Abstract
During Integrated Ocean Drilling Program Expedition 301, we obtained a sample of black rust from a circulation obviation retrofit kit (CORK) observatory at a borehole on the eastern flank of Juan de Fuca Ridge. Due to overpressure, the CORK had failed to seal the borehole. Hot fluids from oceanic crust had discharged to the overlying bottom seawater and resulted in the formation of black rust analogous to a hydrothermal chimney deposit. Both culture-dependent and culture-independent analyses indicated that the black-rust-associated community differed from communities reported from other microbial habitats, including hydrothermal vents at seafloor spreading centers, while it shared phylotypes with communities previously detected in crustal fluids from the same borehole. The most frequently retrieved sequences of bacterial and archaeal 16S rRNA genes were related to the genera Ammonifex and Methanothermococcus, respectively. Most phylotypes, including phylotypes previously detected in crustal fluids, were isolated in pure culture, and their metabolic traits were determined. Quantification of the dissimilatory sulfite reductase (dsrAB) genes, together with stable sulfur isotopic and electron microscopic analyses, strongly suggested the prevalence of sulfate reduction, potentially by the Ammonifex group of bacteria. Stable carbon isotopic analyses suggested that the bulk of the microbial community was trophically reliant upon photosynthesis-derived organic matter. This report provides important insights into the phylogenetic, physiological, and trophic characteristics of subseafloor microbial ecosystems in warm ridge flank crusts.
The upper 500 m of oceanic crust is porous, which allows active circulation of seawater (7, 14). Fluids passing through ridge flank oceanic crust (1 to 65 million years old) account for 2% of the total volume of seawater (30) and flush this entire volume in about 70,000 years (68). Hence, ridge flank oceanic crust may represent a huge yet mostly ignored microbial habitat that has great significance for the global biogeochemical cycle (5, 6, 12, 17, 20, 23). Since hydrothermal alteration of seawater occurs more gently in ridge flanks than in seafloor spreading centers, ridge flank crustal fluids often contain SO42− (66, 69) and even NO3− and O2 (10), which potentially serve as electron acceptors for indigenous microbial communities. Previous studies have revealed the role of microbes in altering cool ridge flank crusts exposed at the seafloor, e.g., via oxidation of S and Fe (12). Additionally, textural and carbon isotopic composition analyses have suggested that microbial alteration of basaltic glass occurs even in high-temperature, deep ridge flank crusts (17, 18). However, very little is known about the diversity, physiology, and activity of microorganisms within the warm ridge flank crusts, in part due to the difficulty of recovering contamination-free samples from deep crust.
Recently, circulation obviation retrofit kits (CORKs) have provided unprecedented opportunities for microbiological investigation of oceanic ridge flanks (6, 7, 23, 35). To date, two microbial diversity studies have been performed using hot crustal fluids collected from a CORK deployed on the eastern flank of Juan de Fuca Ridge (6, 23). The presence of phylogenetically diverse Archaea and Bacteria, potentially transported from ridge flank crust, was revealed by culture-independent methods (6, 23). In addition, the chemical composition of CORK fluids suggested that various microbial processes occur within ridge flanks, including thermophilic sulfate reduction, ammonification of nitrate, and fermentation (6, 69). However, it remains to be determined which microorganisms are responsible for which microbial processes, and the chemical signatures might also be explained by abiotic hydrothermal alteration of seawater and diffusive exchange with the overlying sediment pore waters (6, 67, 69). In addition, the energy sources for the potential microbial activities remain unknown (6). In 2004, we retrieved an old CORK in order to replace it with a new one to prevent further leakage of crustal fluids (16). Visual inspection onboard revealed that there was a black rust deposit on the old CORK (16). As a further step toward understanding the microbial ecosystem within ridge flank crusts, we investigated the microbial community in the black rust by using both culture-dependent and culture-independent methods.
MATERIALS AND METHODS
Study site and sample collection.
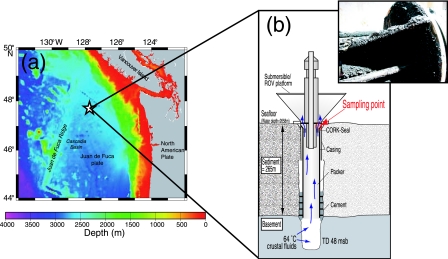
The old CORK was installed in borehole B at Ocean Drilling Program Site 1026 on the eastern flank of the Juan de Fuca Ridge during Ocean Drilling Program Leg 168 in July 1996 (15) (Fig. 1) (47°46′N, 127°46′W; 3.5-million-year-old crust; approximately 100 km from the ridge axis; water depth, 2,658 m; total penetration, 295 m; basement penetration, 48 m). CORKs consist of two parts: (i) a data logger and fluid sampling port sitting on the seafloor and (ii) instruments installed in the sealed borehole below (for a review, see reference 2). When the borehole was drilled for the deployment of CORK 1026B, 64°C crustal fluids flowed up at a rate of 84 liters min−1 (69). CORK 1026B did not completely seal the borehole, which allowed hot fluids to seep out at a rate of 3.1 liters min−1 (7). When we recovered this CORK for replacement on 4 August 2004, we found black rust on the CORK steel surface, immediately above the seal (Fig. 1). The black rust (30 cm3) was scaled and slurried for cultivation with sterile synthetic seawater (50 ml) containing 0.05% (wt/vol) Na2S · 9H2O under an N2 atmosphere (40). Approximately 100 cm3 of the sample was stored at −80°C for other analyses. For optical microscopic observation, 5 cm3 of the sample was fixed with filter-sterilized synthetic seawater (30 ml) containing 3.7% (wt/vol) formaldehyde. A total cell count (cells per cm3) was obtained onboard ship by direct cell counting of the formaldehyde-fixed slurry with DAPI (4′,6-diamidino-2-phenylindole) using epifluorescence microscopy (47).
FIG. 1.
(a) Location of CORK 1026B on the eastern flank of the Juan de Fuca Ridge. (b) Schematic diagram of CORK 1026B and borehole (modified from reference 16). The inset shows the black rust used in this study. msb, meters subbasement.
Mineralogical analysis.
The elemental composition and mineral components of the black rust were determined with a scanning electron microscope or with a microprobe using energy-dispersive X-ray spectroscopy (EDX) (model INCA Energy 200; Oxford Instruments, Oxford, United Kingdom), powder X-ray diffraction pattern analysis (model Rint 2000; Rigaku Corporation, Tokyo, Japan), and selected area electron diffraction (SAED) pattern analysis as previously described (54, 55).
Cultivation test.
To estimate the abundance and diversity of culturable microorganisms, a three-tube most-probable-number (MPN) test using a total of 32 different cultivation conditions (see Table S1 in the supplemental material) was performed onboard as previously described (41, 42). The cultivation conditions that gave positive enrichments are shown in Table 1. Pure cultures were obtained from the highest positive dilution tube by using the dilution-to-extinction technique (1). Sulfate-reducing strains and methanogens were identified by checking production of H2S and by checking autofluorescence by UV microscopy, respectively. The purity was routinely checked by microscopy.
TABLE 1.
Culturable population and phylogenetic characteristics of isolates from the black rust deposit on CORK 1026B
| Strain | MPN (cells per cm3) | Temp (°C) | Medium (reference) | Gas phase | Closest relative (% similarity)a |
|---|---|---|---|---|---|
| Methanogens | |||||
| Mc70 | 1.3 × 103 | 70 | MMJ (61) | 80% H2-20% CO2 (350 kPa) | Methanothermococcus thermolithotrophicus (99.5) |
| Mc55 | 1.3 × 103 | 55 | MMJ (61) | 80% H2-20% CO2 (350 kPa) | Methanothermococcus thermolithotrophicus (99.3) |
| Mc37 | 5.9 × 102 | 37 | MMJ (61) | 80% H2-20% CO2 (350 kPa) | Methanothermococcus thermolithotrophicus (99.1) |
| Ep55 | 5.9 | 55 | MMJHS (61) | 80% H2-20% CO2 (350 kPa) | Methanothermococcus thermolithotrophicus (99.5) |
| Ep70 | 5.9 | 70 | MMJHS (61) | 80% H2-20% CO2 (350 kPa) | Methanothermococcus thermolithotrophicus (99.4) |
| Sulfate reducers | |||||
| Tc37 | 49.6 | 37 | MJYPGS (41) | 100% N2 (200 kPa) | Desulfuromonas michiganensis (92.2) |
| Spi55 | 5.0 | 55 | MEtSR (this study)b | 80% N2-20% CO2 (250 kPa) | Desulfonatronovibrio hydrogenovorans (91.4) |
| Srb55 | 4.9 | 55 | MEtSR2 (this study)c | 80% H2-20% CO2 (350 kPa) | Desulfotomaculum geothermicum (96.1) |
| PM70-1 | NDh | 70 | DSMZ63-2 (this study)d | 80% H2-20% CO2 (200 kPa) | Archaeoglobus veneficus (95.1) |
| Fermenters | |||||
| Ag70 | 49.6 | 70 | MJYS (41) | 80% H2-20% CO2 (350 kPa) | Thermosipho melanesiensis (86.4) |
| Ag55 | 4.9 | 55 | MJYS (41) | 80% H2-20% CO2 (350 kPa) | Thermosipho melanesiensis (87.1) |
| Ag-C55 | 5.9 | 55 | MJYS2 (this study)e | 100% H2 (350 kPa) | Halothermothrix orenii (92.7) |
| Tc55 | 5.9 | 55 | MJYPGS (41) | 100% N2 (200 kPa) | Bacillus sp. strain BR (98.6) |
| Bal55 | 2.0 | 55 | MEtSR (this study) | 80% N2-20% CO2 (250 kPa) | Desulfotomaculum salinum (91.5) |
| Kimo37 | 2.0 | 37 | MJYP (50) | 100% N2 (200 kPa) | Cytophaga sp. strain AN-BI4 (88.6) |
| Yos55 | 2.0 | 55 | MJFM (this study)f | 80% N2-20% CO2 (250 kPa) | Thermohalobacter berrensis (91.5) |
| TH70-3 | ND | 70 | MSTH (this study)g | 100% N2 (200 kPa) | Thermosipho atlanticus (98.1) |
Determined by using the FASTA program of the DDBJ.
MEtSR contained 10 mM ethanol, 0.1 g yeast extract, 1 g NaHCO3, 1.5 g Na2SO4, 0.5 g Na2S, and 1 mg resazurin in 1 liter of MJ synthetic seawater (49).
MEtSR2 contained 0.5 g acetate, 0.5 g lactate, 0.5 g pyruvate, and 0.5 g, citrate instead of ethanol in 1 liter of MEtSR medium.
DSMZ63-2 was DSMZ medium 63 containing 2.5% Fe(NH4)2(SO4)2 instead of FeSO4.
MJYS2 was MJYS medium containing no NaHCO3.
MJFM contained 0.5 g yeast extract, 0.5 g peptone, 0.5 g formate, 0.5 g lactate, 0.5 g acetate, 0.5 g glucose, 0.5 g maltose, and 0.5 g citrate in 1 liter of MJ synthetic seawater (49).
MSTH contained 1 g yeast extract, 1 g peptone, 1 g NaHCO3, 0.5 g Na2S, and 1 mg resazurin in 1 liter of marine medium (70) amended with SL-10 trace mineral solution (71).
ND, not determined.
Phylogenetic analysis of pure cultures.
Phylogenetic analysis of pure cultures was performed as described previously (41, 42). Genomic DNA was extracted from cell pellets of each isolate with a Soil DNA Mini prep kit (MO BIO Laboratories, Inc., Solana Beach, CA). The 16S rRNA gene was amplified by PCR with primers Eubac27F and 1492R (34) for the bacterial rRNA gene or with primers Arch21F (8) and 1492R (34) for the archaeal rRNA gene. Nearly complete sequences of the PCR products were directly determined for both strands. Sequences were used for similarity analysis with databases by using the FASTA algorithm of the DNA Data Bank of Japan (DDBJ) (36).
16S rRNA gene clone library.
Microbial DNA was directly extracted from the microbial community associated with the black rust sample using a Soil DNA Mega prep kit (MO BIO Laboratories). As a negative control to check for experimental contamination, a blank tube was subjected to the same process. The primers described above were used. The PCR conditions used have been described previously (29). The presence of members of the Thermococcaceae was also assessed by PCR using a Thermococcaceae-specific primer set consisting of primers TcPc173F and TcPc589R (52). For a positive control, DNA extracted from a chimney structure in the Iheya North hydrothermal field (41) was used as a template. Each amplicon was purified, and cloning and sequencing were performed by a previously described procedure (29). Primer Eubac27F or Arch21F was used for partial sequencing (approximately 500 bp) of the insert to determine the phylogenetic clone type (phylotype). Clones with ≥97% sequence similarity were assigned to the same phylotype. Approximately 1,400 bp of each representative rRNA gene clone sequence was determined for both strands. Chimeric sequences were searched by checking secondary structure anomalies, using the Bellerophon program (25) and fractional treeing (37). None of the sequences were found to be chimeric.
Construction of phylogenetic tree.
To determine phylogenetic affiliations of isolates and environmental sequences, 16S rRNA gene sequences were compiled by using ARB, version 20030822 (38), and were aligned with a database (26) updated with sequences from the DDBJ. The resulting alignments were manually checked against known secondary structure regions. Phylogenetic analyses were restricted to nucleotide positions that could be unambiguously aligned. Phylogenetic trees were generated by a distance method using PAUP* 4.0b10 (57) and ARB. Distances were estimated with the Jukes-Cantor correction. Bootstrap analyses with 100 trial replications were used to obtain confidence estimates for the tree topologies.
Dissimilatory sulfite reductase (dsrAB) gene clone library.
Amplification, purification, cloning, and sequencing of the dsrAB genes were performed as previously described (44) with DNA obtained as described above. Primers DSR1Fdeg and DSR4Rdeg (32) were used. dsrAB gene clones with ≥95% sequence similarity were assigned to the same clone type. Likewise, the dsrAB genes were also amplified from the sulfate-reducing strains obtained in this study. Approximately 2,000 bp of the dsrAB gene sequence of each representative clone and sulfate-reducing strain were determined for both strands by using internal primers as described by Nakagawa et al. (44). Deduced amino acid sequences were aligned with prokaryotic dissimilatory sulfite reductase amino acid sequences obtained from DDBJ using the CLUSTAL X software (63). A neighbor-joining tree based on Kimura two-parameter distances was constructed by using CLUSTAL X. A bootstrap analysis with 100 trial replications was performed.
Quantification of archaeal 16S rRNA genes.
Quantification of archaeal 16S rRNA genes in the whole microbial 16S rRNA gene assemblage was performed by real-time PCR with TaqMan probes as previously described (58). A dilution series of a DNA sample was prepared and assayed with a universal 16S rRNA gene mixture and an archaeal 16S rRNA gene mixture (58) as standards for quantification of the whole microbial 16S rRNA gene and the archaeal 16S rRNA gene, respectively.
Quantification of dsrAB genes.
The abundance of dsrAB genes in the sample was estimated by real-time PCR with a SYBR Premix Ex Taq kit (TaKaRa, Shiga, Japan) using primers DSR1Fdeg (32) and int350R (5′-GTGCAGCTCGTCCTGGTA-3′). The int350R primer was designed to anneal to the conserved region of the entire dsrAB genes sequenced in this study. PCR-amplified dsrAB genes from the sample were quantified spectrometrically and used as standards. Dilution series of the DNA sample and standards were prepared and assayed by using a real-time PCR system (model 7500; Applied Biosystems, Foster City, CA). The thermal cycle was as follows: initial denaturation of the template DNA at 95°C for 60 s, followed by 40 cycles of amplification in which each cycle consisted of denaturation at 95°C for 5 s, primer annealing, and extension at 60°C for 34 s. Following amplification, a melting curve analysis of the amplicon was performed. Melting temperature analysis confirmed that the specificity of detection was high in all cases.
Bulk sulfur isotopic analysis.
Prior to analysis, the ground sample was washed with distilled water three times and dried. Tin capsules containing a reference (IA-R036, IAEA-S-1, and IA-R025; Iso-Analytical, Cheshire, United Kingdom) or a washed sample plus vanadium pentoxide catalyst were loaded into the automatic sampler of an elemental-analysis isotope ratio mass spectrometer (ANCA-GSL; SerCon, Cheshire, United Kingdom). The capsules were combusted in the presence of oxygen at 1,080°C. The tin capsules flash combusted, raising the temperature in the region of the sample to ∼1,700°C. Sulfur dioxide was separated from N2 and CO2 using a packed gas chromatography (GC) column at 45°C. The resulting SO2 peak entered the ion source of the isotope ratio mass spectrometer, where it was ionized and accelerated. The measured isotopic composition was expressed as δ34SVCDT, which was defined as follows: δ34SVCDT = [(34S/32S)sample/(34S/32S)standard − 1] × 103, where (34S/32S)standard is the abundance ratio for the Canyon Diablo troilite (CDT).
Bulk carbon isotopic analysis.
Bulk carbon isotopic analysis was performed as described previously (56). Ground, lyophilized, and acid-fumed samples were analyzed by using a Thermo Electron DELTAplus Advantage mass spectrometer connected to an elemental analyzer (EA1112) through a ConFlo III interface. The measured isotopic composition was expressed as δ13C, which was defined as follows: δ13CVPDB = [(13C/12C)sample/(13C/12C)standard − 1] × 103, where (13C/12C)standard is the abundance ratio for the Pee Dee belemnite (PDB).
Compound-specific carbon isotopic analysis.
Total lipid was extracted from a lyophilized ground sample using a single-phase chloroform-methanol buffer system of Bligh and Dyer (3). Fatty acid methyl esters (FAMEs) were prepared with anhydrous methanol HCl at 100°C for 3 h (33). They were then analyzed by using a GCQ gas chromatography-mass spectrometry system (Shimadzu, Kyoto, Japan) and GC-carbon isotope ratio mass spectrometry with a Thermo Electron DELTAplus Advantage mass spectrometer connected to a GC (Agilent 6890; Agilent, Mountain View, CA) through a GC/C/C/III interface (56). Standard nomenclature was used for fatty acids, which were designated as follows: CX:Y, where X is the number of carbon atoms and Y is the number of double bonds. The isotopic compositions of the FAMEs were determined with an internal isotopic standard (for C19:0, δ13CVPDB = −29.80‰) with correction for the additional carbon atom from methanol-derivatizing reagents (δ13CVPDB = −39.04‰).
Nucleotide sequence accession numbers.
Sequences obtained in this study have been deposited in the DDBJ/EMBL/GenBank nucleotide sequence databases under the following accession numbers: AB260037 to AB260053 for 16S rRNA genes of isolates, AB260054 to AB260068 for representative 16S rRNA gene clones, AB260069 to AB260071 for dsrAB genes of isolates, and AB260072 to AB260076 for representative dsrAB gene clones.
RESULTS
Sample description.
We collected the black rust from a borehole seal instrument, CORK 1026B, which had been deployed for approximately 8 years on a 295-m-deep borehole in the eastern flank of Juan de Fuca Ridge (Fig. 1). CORK 1026B, like a deep-sea hydrothermal vent, began leaking hot crustal fluids soon after its deployment (7). The temperature of the effluent fluids at the time of recovery was not determined, but previous measurements at the seafloor indicated that it was approximately 54°C in 1997 to 1999 (6) and 62°C in 2002 (23). The black rust was found immediately above the CORK seal, where the leaking hot crustal fluids and cold bottom seawater mixed (Fig. 1). The high temperature surrounding the microbial habitat was clearly confirmed by the dominance of thermophiles (described below). The total cell density was estimated to be 1.9 × 106 cells per cm3.
Mineralogical analyses.
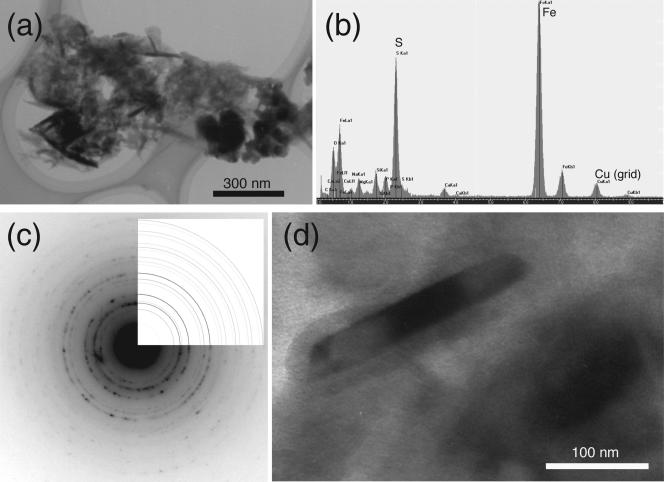
The black rust was attracted to a magnet, indicating that the sample contained magnetic iron-bearing minerals. Scanning electron microscope-EDX analysis revealed that the bulk of the sample consisted of O (65.5 atom%), Si (9.0 atom%), Al (8.2 atom%), C (5.2 atom%), Fe (4.5 atom%), Na (1.8 atom%), Mg (1.8 atom%), S (1.3 atom%), and Cl (1.2 atom%) as the primary (>1 atom%) elements. When the sample was observed with transmission electron microscopy, some cells that contained fine crystals (length, 30 to 350 nm) were found (Fig. 2a and d). EDX and SAED pattern analyses revealed that the crystals were greigite (Fe3S4) (Fig. 2b and c). It is known that sulfate-reducing bacteria form extracellular iron sulfide minerals, such as greigite, during dissimilatory sulfate reduction (48). The formation of greigite crystals thus suggested that there was microbial sulfate reduction in the CORK habitat.
FIG. 2.
(a and b) Transmission electron microscopy image (a) and EDX spectrum (b) of a prokaryotic cell in the black rust that had accumulated iron and sulfur. (c and d) SAED pattern (c), showing that a crystal accumulated on the cell surface (d) is greigite.
Cultivation test.
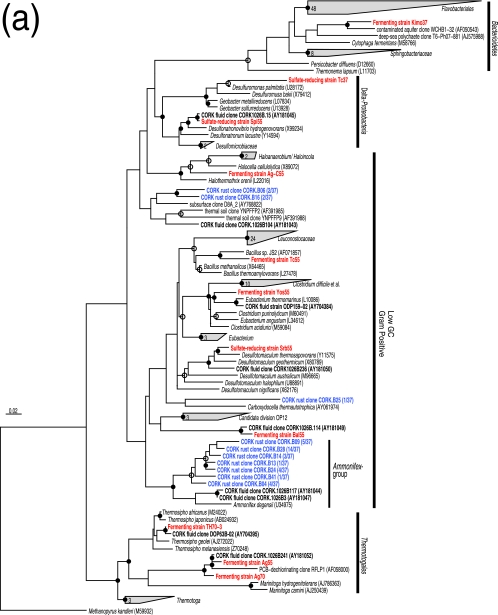
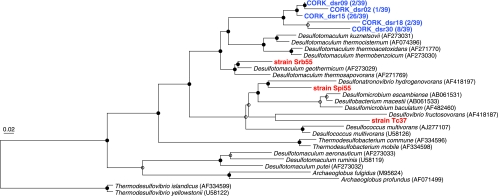
Microorganisms, including both Archaea and Bacteria, were quantitatively cultured and isolated in pure cultures. The overall size of the culturable population was approximately 1.3 × 103 cells per cm3. The numerically most abundant culturable population consisted of thermophilic and hydrogenotrophic methanogens (Table 1). Although the MPNs for other isolates were small, a variety of fermenters and sulfate reducers were also cultured (Table 1). Based on the 16S rRNA gene sequences, most of these isolates were only distantly related to previously cultured microorganisms and represented novel species or genera (Table 1). Among the isolates, thermophilic fermenters belonging to the order Thermotogales (i.e., strains Ag55 and Ag70) exhibited 99.3 and 99.1% 16S rRNA gene similarity with an environmental clone previously detected in fluids from the same borehole (6) (Fig. 3a). Likewise, sulfate-reducing strains Spi55 (a member of the Desulfovibrionales), Srb55 (Desulfotomaculum sp.), and PM70-1 (Archaeoglobus sp.) and fermenting strains Bal55 (a member of the Clostridiales) and TH70-3 (Thermosipho sp.) were closely related to other CORK fluid clones (6, 23) (Fig. 3a and b). None of the enrichments incubated under aerobic, microaerobic, or nitrate-reducing conditions were successful. It should be noted that none of the microorganisms commonly cultured from various deep-sea hydrothermal vent environments (i.e., members of the Thermococcales, Aquificales, and epsilon-Proteobacteria [40]) were cultured, even though the cultivation conditions employed in this study have been successfully used for these microorganisms (41, 42, 43, 59, 60). These results indicated that the black-rust-associated community differed from communities reported for deep-sea hydrothermal vents at seafloor spreading centers, while it shared phylotypes with communities previously detected in crustal fluids from the same borehole (6, 23). All MPNs were small compared to the total cell count. This may have resulted in part from the fact that the recovered CORK was exposed to oxic seawater and even to air for several hours due to the complex ship operation (16).
FIG.3.
Phylogenetic relationships of isolates and representative environmental clones as determined by neighbor-joining analysis of 16S rRNA gene sequences. Trees were constructed by using 419 (a) and 411 (b) sites that could be unambiguously aligned. Some sequences are indicated as follows: red, isolates obtained in this study; blue, clones sequenced in this study; boldface black, clones and isolates obtained in previous fluid surveys (6, 23). The remaining sequences were obtained from DDBJ. The clonal frequency of each representative clone obtained in this study and DDBJ accession numbers are shown in parentheses. Branch points conserved with bootstrap values of >75% (solid circles) and with bootstrap values of 50 to 74% (open circles) are indicated. Some groups are represented by shaded trapezoids that indicate the numbers of sequences. Scale bar = 0.02 expected change per nucleotide position. (a) Tree indicating the phylogenetic relationship among members of the Bacteria. (b) Tree indicating the phylogenetic relationship among members of the Archaea. PCB, polychlorinated biphenyl.
Culture-independent molecular analyses.
DNA was successfully extracted from black rust and used to construct libraries of the 16S rRNA genes of Bacteria and Archaea and the dsrAB genes. Negative controls yielded no amplification in any of the PCRs.
(i) Diversity of bacterial 16S rRNA gene.
Ten different phylotypes were identified from the bacterial 16S rRNA gene library (Fig. 3a). None of these phylotypes have been detected in any natural deep-sea vent environment. As observed for the microbial community in CORK fluid (6), clones most closely related to Ammonifex degensii were detected most frequently (clonal frequency, 86%) (Fig. 3a). Based on multiple alignments with more than 13,000 different rRNA gene sequences in our database, we found that some of the Ammonifex group clones sequenced in this study had characteristic long inserts (up to 93 bp long) between Escherichia coli positions 462 and 469. A. degensii KC4T is an extremely thermophilic, facultatively chemolithoautotrophic, low-G+C-content, gram-positive bacterium that was isolated from a terrestrial hot spring and grows via hydrogen, formate, or pyruvate oxidation coupled with nitrate, sulfate, or S0 reduction or pyruvate fermentation (24). Nevertheless, since the levels of sequence similarity between the Ammonifex group clones and A. degensii KC4T were less than 91% (Fig. 3a), the dominant Ammonifex group bacteria may have different physiological characteristics than A. degensii KC4T. The relatively minor bacterial sequences were also affiliated with other subgroups in the low-G+C-content gram-positive group (Fig. 3a). The close relatives of the less frequently detected bacteria (e.g., Carboxydocella thermautotrophica [53]) have been found in terrestrial thermal habitats but not in deep-sea environments.
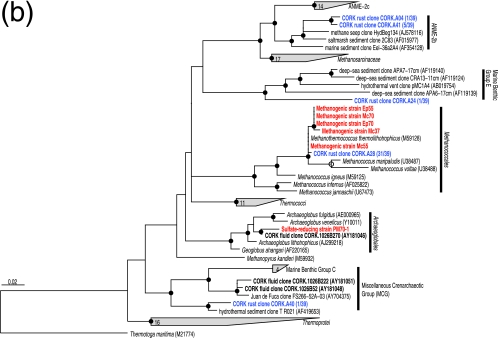
(ii) Diversity and quantification of archaeal 16S rRNA gene.
Five different archaeal phylotypes were identified (Fig. 3b). The 16S rRNA gene sequence of the most frequently detected archaeal phylotype (clonal frequency, 82%) was 99.8 to 99.9% similar to the sequences of thermophilic, hydrogenotrophic methanogens isolated in this study (Fig. 3b). No methanogens were detected in previous CORK fluid surveys (6, 23). The difference in community structure might be related to the fact that we sampled a solid formation, whereas in previous studies fluids were sampled. Archaeal sequences affiliated with ANME-2b, marine benthic group E, and the miscellaneous crenarchaeotic group, all of which are dominant archaeal groups in other deep-sea environments, such as methane seeps (45), pelagic sediments (28, 29, 65), and hydrothermal sediments (62), were detected less frequently (Fig. 3b). The absence of detectable Thermococcaceae members, hyperthermophilic archaea commonly found in natural deep-sea hydrothermal vent environments, was confirmed by PCR using Thermococcaceae-specific primers (52) (see Fig. S1 in the supplemental material).
Whereas the methanogenic archaea represented the most abundant culturable population, the total archaeal 16S rRNA gene population represented only 0.04% (standard deviation, 0.017%; n = 3) of total microbial 16S rRNA gene assemblage. These results indicated that members of the Bacteria with low culturability were numerically dominant and members of the Archaea with high culturability were a minority.
(iii) Diversity and quantification of dsrAB genes.
Five different groups were identified from the dsrAB gene library. The dsrAB gene clones formed a new clade, which was most closely related to a Desulfotomaculum species (Fig. 4). This clade was distantly related to dsrAB genes of sulfate-reducing strains isolated in this study (Fig. 4).
FIG. 4.
Phylogenetic tree based on the deduced amino acid sequences of the dsrAB genes of representative clones and sulfate-reducing strains. The tree was constructed by using 356 amino acid sequences. See the legend to Fig. 3 for additional information.
The abundance of dsrAB genes was estimated to be 9.5 × 107 copies per cm3 (standard deviation, 2.5 × 107 copies per cm3; n = 3). This abundance is approximately 10 times higher than the total cell count estimated by DAPI staining. Assuming that all cells of sulfate reducers have a single copy of the dsrAB genes, as suggested by the genome sequences of a variety of sulfate-reducing bacteria, the total cell count was most probably underestimated, potentially due to the dominance of cells accumulating greigite (Fig. 2) or spore-forming bacteria impermeable to DAPI. Considering the microbial rRNA gene community structure, the abundantly detected dsrAB genes were potentially derived from the Ammonifex group bacteria. This hypothesis is consistent with the ability of A. degensii KC4T to grow via dissimilatory reduction of sulfate (24).
Stable isotopic analysis.
The δ34SVCDT value of the bulk sample was determined to be 5.1‰ (standard deviation, 0.01‰; n = 2), which is depleted in 34S compared to sulfate in seawater (21‰) (51). As the sulfide concentration in the discharged fluids of the borehole was extremely low (<0.0005 mmol per kg) (6), the depletion of 34S probably resulted from microbial sulfate reduction in situ. This conclusion is supported by the detection of abundant dsrAB genes and the formation extracellular greigite, as observed by electron microscopy (Fig. 2).
The δ13CVPDB value of total organic carbon in the sample was −22.1‰. The carbon isotopic compositions of FAMEs from the sample were as follows: for C16:1, 2.7 area% and δ13CVPDB value of −45.4‰ ± 1.7‰ (mean ± standard deviation; n = 3); for C16:0, 46.4 area% and δ13CVPDB value of −27.4‰ ± 0.6‰ (mean ± standard deviation; n = 3); for C17:0, 10.6 area% and δ13CVPDB value of −44.1‰ ± 0.5‰ (mean ± standard deviation; n = 3); for C18:1, 14.1 area% and δ13CVPDB value of −24.4‰ ± 1.0‰ (mean ± standard deviation; n = 3); and for C18:0, 26.2 area% and δ13CVPDB value of −24.9‰ ± 1.3‰ (mean ± standard deviation; n = 3). Of the FAMEs, C16:1 and C17:0 were minor but significantly depleted in 13C relative to other FAMEs, indicating that there were at least two carbon sources or two types of carbon metabolism in the microbial community.
DISCUSSION
We characterized the microbial community inhabiting a black rust deposit on a CORK borehole observatory on the eastern flank of Juan de Fuca Ridge. Although the black rust had been exposed to hot ridge flank crustal fluids, this habitat cannot be considered a natural microbial habitat for obvious reasons. Hence, it is unlikely that the microbial community in ridge flank crust is identical to the community characterized in this study. However, many relatives of microorganisms found in black rust were previously detected in crustal fluids discharged from the CORK but not in bottom seawater or sediments (6, 23). It appears very likely that the microorganisms were transported and supported by the hot ridge flank crustal fluids, although the crustal fluids might be affected by corrosion of casing and CORK steel (6, 23, 69). Efforts have been made to develop technologies for collecting contamination-free ridge flank crustal fluids (2). This study is the first study in which potentially dominant and indigenous inhabitants of warm ridge flank crust were isolated in pure culture by using conventional cultivation approaches, indicating that the CORKs provide outstanding opportunities for retrieving microbes from this hard-to-access environment.
Porous oceanic crust may represent a significant microbial biosphere, which is often referred to as the “subseafloor ocean” (5, 12, 17, 30). Yet microbial life within oceanic crust remains largely unknown due to the difficulty of recovering intact, contamination-free oceanic crust. In hydrologically active regions, fluids emanating from crust have yielded important clues (6, 9, 23). Hydrothermal circulation of large volumes of seawater occurs in two different regions, seafloor spreading centers and ridge flanks (30). Based on evidence from various deep-sea hydrothermal systems located in the seafloor spreading centers, there is a growing consensus that the hydrothermal circulation of seawater and concomitant supply of inorganic energy and carbon sources might sustain a prosperous subseafloor microbial ecosystem (9, 19, 22, 41, 59). In seafloor spreading centers, hydrothermal alteration completely removes potential electron acceptors for microbial energy metabolism from intruded seawater and instead provides volatile energy and carbon sources (e.g., H2 and CO2). Therefore, thermophilic methanogens and thermophilic fermenters are likely to be the dominant organisms in habitats supported by altered hot fluids (9, 59).
In ridge flank crusts, the fluid-rock interaction and the supply of magmatic volatile species are less intense, and significant concentrations of electron acceptors, such as sulfate, have been found in circulating fluids (10, 66, 69). Similar fluids might exist even in crust in seafloor spreading centers where intruded seawater has not reached the high-temperature reaction zone. The flux of hydrothermal circulation through ridge flanks is increasingly recognized to be much greater than that in seafloor spreading centers (13, 39, 68). Therefore, hydrothermal circulation in warm ridge flank crusts may produce significant chemical fluxes to the overlying ocean (66, 68). Microbial effects on this potentially important geochemical process are poorly understood. The isolates obtained in this study are useful for evaluating the microbial roles in the warm ridge flank crust.
Microbial communities in ridge flank crusts may provide a key to a better understanding of the propagation of microorganisms inhabiting deep-sea hydrothermal vent environments in seafloor spreading centers. In vent fields located in seafloor spreading centers, there are cosmopolitan microbes, including members of the Thermococcales (hyperthermophilic fermenters), Archaeoglobales (mainly hyperthermophilic sulfate reducers), Aquificales (thermophilic hydrogen and/or sulfur oxidizers), Methanococcales (mesophilic to hyperthermophilic methanogens), and epsilon-Proteobacteria (mesophilic to thermophilic hydrogen and/or sulfur oxidizers) (40), suggesting that these microbial populations are less geographically isolated than macrofaunal communities living together (64). Especially, members of the Thermococcales have been found even in low-temperature diffusing fluids, suggesting that they are distributed globally in hot subseafloor environments (9, 19, 22). Although little is known about how these extremophiles could be distributed globally, hydrothermal circulation in ridge flanks is a potential stepping stone for the dispersal of microbial populations. The absence of these microorganisms, except for members of the Methanococcales and Archaeoglobales, was thus highly surprising. The most dominant members of the Bacteria and Archaea detected in the microbial community characterized in this study were Ammonifex group bacteria (potentially thermophilic sulfate reducers) and Methanothermococcus (thermophilic methanogens), respectively. Although more investigations of ridge flanks are necessary for further verification, recent studies, including this report, strongly suggest that oceanic crusts in hydrologically active ridge flanks might harbor subseafloor microbial communities that are distinct from the communities in pelagic sediments or crusts in seafloor spreading centers (6, 31).
The δ13CVPDB value of total organic carbon in the bulk sample was −22.1‰, which was 13C depleted relative to both CO2 dissolved in seawater (1‰) and CO2 in typical vent fluids in natural deep-sea hydrothermal fields (−1 to −10‰) (51). The energy sources available in hot ridge flank crustal fluids have been a subject of debate. Two different potential energy sources have been proposed: (i) organic matter released from sediments and (ii) H2 produced by seawater-basalt reactions at high temperatures (6, 23). Considering that neither the isotopic fractionation nor the potential carbon fixation pathway of the most dominant phylotype, Ammonifex group bacteria, has been determined due to the resistance of the organisms to cultivation, the δ13CVPDB value should be interpreted with caution. However, the carbon isotopic fractionation to biomass measured for A. degensii grown chemolithoautotrophically was reported to be only 2.8‰ (21). In addition, the concentration of total CO2 in fluids circulating through this ridge flank is very low (66, 69), suggesting that CO2 in the effluent fluids might be unable to serve as the major carbon source for the microbial community characterized. Considering that the δ13CVPDB value of the total organic carbon in the sample was comparable to that in ocean sediments (−27 to ca. −20 ‰) (51), it is most likely that the major carbon source is photosynthesis-derived organic matter released from sediments. Even in a CO2-rich hydrothermal field, it was recently demonstrated that photosynthesized organic matter released from deep sediments is the major carbon source of a bacterial mat (46).
It is well accepted that steel corrosion is stimulated by anaerobic microorganisms in the absence of oxygen (4), although the mechanism for this microbially influenced corrosion is still hotly debated. Molecular hydrogen, called cathodic hydrogen, which is formed on the steel surface by the dissociation of water, may serve as an energy source for hydrogen-oxidizing microorganisms, including sulfate reducers and methanogens (4). Especially sulfate reducers are notorious for stimulating steel corrosion by producing the corrosive agent hydrogen sulfide, either by utilizing cathodic hydrogen or organic matter as an energy source (4) or by directly utilizing electrons from steel (11). Although microbially influenced corrosion at high temperatures has been poorly investigated, the microbial community investigated in this study might be associated with the formation of black rust. Based on the results obtained in this study, we propose the following process for black rust formation and the concomitant succession of the microbial community: (i) colonization of microorganisms that are indigenous to ridge flank crust and are transported by effluent crustal fluids (precipitation of silicate from the hot fluids might help the microbial colonization [27]), (ii) activity of sulfate reducers, including Ammonifex group bacteria, is stimulated by sulfate supplied from bottom seawater, and (iii) increased production of hydrogen sulfide creates reducing microhabitats where methanogens can grow. This process might explain the absence of detectable methanogens in the effluent fluids (6, 23).
Much remains to be learned about biogeochemical processes within warm ridge flank oceanic crusts. The vertical distribution and horizontal distribution of microorganisms within ridge flank oceanic crusts have been poorly explored. The minimum energy, carbon, and nitrogen fluxes and the sources required to sustain microbial activities within the ridge flank crusts are of considerable interest but remain to be determined. Both further explorations of this substantial biosphere and further physiological characterizations of the retrieved microorganisms under in situ conditions are necessary.
Supplementary Material
Acknowledgments
This research used samples provided by the Integrated Ocean Drilling Program. We thank the captain and the crew of R/V JOIDES RESOLUTION for helping us to obtain samples. We thank Julie Huber for providing clone sequences and their frequencies. We also thank Anna-Louise Reysenbach for providing laboratory facilities.
S.N. was supported by a research fellowship from the Japan Society for the Promotion of Science.
Footnotes
Published ahead of print on 21 August 2006.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Baross, J. A. 1995. Isolation, growth and maintenance of hyperthermophiles, p. 15-23. In F. T. Robb and A. R. Place (ed.), Archaea: a laboratory manual. Thermophiles, Cold Spring Harbor Laboratory, Cold Springer Harbor, N.Y.
- 2.Becker, K., and E. E. Davis. 2006. A review of CORK designs and operations during the Ocean Drilling Program. In A. T. Fisher, T. Urabe, A. Klaus, and the Expedition 301 Scientists (ed.), Proceedings of the Integrated Ocean Drilling Program, vol. 301. Ocean Drilling Program, College Station, Tex. [Online.] http://iodp.tamu.edu/publications/exp301/EXP_REPT/CHAPTERS/301_104.PDF.
- 3.Bligh, E. G., and W. J. Dyer. 1954. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 4.Cord-Ruwisch, R. 2000. Microbially influenced corrosion of steel, p. 159-173. In D. R. Lovley (ed.), Environmental microbe-metal interactions. ASM Press, Washington, D.C.
- 5.Cowen, J. P. 2004. The microbial biosphere of sediment-buried oceanic basement. Res. Microbiol. 155:497-506. [DOI] [PubMed] [Google Scholar]
- 6.Cowen, J. P., Giovannoni, S. J., Kenig, F., Johnson, H. P., Butterfield, D., Rappe, M. S., Hutnak, M., and Lam, P. 2003. Fluids from aging ocean crust that support microbial life. Science 299:120-123. [DOI] [PubMed] [Google Scholar]
- 7.Davis, E. E., and K. Becker. 2002. Observations of natural-state fluid pressures and temperatures in young oceanic crust and inferences regarding hydrothermal circulation. Earth Planet. Sci. Lett. 204:231-248. [Google Scholar]
- 8.DeLong, E. F. 1992. Archaea in coastal marine environments. Proc. Natl. Acad. Sci. USA 89:5685-5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deming, J. W., and J. A. Baross. 1993. Deep-sea smokers: windows to a subsurface biosphere? Geochim. Cosmochim. Acta 57:3219-3230. [DOI] [PubMed] [Google Scholar]
- 10.D'Hondt, S., B. B. Jørgensen, D. J. Miller, A. Batzke, R. Blake, B. A. Cragg, H. Cypionka, G. R. Dickens, T. Ferdelman, K. U. Hinrichs, N. G. Holm, R. Mitterer, A. Spivack, G. Wang, B. Bekins, B. Engelen, K. Ford, G. Gettemy, S. D. Rutherford, H. Sass, C. G. Skilbeck, I. W. Aiello, G. Guèrin, C. H. House, F. Inagaki, P. Meister, T. Naehr, S. Niitsuma, R. J. Parkes, A. Schippers, D. C. Smith, A. Teske, J. Wiegel, C. N. Padilla, and J. L. Acosta. 2004. Distributions of microbial activities in deep subseafloor sediments. Science 306:2216-2221. [DOI] [PubMed] [Google Scholar]
- 11.Dihn, H. T., J. Kuever, M. Mußmann, A. W. Hassel, M. Stratmann, and F. Widdel. 2004. Iron corrosion by novel anaerobic microorganisms. Nature 427:829-832. [DOI] [PubMed] [Google Scholar]
- 12.Edwards, K. J., D. R. Rogers, C. O. Wirsen, and T. M. McCollom. 2003. Isolation and characterization of novel psychrophilic, neutrophilic, Fe-oxidizing chemolithoautotrophic α- and γ-Proteobacteria from the deep sea. Appl. Environ. Microbiol. 69:2906-2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elderfield, H., and Shultz, A. 1996. Mid-ocean ridge hydrothermal fluxes and the chemical composition of the ocean. Annu. Rev. Earth Planet. Sci. 24:191-224. [Google Scholar]
- 14.Fisher, A. T. 1998. Permeability within basaltic oceanic crust. Rev. Geophys. 36:143-182. [Google Scholar]
- 15.Fisher, A. T., E. E. Davis, and C. Escutia (ed.). 2000. Proceedings of Ocean Drilling Program, scientific results, vol. 168. Ocean Drilling Program, College Station, Tex.
- 16.Fisher, A. T., T. Urabe, A. Klaus, and the Expedition 301 Scientists (ed.). 2005. Proceedings of the Integrated Ocean Drilling Program, vol. 301. [Online.] Integrated Ocean Drilling Program Management International, Inc., College Station, Tex. http://iodp.tamu.edu/publications/exp301/301toc.htm.
- 17.Fisk, M. R., S. J. Giovannoni, and I. H. Thorseth. 1998. Alteration of oceanic volcanic glass: textual evidence of microbial activity. Science 281:978-980. [DOI] [PubMed] [Google Scholar]
- 18.Furnes, H., K. Muehlenbachs, T. Torsvik, I. H. Thorseth, and O. Tumyr. 2001. Microbial fractionation of carbon isotopes in altered basaltic glass from the Atlantic Ocean, Lau Basin and Costa Rica Rift. Chem. Geol. 173:313-330. [Google Scholar]
- 19.Holden, J. F., M. Summit, and J. A. Baross. 1998. Thermophilic and hyperthermophilic microorganisms in 3-30°C hydrothermal fluids following a deep-sea volcanic eruption. FEMS Microbiol. Ecol. 25:33-41. [Google Scholar]
- 20.Holland, H. D. 1984. The chemical evolution of the atmosphere and oceans. Princeton University Press, Princeton, NJ.
- 21.House, C. H., J. W. Schopf, and K. O. Stetter. 2003. Carbon isotopic fractionation by archaeans and other thermophilic prokaryotes. Org. Geochem. 34:345-356. [Google Scholar]
- 22.Huber, J. A., D. A. Butterfield, and J. A. Baross. 2002. Temporal changes in archaeal diversity and chemistry in a mid-ocean ridge subseafloor habitat. Appl. Environ. Microbiol. 68:1585-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huber, J. A., H. P. Johnson, D. A. Butterfield, and J. A. Baross. 2006. Microbial life in ridge flank crustal fluids. Environ. Microbiol. 8:88-99. [DOI] [PubMed] [Google Scholar]
- 24.Huber, R., P. Rossnagel, C. R. Woese, R. Rachel, T. A. Langworthy, and K. O. Stetter. 1996. Formation of ammonium from nitrate during chemolithoautotrophic growth of the extremely thermophilic bacterium Ammonifex degensii gen. nov. sp. nov. Syst. Appl. Microbiol. 19:40-49. [DOI] [PubMed] [Google Scholar]
- 25.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon; a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 26.Hugenholtz, P. 2002. Exploring prokaryotic diversity in the genomic era. Genome Biol. 3:003.1-003.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inagaki, F., Y. Motomura, and S. Ogata. 2003. Microbial silica deposition in geothermal hot waters. Appl. Microbiol. Biotechnol. 60:605-611. [DOI] [PubMed] [Google Scholar]
- 28.Inagaki, F., T. Nunoura, S. Nakagawa, A. Teske, M. Lever, A. Lauer, M. Suzuki, K. Takai, M. Delwiche, F. S. Colwell, K. H. Nealson, K. Horikoshi, S. D'Hondt, and B. B. Jørgensen. 2006. Biogeographical distribution and diversity of microbes in methane hydrate-bearing deep marine sediments on the Pacific Ocean Margin. Proc. Natl. Acad. Sci. USA 103:2815-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inagaki, F., M. Suzuki, K. Takai, H. Oida, T. Sakamoto, K. Aoki, K. H. Nealson, and K. Horikoshi. 2003. Microbial communities associated with geological horizons in coastal subseafloor sediments from the Sea of Okhotsk. Appl. Environ. Microbiol. 69:7224-7235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson, H. P., and M. J. Pruis. 2003. Fluxes of fluid and heat from the oceanic crustal reservoir. Earth Planet. Sci. Lett. 216:565-574. [Google Scholar]
- 31.Kelley, D. S., J. A. Karson, D. K. Blackman, G. L. Früh-Green, D. A. Butterfield, M. D. Lilley, E. J. Olson, M. O. Schrenk, K. K. Roe, G. T. Lebon, P. Rivizzigno, and AT3-60 shipboard party. 2001. An off-axis hydrothermal vent field near the Mid-Atlantic Ridge at 30°N. Nature 412:145-149. [DOI] [PubMed] [Google Scholar]
- 32.Klein, M., M. Friedrich, A. J. Roger, P. Hugenholtz, S. Fishbain, H. Abicht, L. L. Blackall, D. A. Stahl, and M. Wagner. 2001. Multiple lateral transfers of dissimilatory sulfite reductase genes between major lineages of sulfate-reducing prokaryotes. J. Bacteriol. 183:6028-6035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Komagata, K., and K. Suzuki. 1987. Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol. 19:161-207. [Google Scholar]
- 34.Lane, D. J. 1985. 16S/23S sequencing, p. 115-176. In E. Stackbrandt and M. Goodfellow (ed.), Bacterial systematics. Wiley, New York, N.Y.
- 35.Lanoil, B. D., M. T. La Duc, M. Wright, M. Kastner, K. H. Nealson, and D. Bartlett. 2005. Archaeal diversity in ODP legacy borehole 892b and associated seawater and sediments of the Cascadia Margin. FEMS Microbiol. Ecol. 54:167-178. [DOI] [PubMed] [Google Scholar]
- 36.Lipman, D. J., and W. R. Pearson. 1985. Rapid and sensitive protein similarity searches. Science 227:1435-1441. [DOI] [PubMed] [Google Scholar]
- 37.Ludwig, W., S. H. Bauer, M. Bauer, I. Held, G. Kirchhof, R. Schulze, I. Huber, Spring, S., Hartmann, A., and Schleifer, K. H. 1997. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol. Lett. 153:181-190. [DOI] [PubMed] [Google Scholar]
- 38.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüßmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mottl, M. J., and C. G. Wheat. 1994. Hydrothermal circulation through mid-ocean ridge flanks: fluxes of heat and magnesium. Geochim. Cosmochim. Acta 58:2225-2237. [Google Scholar]
- 40.Nakagawa, S., and K. Takai. 2006. Methods for the isolation of thermophiles from deep-sea hydrothermal environments. Methods Microbiol. 35:55-91. [Google Scholar]
- 41.Nakagawa, S., K. Takai, F. Inagaki, H. Chiba, J. Ishibashi, S. Kataoka, H. Hirayama, T. Nunoura, K. Horikoshi, and Y. Sako. 2005. Variability in microbial community and venting chemistry in a sediment-hosted backarc hydrothermal system: impacts of subseafloor phase-separation. FEMS Microbiol. Ecol. 54:141-155. [DOI] [PubMed] [Google Scholar]
- 42.Nakagawa, S., K. Takai, F. Inagaki, H. Hirayama, T. Nunoura, K. Horikoshi, and Y. Sako. 2005. Distribution, phylogenetic diversity and physiological characteristics of epsilon-Proteobacteria in a deep-sea hydrothermal field. Environ. Microbiol. 7:1619-1632. [DOI] [PubMed] [Google Scholar]
- 43.Nakagawa, S., K. Takai, F. Inagaki, K. Horikoshi, and Y. Sako. 2005. Nitratiruptor tergarcus gen. nov., sp. nov. and Nitratifractor salsuginis gen. nov., sp. nov., nitrate-reducing chemolithoautotrophs of the ɛ-Proteobacteria isolated from a deep-sea hydrothermal system in the Mid-Okinawa Trough. Int. J. Syst. Evol. Microbiol. 55:925-933. [DOI] [PubMed] [Google Scholar]
- 44.Nakagawa, T., S. Nakagawa, F. Inagaki, K. Takai, and K. Horikoshi. 2004. Phylogenetic diversity of sulfate-reducing prokaryotes in active deep-sea hydrothermal vent chimney structures. FEMS Microbiol. Lett. 232:145-152. [DOI] [PubMed] [Google Scholar]
- 45.Orphan, V. J., K.-U. Hinrichs, W. Ussler, C. K. Paull, L. T. Taylor, S. P. Sylva, J. M. Hayes, and E. F. DeLong. 2001. Comparative analysis of methane-oxidizing archaea and sulfate-reducing bacteria in anoxic marine sediments. Appl. Environ. Microbiol. 67:1922-1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pearson, A., J. S. Seewald, and T. I. Eglinton. 2005. Bacterial incorporation of relict carbon in the hydrothermal environment of Guaymas Basin. Geochim. Cosmochim. Acta 69:5477-5486. [Google Scholar]
- 47.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 48.Rickard, D. T. 1969. The microbiological formation of iron sulfides. Stockh. Contrib. Geol. 20:50-66. [Google Scholar]
- 49.Sako, Y., K. Takai, Y. Ishida, A. Uchida, and Y. Katayama. 1996. Rhodothermus obamensis sp. nov., a modern lineage of extremely thermophilic marine bacteria. Int. J. Syst. Bacteriol. 46:1099-1104. [DOI] [PubMed] [Google Scholar]
- 50.Sako, Y., S. Nakagawa, K. Takai, and K. Horikoshi. 2003. Marinithermus hydrothermalis gen. nov., sp. nov., a strictly aerobic, thermophilic bacterium from a deep-sea hydrothermal vent chimney. Int. J. Syst. Evol. Microbiol. 53:59-65. [DOI] [PubMed] [Google Scholar]
- 51.Shanks, W. C., III, J. K. Böhlke, and R. R. Seal II. 1995. Stable isotopes in mid-ocean ridge hydrothermal systems: interactions between fluids, minerals, and organisms, p. 194-221. In S. E. Humphris, R. A. Zierenberg, L. S. Mullineaux, and R. E. Thompson (ed.), Geophysical monograph, vol. 91. Seafloor hydrothermal systems. American Geophysical Union, Washington, D.C. [Google Scholar]
- 52.Slobodkina, G. A., N. A. Chernyh, A. I. Slobodkin, I. V. Subbotina, E. A. Bonch-Osmolovskaya, and A. V. Lebedinsky. 2004. PCR-based identification of hyperthermophilic archaea of the family Thermococcaceae. Appl. Environ. Microbiol. 70:5701-5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sokolova, T. G., N. A. Kostrikina, N. A. Chernyh, T. P. Tourova, T. V. Kolganova, and E. A. Bonch-Osmolovskaya. 2002. Carboxydocella thermautotrophica gen. nov., sp. nov., a novel anaerobic, CO-utilizing thermophile from a Kamchatkan hot spring. Int. J. Syst. Evol. Microbiol. 52:1961-1967. [DOI] [PubMed] [Google Scholar]
- 54.Suzuki, Y., F. Inagaki, K. Takai, K. H. Nealson, and K. Horikoshi. 2004. Microbial diversity in inactive chimney structures from deep-sea hydrothermal systems. Microb. Ecol. 47:186-196. [DOI] [PubMed] [Google Scholar]
- 55.Suzuki, Y., R. E. Kopp, T. Kogure, A. Suga, K. Takai, S. Tsuchida, N. Ozaki, K. Endo, J. Hashimoto, Y. Kato, C. Mizota, T. Hirata, H. Chiba, K. H. Nealson, K. Horikoshi, and J. L. Kirschvink. 2006. Sclerite formation in the hydrothermal-vent “scaly-foot” gastropod—possible control of iron sulfide biomineralization by the animal. Earth Planet. Sci. Lett. 242:39-50. [Google Scholar]
- 56.Suzuki, Y., T. Sasaki, M. Suzuki, Y. Nogi, T. Miwa, K. Takai, K. H. Nealson, and K. Horikoshi. 2005. Novel chemoautotrophic endosymbiosis between a member of the Epsilonproteobacteria and the hydrothermal-vent gastropod Alviniconcha aff. hessleri (Gastropoda: Provannidae) from the Indian Ocean. Appl. Environ. Microbiol. 71:5440-5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swofford, D. L. 2000. PAUP*. Phylogenetic analysis using parsimony (and other methods), version 4. Sinauer Associates, Sunderland, Mass.
- 58.Takai, K., and K. Horikoshi. 2000. Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl. Environ. Microbiol. 66:5066-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takai, K., T. Gamo, U., Tsunogai, N. Nakayama, H. Hirayama, K. H. Nealson, and K. Horikoshi. 2004. Geochemical and microbiological evidence for a hydrogen-based, hyperthermophilic subsurface lithoautotrophic microbial ecosystem (HyperSLiME) beneath an active deep-sea hydrothermal field. Extremophiles 8:269-282. [DOI] [PubMed] [Google Scholar]
- 60.Takai, K., F. Inagaki, S. Nakagawa, H. Hirayama, T. Nunoura, Y. Sako, K. H. Nealson, and K. Horikoshi. 2003. Isolation and phylogenetic diversity of members of previously uncultivated ɛ-Proteobacteria in deep-sea hydrothermal fields. FEMS Microbiol. Lett. 208:167-1174. [DOI] [PubMed] [Google Scholar]
- 61.Takai, K., A. Inoue, and K. Horikoshi. 2002. Methanothermococcus okinawensis sp. nov., a thermophilic, methane-producing archaeon isolated from a Western Pacific deep-sea hydrothermal system. Int. J. Syst. Evol. Microbiol. 52:1089-1095. [DOI] [PubMed] [Google Scholar]
- 62.Teske, A., K.-U. Hinrichs, V. Edgcomb, A. V. Gomez, D. Kysela, S. P. Sylva, M. L. Sogin, and H. W. Jannasch. 2002. Microbial diversity of hydrothermal sediments in the Guaymas Basin: evidence for anaerobic methanotrophic communities. Appl. Environ. Microbiol. 68:1994-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Thompson, T. D., T. J. Gibosn, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van Dover, C. L. 2000. The ecology of deep-sea hydrothermal vents. Princeton University Press, Princeton, N.J.
- 65.Vetriani, C., H. W. Jannasch, B. J. MacGregor, D. A. Stahl, and A.-L. Reysenbach. 1999. Population structure and phylogenetic characterization of marine benthic archaea in deep-sea sediments. Appl. Environ. Microbiol. 65:4375-4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wheat, C. G., and M. J. Mottl. 2000. Composition of pore and spring waters from Baby Bare: global implications of geochemical fluxes from a ridge flank hydrothermal system. Geochim. Cosmochim. Acta 64:629-642. [Google Scholar]
- 67.Wheat, C. G., H. Elderfield, M. J. Mottl, and C. Monnin. 2000. Chemical composition of basement fluids within an oceanic ridge flank: implications for along-strike and across-strike hydrothermal circulation. J. Geophys. Res. 105:13437-13447. [Google Scholar]
- 68.Wheat, C. G., J. McManus, M. J. Mottl, and E. Giambalvo. 2003. Oceanic phosphorous imbalance: the magnitude of the ridge-flank hydrothermal sink. Geophys. Res. Lett. 30:1895. [Online.] doi: 10.1029/2003GL017318. [DOI] [Google Scholar]
- 69.Wheat, C. G., H. W. Jannasch, M. Kastner, J. N. Plant, E. H. DeCarlo, and G. Lebon. 2004. Venting formation fluids from deep-sea boreholes in a ridge flank setting: ODP sites 1025 and 1026. Geochem. Geophys. Geosys. 5:Q08007. [Online] doi: 10.1029/2004GC000710. [DOI] [Google Scholar]
- 70.Widdel, F., and N. Pfennig. 1981. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. I. Isolation of new sulfate-reducing bacteria enriched with acetate from saline environments. Description of Desulfobacter postgatei gen. nov. sp. nov. Arch. Microbiol. 129:395-400. [DOI] [PubMed] [Google Scholar]
- 71.Widdel, F., G. W. Kohring, and F. Mayer. 1983. Studies on dissimilatory sulfate reducing bacteria that decompose fatty acids. III. Characterization of the filamentous gliding Desulfonema limicola gen. nov., sp. nov., and Desulfonema magnum sp. nov. Arch. Microbiol. 134:286-294. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.