complementary to catR primer and then 38 bp before 3′ end of hp0105 (luxS) |
| 3. catR | 5′-TTATCAGTGCGACAAACTGGG | 24 bp after 3′ end of cat |
| 4. catF | 5′-GATATAGATTGAAAAGTGGAT | 94 bp before 5′ end of cat |
| 5. HP105F3 | 5′-ATCCACTTTTCAATCTATATCGGTGTTTTCATGTTTTTAACTCC | Complementary to catF primer and then 10 bp after 5′ end of hp0105 (luxS) |
| 6. HP105R2 | 5′-ATACTTAGGCGGGCATAGCGATG | 558 bp before 3′ end of hp0106 |
| Insertion of erm determinant downstream of luxS locus and restoration of luxSwt or ΔluxS | ||
| 7. HP105CF2 | 5′-ATCCTTGTCAAGCCGTTATTGG | 160 bp after 5′ end of hp0104 |
| 8. HP105CR3 | 5′-TTCAATAGCTATAAATTATTTAATAAGTAAGTGGTCTGAAGTGGGGGTTTGA | Complementary to eryR primer and then 21 bp before 3′ end of hp0105 (luxS) |
| 9. eryR | 5′-TTACTTATTAAATAATTTATAGCTATTGAA | Coincides with 3′ end of erm |
| 10. eryF | 5′-CAATAATCGCATCAGATTGCAGTA | 118 bp before 5′ end of erm |
| 11. HP105CF3 | 5′-TACTGCAATCTGATGCGATTATTGTCAAACCCCCACTTCAGACCAC | Complementary to eryF primer and then coincides with 3′ end of hp0105 (luxS) |
| 12. HP105CR2 | 5′-GGAGTTAAAAACATGAAAACACC | 12 bp before 5′ end of hp0105 |
Overlaps between primers that allow assembly of separate PCR products are indicated by underlining: primers 2 with 3 and 5 with 6 for assembly of ΔluxS-cat allele by PCR with primers 1 and 6; primers 8 with 9 and 11 with 10 for assembly of the erm-linked luxSwt or ΔluxS-cat alleles with primers 7 and 6 or 7 and 12, respectively (see Fig. 2).
Two precautions were taken to guard against inadvertent attenuation of mouse-colonizing ability. First, cultures used for transformation with DNA containing the ΔluxS allele were derived from pools of about 10 colonies that had been recovered by culture on H. pylori selective agar from infected mice. Second, cells from pools of about 10 ΔluxS transformant colonies were then used to infect mice, and another set of pools of 10 colonies recovered by culture 2 weeks later (all carrying the ΔluxS allele, as expected) was then tested for ability to compete with isogenic mouse-passaged luxS wild-type (luxSwt) strains. No effect of mouse passage on the chloramphenicol or erythromycin resistance phenotype of the various transformants used here was detected. This use of pooled colonies and frequent mouse passage avoided the risk of inadvertently using a single colony that might have lost colonization ability due to mutation at loci unrelated to the luxS gene under study (see also reference 23).
The ability of conditioned medium from H. pylori cultures to induce luminescence was tested with the Vibrio harveyi reporter strain BB170 (10, 21). H. pylori cells were grown overnight in a 24-well dish (1 ml per well) and then subcultured into fresh brucella broth (20 ml) with shaking for various lengths of time. At each time point, aliquots of culture were collected, most cells were removed by centrifugation, and the supernatant-conditioned medium was sterilized by passage through a 0.22-μm filter. Supernatants were then stored until assayed at −80°C.
To detect AI-2-type compounds produced by H. pylori, V. harveyi BB170 was grown overnight in autoinducer bioassay (AB) medium (10), washed with fresh AB medium, and inoculated (1:5,000) into fresh AB medium containing 10% (vol/vol) of either H. pylori conditioned medium or, as a control, sterile brucella broth or medium conditioned by growth of this same V. harveyi strain. The V. harveyi reporter cultures were then grown at room temperature with shaking (150 rpm) for 6 h, and luminescence was measured using a Monolight 3010 luminometer. n-fold luminescence induction values were calculated as values obtained after adding conditioned medium versus values obtained after adding sterile brucella broth. The analysis was done five times with conditioned medium prepared on four different occasions.
RESULTS
To test if luxS is important in vivo, we constructed a PCR product containing a deletion of the luxS gene marked with a nonpolar cat resistance marker (Fig. 2). Wild-type strains were then transformed with the DNA containing this ΔluxS allele, and chloramphenicol-resistant (ΔluxS) transformant colonies were selected. Initial tests indicated that deletion of luxS from SS1 or X47 did not severely impair the ability of either strain to colonize mice: H. pylori densities were in the range of ∼106 CFU/stomach 2 weeks after inoculation in each of five mice tested with each H. pylori strain, which is similar to results obtained with isogenic wild-type parents (1).
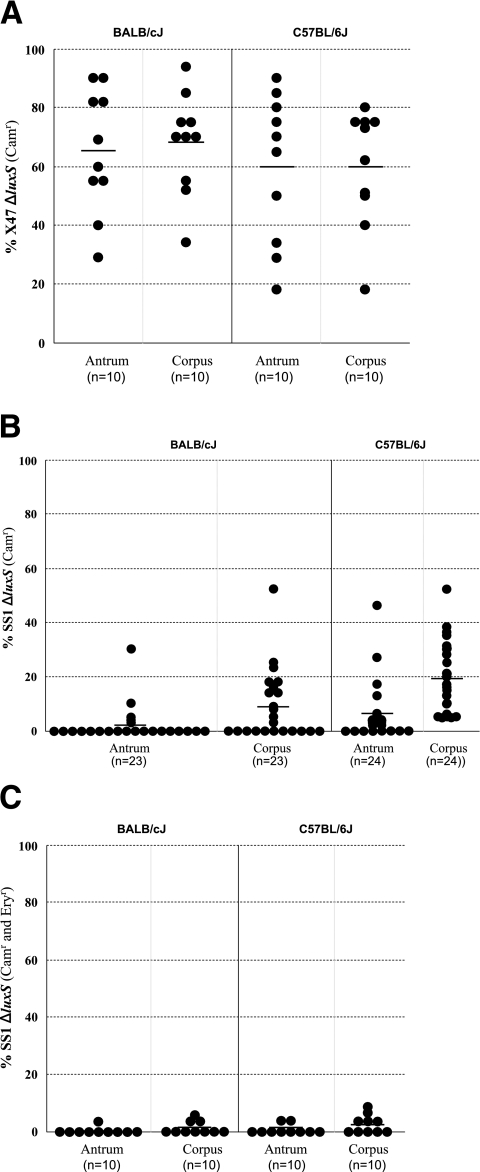
Competition tests were used to examine more critically the possibility of an effect of luxS inactivation on colonization ability. Mice were each inoculated with 1:1 mixtures of ΔluxS mutant and isogenic luxSwt parent strains and then sacrificed 2 weeks later. H. pylori was cultured from separated antrum and corpus tissues, and at least 20 separate colonies from each tissue from each mouse were tested for chloramphenicol resistance (ΔluxS) versus susceptibility (luxSwt). In the case of X47, the ΔluxS derivative comprised on average ∼63% of H. pylori from the antrum and corpus of each mouse line (Fig. 3A; see legend for details). In no case was there any indication that the ΔluxS allele decreased X47 fitness. Indeed, the X47 ΔluxS strain was slightly more abundant than its wild-type parent in the corpus of BALB/c mice (68% [±17%]; significantly greater than 50% [P = 0.01], one-sample sign test).
FIG. 3.
Competition tests of the importance of a functional luxS gene for mouse colonization. Mice of two different inbred lines (C57BL/6J and BALB/cJ) were inoculated with 1:1 mixtures of isogenic luxSwt and ΔluxS derivative strains (chloramphenicol sensitive and resistant, respectively) shown in Fig. 2. Mice were sacrificed 2 weeks later, antrum and corpus were separated, and H. pylori was recovered from each tissue by colony formation (see Materials and Methods). At least 20 single colonies from each tissue from each mouse were scored as resistant (ΔluxS) or susceptible (luxSwt) to chloramphenicol. Each point represents the ratio of two types from a different mouse. Horizontal lines depict mean ratios. Panel A. Strain X47 derivatives diagrammed in Fig. 2, lines A and B. Panel B. Strain SS1 derivatives diagrammed in Fig. 2, lines A and B. Panel C. Derivatives of the SS1 ΔluxS strain used in panel B that had been transformed with DNA containing a luxSwt allele linked to a downstream erm (resistance) insertion or with DNA containing the original ΔluxS allele linked to this erm cassette at the same site (see Fig. 2, lines C and D).
With SS1, in contrast, the ΔluxS mutant comprised, on average, only 2% (±7%) and 6% (±11%) of colonies from the antrum of 23 BALB/cJ mice and 24 C57BL/6J mice, respectively, and only 9% (±13%) and 20% (±13%) of colonies from the corpus of 23 BALB/cJ mice and 24 C57BL/6J mice, respectively (Fig. 3B). These yields were, in each case, far less than the 50% expected if ΔluxS had been neutral in the SS1 genetic background (P < 0.001).
Restoration of wild-type luxS.
In principle the lower fitness of SS1 ΔluxS could have been due to secondary mutations, selected if luxS inactivation had decreased bacterial fitness in culture, rather than to an effect of loss of luxS function itself. To test such a possibility, we first made PCR products in which a selectable erythromycin resistance marker (erm) had been inserted in the intergenic space just downstream of luxSwt and also at the same site downstream of a ΔluxS allele (Fig. 2C and D). These DNAs were each used to transform the SS1 ΔluxS strain (Fig. 2B) that had competed poorly with its luxSwt parent. This generated a new pair of isogenic luxSwt and ΔluxS derivatives of strain SS1, each marked with the erm gene just downstream of the luxS locus but again distinguishable by chloramphenicol susceptibility versus resistance. Preliminary tests indicated that each type of Ermr transformant colonized mice efficiently when inoculated alone, as expected. A mixture of these new isogenic luxSwt and ΔluxS strains was then used to inoculate 10 C57BL/6J mice and 10 BALB/cJ mice, as described above. Analyses of colonies recovered 2 weeks later showed that the strain that had retained the ΔluxS allele was less fit than its sibling, in which luxSwt had been restored (Fig. 3C). Indeed, it seemed that the cost of the ΔluxS mutation was more severe in the strain carrying the erm insertion than in a strain without the erm gene (compare Fig. 3B and C). One possible explanation invokes perturbation by erm in expression of downstream genes, which putatively encode a cyclic-nucleotide phosphodiesterase and a methyl-accepting chemotaxis protein (2, 24), despite this cassette having been engineered to remove likely transcription termination sequences (23). This model would invoke synergism between effects of the erm insertion on downstream genes and those caused by the ΔluxS allele. The ΔluxS allele is marked with cat, but studies using other chromosomal loci had indicated that this resistance determinant itself does not decrease strain SS1's fitness in mice (23). Hence, we conclude that SS1 ΔluxS's lower fitness in vivo stems primarily from loss of luxS activity per se, not the resistance marker used for selection.
Motility.
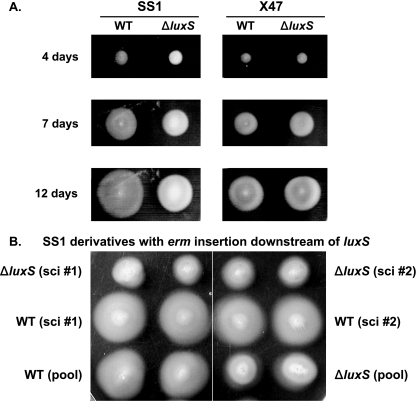
No effect of inactivation of luxS on H. pylori motility in broth was found by light microscopy (as in reference 12), although other studies using different strains had indicated that luxS inactivation decreased expression of flaA (14), which encodes one of the two H. pylori flagellins. We tested for effects of ΔluxS on motility with strains SS1 and X47, using a soft agar assay that detects changes in the strength of flagellum-driven swimming in a viscous environment, and also in chemotaxis along gradients that arise as nutrients are depleted by growth of bacterial colonies (18, 19). Figure 4A shows that ΔluxS mutant strains did indeed produce ever-expanding halos of growth, indicative of motility, although the SS1 ΔluxS halos were smaller than those of the isogenic SS1 wild type. In contrast, the halo sizes of X47 ΔluxS and isogenic X47 wild-type strains were not distinguishable either early or late during this growth period (Fig. 4A). Further tests showed that normal motility was restored if the ΔluxS allele was replaced with luxSwt linked to the erm resistance marker, whereas it was not restored in cells that had received the ΔluxS allele linked to this same erm insertion (Fig. 2C and D and 4B).
FIG. 4.
Motility assays. Panel A. Colonies of SS1 luxSwt, X47 luxSwt, or ΔluxS derivatives of these two strains were stabbed once into soft agar and incubated for 4, 7, or 12 days, as indicated on the left. Panel B. Derivatives of strain SS1 ΔluxS used in panel A that had been transformed with DNA containing an erm insertion downstream of either the ΔluxS or luxSwt allele (see Fig. 2, lines C and D) were tested. Two single colonies (sci #1 and sci #2) and also a pool from each transformation (separated by white vertical line) were tested by stabbing in duplicate into soft agar and incubation for 7 days.
To test for possible intercellular “complementation” of the ΔluxS strain's motility defect by AI-2 or any other metabolite produced by SS1 luxSwt, the ΔluxS-cat and luxSwt SS1 strains were mixed in a ratio of 1:10, 1:1, or 10:1, and these strain mixtures were inoculated in soft agar as for Fig. 4 and incubated for 7 days. Cell populations from the edges and centers of halos of growth were streaked to form single colonies, and these colonies were tested on chloramphenicol agar to estimate ratios of ΔluxS versus luxSwt strain types at each site. Both strain types were found at the centers of halos after 7 days in ratios equivalent to those used in the original inoculations (data not shown). In contrast, the ΔluxS strain was found at the halo edge only when ΔluxS had been in a 10-fold excess in the original inoculum, and then it comprised 27% of isolates on average. In contrast, the ΔluxS strain comprised <0.01% of bacteria found at the halo edge when the starting inoculum consisted of a 1:1 or 1:10 (ΔluxS:luxSwt) mixture (each of two separate trials). This indicated that the defect caused by luxS inactivation was not restored by metabolites from the wild-type strain. Based on transmission electron microscopy (as described in reference 23), SS1 luxSwt and SS1 ΔluxS cells did not seem to be different morphologically: most cells of each type were slightly curved and rod shaped and typically bore several flagella of similar lengths at one pole (data not shown).
luxS integrity and AI-2 synthesis.
Given the reproducible quantitative effects of luxS inactivation on fitness in vivo and motility in strain SS1, one class of explanations for the lack of effect of the ΔluxS allele in strain X47 invokes a naturally occurring mutation in luxS or elsewhere that prevents AI-2 synthesis or speeds its removal. PCR amplification and DNA sequence analysis of the X47's luxS gene revealed a complete 155-codon open reading frame (GenBank accession no. DQ777750), with 96% amino acid sequence identity to luxS genes in reference strains 26695 and J99 (2, 24). This suggested that luxS should be functional.
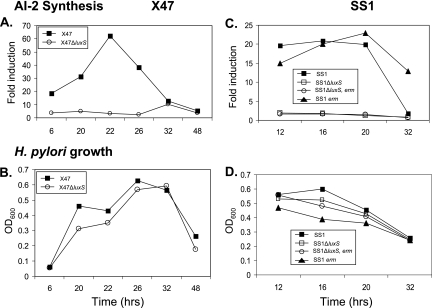
More critically, we bioassayed AI-2 levels in filter-sterilized culture supernatants by testing the abilities of these supernatants to induce luminescence in the Vibrio harveyi reporter strain BB170. Figure 5A and B show that the inducing activity from X47 wild type increased severalfold as cells grew from mid- to late log phase, ultimately attaining levels that were about 12-fold higher than the nonspecific background seen with media from cultures of the isogenic ΔluxS mutant. This level was about 30% of that seen using conditioned media from the canonical AI-2-producing Vibrio harveyi grown in parallel (J. T. Loh and T. L. Cover, unpublished data). The level of AI-2 activity in H. pylori-conditioned media decreased sharply after prolonged (>∼22 h) incubation, much as had been seen previously with an unrelated H. pylori strain (10). This might reflect intrinsic instability or degradation of AI-2 in culture. Data similar to those shown here were obtained in three other experiments, involving aliquots taken from fewer points in the growth curve (data not shown). It was also noteworthy that the maximum levels of AI-2-type activity produced by strain SS1 (in which the vigor of colonization is affected by luxS status) were substantially lower than those produced by strain X47 (Fig. 5A and C). Thus, the lack of effect of luxS on motility or colonization by X47 is not likely to be due to a defect in AI-2 accumulation.
FIG. 5.
Representative analyses of AI-2 production. AI-2 activity in H. pylori broth supernatants (i.e., conditioned medium) was measured in a bioluminescence assay using V. harveyi BB170 as an AI-2 reporter strain (Materials and Methods). Panels A and C show induction by conditioned brucella broth medium relative to results with sterile brucella broth. Panels B and D show optical densities at 600 nm (OD600) of the corresponding H. pylori cultures at the times when aliquots were withdrawn. The ΔluxS allele used was marked with a cat (resistance) gene (see Fig. 2). luxS status, when not specified in graph caption, implies a wild-type allele.
DISCUSSION
We found that deletion of luxS from H. pylori reference strain SS1 caused quantitative decreases in halo size (motility) in soft agar and in the ability to compete with isogenic SS1 wild type during mouse infection. Restoration of luxSwt by cotransformation with an erm resistance determinant inserted just downstream of luxS restored full motility and vigor in vivo relative to its isogenic ΔluxS sibling, also carrying erm at the same site. Thus, the ΔluxS-associated decrease in SS1 fitness likely stems from loss of luxS itself, not altered expression of downstream genes (predicted to encode a cyclic phosphodiesterase and a methyl-accepting chemotaxis protein [2, 24]) or mutation elsewhere in the genome. With the unrelated strain X47, in contrast, deletion of luxS did not significantly affect motility or vigor in vivo. DNA sequencing revealed a complete luxS open reading frame in this strain, and bioassays revealed normal luxS-dependent AI-2 production. Thus, the lack of effect of the ΔluxS allele on X47 phenotypes stems from this strain's tolerance of luxS inactivation, rather than a preexisting luxS deficiency in X47 wild type.
The decrease in SS1 halo size caused by the ΔluxS allele in soft agar is in accord with the previously reported ∼2- to ∼10-fold decrease in expression of flaA-lacZ or flaA-cat transcription fusions after luxS inactivation in other H. pylori strains (14). However, transmission electron microscopy (as described in reference 23) indicated that most cells in SS1 ΔluxS cultures carried several flagella of apparently normal length, as did their SS1 wild-type parents (data not shown). Alternatively, this decrease in halo size might reflect altered chemotaxis, a bacterial behavior linked to the activated methyl cycle via methylation of key chemotactic regulators (5, 16). In either case, it will be interesting to test if the thicker biofilms at a glass-broth-air interface found after luxS inactivation truly stem from direct suppression of biofilm formation by AI-2 (9) or less-efficient swimming or chemotaxis away from the biofilms into planktonic phase. That others had not noticed an effect of luxS inactivation on motility (12) could be ascribed to methodologic differences in assays (liquid culture versus soft agar) or a feature of background genotype in the “Aston” strain they used that could have resulted in tolerance of a luxS deficiency, much as invoked here with strain X47.
In terms of possible fitness mechanisms, the in vivo cost of deleting luxS in SS1 could be ascribed to a lack of AI-2-directed signaling in this strain when at high density. We do not favor this explanation, however, because (i) the sequenced H. pylori genomes each seemed to lack homologs of genes that in other systems participate in responses to AI-2 signals (2, 20, 24) (although, given diversity in gene content in H. pylori, the possibility of unrecognized AI-2 response genes in certain strains, SS1 included, is not completely excluded); and (ii) density-dependent flaA gene transcription, one of the few events affected by luxS inactivation, was only weakly stimulated by adding conditioned medium to a culture of a luxS-null strain (14). Alternatively, the decreased fitness of SS1 ΔluxS might stem from metabolic disturbances caused by the loss of luxS, specifically disruption of the cycle of SRH consumption and homocysteine synthesis (Fig. 1): e.g., if rates of homocysteine synthesis were limiting or SRH consumption was needed to deplete SAH, its potentially toxic precursor (Fig. 1) (8). The possibility of less-direct metabolic network explanations is well illustrated by the finding with Escherichia coli of a third quorum sensing signal, AI-3, that is chemically distinct from AI-2 but whose synthesis is also luxS dependent (26). AI-3's synthesis was traced to oxaloacetate, a metabolite also used in a second luxS-independent path for homocysteine biosynthesis. AI-3's luxS dependence was ascribed to siphoning of substrates from its synthesis into the alternative homocysteine biosynthesis pathway in ΔluxS strains (23). Although the types of metabolic connections and their relative importances vary among species, this study emphasizes the significance of network architecture and the potential of seemingly indirect effects to shape quantitative strain-variable phenotypes.
In summary, although previous reports of luxS-mutant-associated H. pylori phenotypes had tended to favor AI-2 signaling-based explanations, alternative physiologic explanations, such as those just outlined, seem more parsimonious to us at present. Further studies will be needed to define mechanisms underlying the differences in effects of ΔluxS alleles in strains X47 and SS1. Possibilities include differences between them in resistance to SAH and/or SRH intermediates, in LuxS-independent pathways for disposing of these intermediates, or in luxS-influenced pathways for generating other important metabolites. Also not excluded are models in which differences in dependence on AI-2 (if such metabolites are ever used by H. pylori) reflect interactions with coexisting bacterial species, some of which are AI-2 responsive and whose own activities affect host permissiveness for particular H. pylori strains. Formally, these luxS results are reminiscent of our finding that consequences of a polyphosphate kinase 1 deficiency varied with H. pylori genetic background. However, a functional polyphosphate kinase 1 gene was more important in vivo for X47 than for SS1 (23), the reverse of the luxS dependence seen here. Such contrasting outcomes illustrate that the spectra of potentially limiting metabolic factors vary among H. pylori strains and that no one strain is fully representative of this genetically diverse species. This consideration will become increasingly important as more H. pylori genomes are sequenced and as metabolic reconstructions and the discipline of systems biology (11, 13) become more refined.
Acknowledgments
We thank Mark Forsyth for stimulating discussions.
This work was supported by a fellowship from the Sankyo Foundation of Science (K. Ogura); by grants RO1 DK063041, P30 DK52574, and RO1 DK53623 from the National Institutes of Health; and by a grant from the Department of Veterans Affairs (T. L. Cover).
Footnotes
Published ahead of print on 25 August 2006.
REFERENCES
- 1.Akada, J. K., K. Ogura, D. Dailidiene, G. Dailide, J. M. Cheverud, and D. E. Berg. 2003. Helicobacter pylori tissue tropism: mouse-colonizing strains can target different gastric niches. Microbiology 149:1901-1909. [DOI] [PubMed] [Google Scholar]
- 2.Alm, R. A., L. S. Ling, D. T. Moir, B. L. King, E. D. Brown, P. C. Doig, D. R. Smith, B. Noonan, B. C. Guild, B. L. deJonge, G. Carmel, P. J. Tummino, A. Caruso, M. Uria-Nickelsen, D. M. Mills, C. Ives, R. Gibson, D. Merberg, S. D. Mills, Q. Jiang, D. E. Taylor, G. F. Vovis, and T. J. Trust. 1999. Genomic-sequence comparison of two unrelated isolates of the human gastric pathogen Helicobacter pylori. Nature 14:176-180. [DOI] [PubMed] [Google Scholar]
- 3.Bik, E. M., P. B. Eckburg, S. R. Gill, K. E. Nelson, E. A. Purdom, F. Francois, G. Perez-Perez, M. J. Blaser, and D. A. Relman. 2006. Molecular analysis of the bacterial microbiota in the human stomach. Proc. Natl. Acad. Sci. USA 103:732-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaser, M. J., and J. C. Atherton. 2004. Helicobacter pylori persistence: biology and disease. J. Clin. Investig. 113:321-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourret, R. B., and A. M. Stock. 2002. Molecular information processing: lessons from bacterial chemotaxis. J. Biol. Chem. 277:9625-9628. [DOI] [PubMed] [Google Scholar]
- 6.Camilli, A., and B. L. Bassler. 2006. Bacterial small-molecule signaling pathways. Science 311:1113-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chalker, A. F., H. W. Minehart, N. J. Hughes, K. K. Koretke, M. A. Lonetto, K. K. Brinkman, P. V. Warren, A. Lupas, M. J. Stanhope, J. R. Brown, and P. S. Hoffman. 2001. Systematic identification of selective essential genes in Helicobacter pylori by genome prioritization and allelic replacement mutagenesis. J. Bacteriol. 183:1259-1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Challan Belval, S., L. Gal, S. Margiewes, D. Garmyn, P. Piveteau, and J. Guzzo. 2006. Assessment of the roles of LuxS, S-ribosyl homocysteine, and autoinducer 2 in cell attachment during biofilm formation by Listeria monocytogenes EGD-e. Appl. Environ. Microbiol. 72:2644-2650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cole, S. P., J. Harwood, R. Lee, R. She, and D. G. Guiney. 2004. Characterization of monospecies biofilm formation by Helicobacter pylori. J. Bacteriol. 186:3124-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Forsyth, M. H., and T. L. Cover. 2000. Intercellular communication in Helicobacter pylori: luxS is essential for the production of an extracellular signaling molecule. Infect. Immun. 68:3193-3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Francke, C., R. J. Siezen, and B. Teusink. 2005. Reconstructing the metabolic network of a bacterium from its genome. Trends Microbiol. 13:550-558. [DOI] [PubMed] [Google Scholar]
- 12.Joyce, E. A., B. L. Bassler, and A. Wright. 2000. Evidence for a signaling system in Helicobacter pylori: detection of a luxS-encoded autoinducer. J. Bacteriol. 182:3638-3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kell, D. B., M. Brown, H. M. Davey, W. B. Dunn, I. Spasic, and S. G. Oliver. 2005. Metabolic footprinting and systems biology: the medium is the message. Nat. Rev. Microbiol. 3:557-565. [DOI] [PubMed] [Google Scholar]
- 14.Loh, J. T., M. H. Forsyth, and T. L. Cover. 2004. Growth phase regulation of flaA expression in Helicobacter pylori is luxS dependent. Infect. Immun. 72:5506-5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mobley, H. L. T., G. L. Mendz, and S. L. Hazell (ed.). 2001. Helicobacter pylori. ASM Press, Washington, D.C. [PubMed]
- 16.Parkinson, J. S. 2003. Bacterial chemotaxis: a new player in response regulator dephosphorylation. J. Bacteriol. 185:1492-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parsek, M. R., and E. P. Greenberg. 2005. Sociomicrobiology: the connections between quorum sensing and biofilms. Trends Microbiol. 13:27-33. [DOI] [PubMed] [Google Scholar]
- 18.Pittman, M. S., M. Goodwin, and D. J. Kelly. 2001. Chemotaxis in the human gastric pathogen Helicobacter pylori: different roles for CheW and the three CheV paralogues, and evidence for CheV2 phosphorylation. Microbiology 147:2493-2504. [DOI] [PubMed] [Google Scholar]
- 19.Suerbaum, S., C. Josenhans, and A. Labigne. 1993. Cloning and genetic characterization of the Helicobacter pylori and Helicobacter mustelae flaB flagellin genes and construction of H. pylori flaA- and flaB-negative mutants by electroporation-mediated allelic exchange. J. Bacteriol. 175:3278-3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun, J., R. Daniel, I. Wagner-Döbler, and A. P. Zeng. 2004. Is autoinducer-2 a universal signal for interspecies communication: a comparative genomic and phylogenetic analysis of the synthesis and signal transduction pathways. BMC Evol. Biol. 4:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Surette, M. G., and B. L. Bassler. 1998. Quorum sensing in Escherichia coli and Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 95:7046-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan, S., and D. E. Berg. 2004. Motility of urease-deficient derivatives of Helicobacter pylori. J. Bacteriol. 186:885-888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan, S., C. D. Fraley, M. Zhang, D. Dailidiene, A. Kornberg, and D. E. Berg. 2005. Diverse phenotypes resulting from polyphosphate kinase gene (ppk1) inactivation in different strains of Helicobacter pylori. J. Bacteriol. 187:7687-7695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, E. K. Hickey, D. E. Berg, J. D. Gocayne, T. R. Utterback, J. D. Peterson, J. M. Kelley, M. D. Cotton, J. M. Weidman, C. Fujii, C. Bowman, L. Watthey, E. Wallin, W. S. Hayes, M. Borodovsky, P. D. Karp, H. O. Smith, C. M. Fraser, and J. C. Venter. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 25.Vendeville, A., K. Winzer, K. Heurlier, C. M. Tang, and K. R. Hardie. 2005. Making ‘sense’ of metabolism: autoinducer-2, LuxS and pathogenic bacteria. Nat. Rev. Microbiol. 3:383-396. [DOI] [PubMed] [Google Scholar]
- 26.Walters, M., M. P. Sircili, and V. Sperandio. 2006. AI-3 synthesis is not dependent on luxS in Escherichia coli. J. Bacteriol. 188:5668-5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Walters, M., and V. Sperandio. 2006. Quorum sensing in Escherichia coli and Salmonella. Int. J. Med. Microbiol. 296:125-131. [DOI] [PubMed] [Google Scholar]