Abstract
We investigated seasonal differences in community structure and activity (leucine incorporation) of the planktonic bacterial assemblage in the freshwater and brackish-water zones of a shallow coastal lagoon of the southwestern Atlantic Ocean. Alphaproteobacteria formed the dominant microbial group in both zones throughout the sampling period. After an intrusion of marine water, members of the SAR11 lineage became abundant in the brackish-water zone. These bacteria were apparently distributed over the lagoon during the following months until they constituted almost 30% of all prokaryotic cells at both sampling sites. At the first sampling date (March 2003) a single alphaproteobacterial species unrelated to SAR11, Sphingomonas echinoides, dominated the microbial assemblages in both zones of the lagoon concomitantly with a bloom of filamentous cyanobacteria. Pronounced maxima of leucine incorporation were observed once in each zone of the lagoon. In the freshwater zone, this highly active microbial assemblage was a mix of the typical bacteria lineages expected in aquatic systems. By contrast, a single bacterial genotype with >99% similarity to the facultative pathogen gammaproteobacterial species Stenotrophomonas maltophilia formed >90% of the bacterial assemblage (>107 cell ml−1) in the brackish-water zone at the time point of highest bacterial leucine incorporation. Moreover, these bacteria were equally dominant, albeit less active, in the freshwater zone. Thus, the pelagic zone of the studied lagoon harbored repeated short-term blooms of single bacterial species. This finding may have consequences for environmental protection.
Shallow coastal lagoons are among the most productive natural ecosystems on Earth (1). These relatively closed basins are extremely vulnerable to nutrient input from the surrounding catchment. Therefore, they are highly susceptible to anthropogenic influence and pollution (8). Coastal lagoons, moreover, exhibit great spatial and temporal variability in their physicochemical water characteristics due to the sporadic mixing of freshwater with marine influx. They are commonly surrounded by extensive areas of freshwater and salt marshes, which are important sources of the dissolved organic matter (DOM) that drives the ecosystem metabolism (44). The exchange of water masses of freshwater and marine origin is crucial to the natural functioning of coastal lagoons and their associated littoral wetlands because basic ecological processes are controlled by this mixing. For example, it determines the gradient of DOM and nutrients between the limnetic and brackish areas, which in turn controls the productivity of the system (12, 13).
The biotic mineralization of DOM in aquatic ecosystems is carried out by a diverse community of heterotrophic microbes from various phylogenetic lineages (15, 36). Some of these groups differ in their distributions across freshwater and marine habitats (27, 32, 62). For example, Alphaproteobacteria related to “Candidatus Pelagibacter” (SAR11) are among the most abundant bacterial group in open oceans (42), but they are typically absent in freshwater lakes. The opposite has been found for several phylogenetic lineages of Actinobacteria (59). Microbial communities in estuarine environments are believed to be more diverse than those in other water bodies due to the great physicochemical variety of estuarine systems (16). Estuarine microbial assemblages harbor microbial taxa from both freshwater and marine habitats, groups that are introduced from soil during periods of high runoff, and bacteria that are specific to estuarine environments (16). This may also be the case for habitats that are shaped by similarly complex processes, such as coastal lagoons.
The relationship between the composition of bacterioplankton communities and their activity is poorly understood. It is conceivable that productive environments might favor greater microbial diversity, for example, by providing more spatial heterogeneity and microniches, such as organic aggregates (28). Alternatively, high microbial activity might also reflect the dominance of a few rapidly growing bacterial populations that profit from sudden changes in growth conditions or the availability of a specific substrate. The latter is typically observed in enclosures after experimental disturbance (37) or during short-term incubations (56). Microbial assemblages in unmanipulated aquatic systems are believed to be more resilient against such invasive species due to feedbacks within the microbial food web (6). In fact, dominance by a single population of heterotrophic bacteria (i.e., 50% of total cell counts or more) has rarely been reported for a natural body of water. A recent investigation by Miller et al. suggests that an instance of milky sea detected by satellite remote sensing in the Indian Ocean may have been caused by massive blooms (>1022 cells) of bioluminescent bacteria growing on phytoplankton (41).
Bacterial diversity and community structure in estuaries have been studied mainly in the temperate zones (9, 16, 32). By contrast, little is known about the composition and activity of microbial assemblages in other habitats with pronounced salinity gradients, such as coastal lagoons from subtropical zones. Thus, the aim of this study was to examine the activity and composition of the bacterioplankton community in a coastal lagoon of the southwestern Atlantic Ocean during different seasons.
MATERIALS AND METHODS
Study site.
Laguna de Rocha is a highly productive shallow coastal lagoon (mean depth, 0.6 m; area, 72 km2) influenced by freshwater and by occasional marine intrusions. It is located in Uruguay, on the southeastern coast of South America (34°33′S, 54°22′W). The system is linked with the sea through a single-mouth inlet across a sand barrier that opens either naturally (when the water depth exceeds approximately 1.4 m) or by human action. When the sand bar is opened, the marine intrusion gradually divides the lagoon into a zone of brackish water and a freshwater zone that is characterized by high turbidity and nutrient concentration (12).
Sampling and in situ measurements.
Samples were taken from two sites corresponding to the typical freshwater- and seawater-influenced areas of the lagoon. The selection of these sites was based on previous studies about the abiotic and biological characteristics of the system (12, 13). Six samplings were performed, in March 2003 and in bimonthly intervals between August 2003 and April 2004. Triplicate water samples (250 ml each) were collected in acid (10% HCl)-washed and autoclaved flasks (Nalgene) from an area of ca. 10 m2 at each site. Two hundred milliliters of each replicate sample was fixed with buffered paraformaldehyde (PFA [pH 7.2]; final concentration, 1% [vol/vol]) at room temperature for 1 h and then stored at 4°C for 6 to 12 h. The remaining 50 ml was used to determine [3H]leucine incorporation into bacterial biomass (see below). In December 2003, additional samples were obtained from the brackish-water zone in order to establish that the sampling scheme was sufficiently representative. Five samples instead of three were collected from the sampling site, and two more samples from this site were obtained on the following two days.
On each sampling date, pH, temperature, and conductivity were measured with a portable meter (ES-12; Horiba Inc., Irvine, CA). The concentrations of soluble reactive phosphorus (SRP) and ammonium (NH4) were determined by standard chemical methods (3). For chlorophyll a analyses, 0.1 to 0.5 liter of water was gently filtered through glass fiber filters (type GF/F; Whatman Inc., Florham Park, NJ), which then were sonicated for 1 min and extracted overnight in the dark at 4°C with 90% (vol/vol) alkaline acetone. Extracts were cleared with glass fiber filters (type GF/C; Whatman) and scanned against an acetone reference in a spectrophotometer (range, 400 to 750 nm; model DU-6; Beckman Coulter, Brea, CA). The concentration of pigments was estimated according to the method of Jeffrey and Humphrey (29).
Bacterial abundances and biomass production.
Total numbers of bacteria in both zones of the lagoon were determined from the PFA-fixed triplicate samples described above by staining 1 to 2 ml of these samples with 4′,6-diamino-2-phenylindole (DAPI; final concentration, 1 μg ml−1) and by epifluorescence microscopy. After staining, samples were filtered onto polycarbonate filters (diameter, 25 mm; pore size, 0.2 μm; type GTTP; Millipore) using gentle vacuum and cellulose nitrate support filters (pore size, 0.45 μm; Sartorius) to optimize the distribution of cells on the filters. Bacterial enumeration was done semiautomatically as previously described (49) on a motorized epifluorescence microscope (Axioplan II Imaging; Carl Zeiss, Jena, Germany) equipped with a charge-coupled-device camera (ORCA; Hamamatsu, Herrsching, Germany), using the KS400 image analysis software (Carl Zeiss).
Bacterial activity was estimated via the incorporation of [3H]leucine (Amersham, Little Chalfont, England). Radiolabeled leucine was added at saturating concentrations (20 nM) to triplicate subsamples (5 ml) and to one control prefixed with PFA (final concentration, 3% [vol/vol]). The vials were incubated for 1 h at in situ temperature in the dark. The incubation was subsequently stopped by addition of PFA (final concentration, 3% [vol/vol]). Filtration onto cellulose mixed-ester filters (type GSWP; Millipore) and macromolecule extractions were performed as previously described (33). Incorporation of radiolabel material into bacterial biomass was determined using a Beckman (Fullerton, CA) LS5000TD liquid scintillation counter. Measurements were corrected for quenching (external standard method) and by subtraction of counts from prefixed controls.
Microbial community composition.
Ten microliters from the PFA-fixed samples was filtered onto 0.2-μm-pore-size polycarbonate filters (diameter, 47 mm; type GTTP; Millipore). The filters were rinsed twice with 1× phosphate-buffered saline and once with deionized water, air-dried, and stored at −20°C until further processing. Fluorescence in situ hybridization with horseradish peroxidase-labeled probes and catalyzed reporter deposition (CARD-FISH) was performed as described previously (47). The following group-specific probes were applied: EUB I-III (most Bacteria) (17), ALF968 (Alphaproteobacteria) (27), BET42a (Betaproteobacteria), GAM42a (Gammaproteobacteria) (40), CF319a (members of the Cytophaga-Flavobacteria lineage of Bacteroidetes) (39), and HGC69a (Actinobacteria) (52). Bacteria from the SAR11 clade of Alphaproteobacteria were quantified by probe SAR11-441 (42), and cells affiliated with Sphingomonas echinoides and Stenotrophomonas maltophilia were detected with the newly designed probes SphEch_1249 and SteMal_439 (see below). Filter sections were stained with DAPI (1 μg ml−1), rinsed with deionized water and 80% (vol/vol) ethanol, and embedded in mountant (47). Double-stained cells were quantified on an Axioplan II Imaging epifluorescence microscope (Carl Zeiss) by semiautomated image analysis (49). All FISH determinations were done in triplicate using independently collected water samples.
PCR amplification, cloning, and sequencing of 16S rRNA genes.
16S rRNA gene clone libraries were constructed from samples obtained from the brackish zone (March 2003 and December 2003) and the freshwater zone (March 2003). Different volumes of fixed and unfixed bacterial biomass were filtered onto polycarbonate membrane filters (type GTTP; Millipore). Pieces of approximately 6 mm2 were cut out from these filters and used directly as a template for amplification by PCR without previous DNA extraction (34). DNA-free purified water served as a negative control. PCR amplification with the general bacterial primers GM3F and GM4R (43) was performed with a Mastercycler (Eppendorf, Hamburg, Germany) using the following conditions: an initial denaturation step of 5 min at 94°C, 1 min of denaturation at 94°C, 1.5 min of annealing at 48°C, and 2 min of primer extension at 72°C. This cycle was repeated 30 times, followed by a final step of 2 min at 72°C. The PCR products were checked by agarose (1%, wt/vol) gel electrophoresis.
The amplified 16S rRNA gene fragments were purified with a QIAquick PCR purification kit (QIAGEN, Hilden, Germany), inserted into the TOPO vector (TOPO TA cloning kit; Invitrogen, Karlsruhe, Germany), and cloned into competent cells of Escherichia coli as described by the manufacturer. The transformed cells were plated on Luria-Bertani agar plates containing 50 μg of ampicillin ml−1 and incubated overnight at 37°C. The clones were screened for correctly sized inserts by agarose gel electrophoresis (1%). Plasmids were prepared with a QIAprep Spin Miniprep kit (QIAGEN) according to the manufacturer's specifications. For a first screening, the plasmid DNAs were sequenced using the primer M13F (5′-GTA AAA CGA CGG CCA G-3′; located on the vector pCR4-TOPO) on an ABI PRISM 3100 genetic analyzer (Applied Biosystems, Foster City, Calif.). Nearly full-length 16S rRNA sequences were obtained from selected inserts by additional sequencing with the primers GM1 (43) and M13R (5′-CAG GAA ACA GCT ATG AC-3′).
Phylogenetic analysis and probe design.
Sequences were assembled from three to five partial sequences using the software Sequencher (Gene Codes Corp., Ann Arbor, Mich.). The resulting contigs were tested for chimeric origin with the free software Mallard (http://www.cf.ac.uk/biosi/research/biosoft/) based on the Pintail algorithm (4). All sequences were subsequently analyzed by BLAST (2) to identify their closest relatives and to establish tentative phylogenetic affiliations. Phylogenetic analyses were performed using the ARB software package (38). For the reconstruction of a phylogenetic tree depicting the affiliations of newly obtained alphaproteobacterial sequences, only nearly complete (i.e., longer than 1,400-nucleotide) 16S rRNA sequences were considered. A specific 50% base frequency filter was used to exclude highly variable positions, and maximum-parsimony, neighbor-joining, and maximum-likelihood analyses were performed on various subsets of sequences. The branching patterns of the resulting tree were compared manually, and a consensus tree was constructed that showed bifurcations only if branchings were stable in the majority of analyses. Multifurcations were introduced if tree topology could not be unambiguously resolved.
The appropriate ARB tool was used to design oligonucleotide probes for sequence types related to Sphingomonas echinoides (SphEch_1249 [5′-TGC GAG ATT GCT GCC CAC TG-3′]) and Stenotrophomonas maltophilia (SteMal_439 [5′-GCT GGA TTT CTT TCC CAA CA-3′]). The theoretical specificity of the probes was established by checking them against the latest release of the 16S rRNA gene database of Ribosomal Database Project II (11). The probes were subsequently tested by CARD-FISH. Stringent hybridization conditions were established by comparing the fluorescence intensities of target versus nontarget organisms after FISH at increasing concentrations of formamide in the hybridization buffer. Bacterial target strains were Sphingomonas echinoides DSM 1805 and Stenotrophomonas maltophilia DSM 50170. Bacterial strains with one mismatch to the probe target sequence were Sphingomonas pituitosa DSM 13101 and Pseudoxanthomonas broegbernensis DSM 12573, respectively. All bacterial strains were grown according to the recommendations of the supplier (Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany). The specificity of probe SphEch_1249 at the level of single-mismatch discrimination could only be established by addition of two unlabeled competitors (SpEch_C1 [5′-TGC GGG ATT GCT GCC CAC TG-3′] and SpEch_C2 [5′-TGC GAG KTT GCT GCC CAC TG-3′]).
Nucleotide sequence accession numbers.
Sequences were deposited in GenBank under the accession numbers DQ435783, DQ435784, DQ450165 to DQ450197, and DQ655707 to DQ655709.
RESULTS
Environmental characteristics.
Water temperatures ranged between 14 and 17.5°C in both zones during the winter (August), spring (October), and autumn (April) months and rose to >25°C in late summer (February, March) (Table 1). Conductivity clearly differed in the two areas of the lagoon (Table 1). After a marine intrusion in October 2003, conductivity gradually increased in the freshwater zone, from 0.19 to a maximum of 7.8 mS cm−1 in April. The concentration of NH4 varied by a factor of 30, and distinct maxima were observed in October and December 2003 in the freshwater and brackish-water zones, respectively. SRP concentrations ranged from undetectable (March 2003) to >30 μg P liter−1 (December 2003). During the late autumn and early summer months (April and December), SRP concentrations were higher in the freshwater than in the brackish-water zone, whereas the opposite was found in late summer (February 2004). The first sampling date (March 2003) was characterized by a pronounced bloom of mainly filamentous cyanobacteria in both areas of the lagoon and by corresponding high concentrations of chlorophyll a (Table 1). An analysis of environmental 16S rRNA sequences from this period indicated that this bloom was composed of cyanobacteria affiliated with the genera Prochlorothrix, Cylindrospermopsis, and Oscillatoria (i.e., Planktothrix) (Table 2), whereas microscopic inspection suggested the dominance of Pseudanabaena cf. moniliformis (V. Heinz, personal communication). However, Prochlorothrix filaments are morphologically very similar to those of Pseudanabaena and Planktothrix, rendering correct microscopic identification very difficult (25).
TABLE 1.
Basic physicochemical characteristics of lagoon water at the indicated sampling time pointsa
| Sampling date and sample origin | Temp (°C) | Conductivity (mS cm−1) | Concn of NH4 (μg liter−1) | Concn of SRP (μg liter−1) | Concn of chlorophyll a (μg liter−1) |
|---|---|---|---|---|---|
| 8 March 2003 | |||||
| FW | 28.5 | 0.3 | 2.70 | BD | 9.8 |
| BW | 25.5 | 12.1 | 2.70 | BD | 16.8 |
| 05 August 2003 | |||||
| FW | 14.9 | 0.3 | 13.4 | 18.9 | ND |
| BW | 14.1 | 15.6 | 23.2 | 4.5 | ND |
| 09 October 2003 | |||||
| FW | 16.2 | 0.2 | 92.0 | 29.8 | 3.9 |
| BW | 17.4 | 10.3 | 24.0 | 25.9 | 13.2 |
| 03 December 2003 | |||||
| FW | 24.1 | 1.4 | 19.7 | 33.1 | 5.0 |
| BW | 22.6 | 17.0 | 84.0 | 15.2 | 12.0 |
| 04 February 2004 | |||||
| FW | 25.5 | 5.8 | 10.2 | 22.4 | 12.0 |
| BW | 25.6 | 20.3 | 32.8 | 27.7 | 9.3 |
| 20 April 2004 | |||||
| FW | 17.3 | 7.8 | 24.7 | 29.6 | 6.2 |
| BW | 17.4 | 19.0 | 21.3 | 18.1 | 4.4 |
FW, freshwater; BW, brackish water; BD, below detection limit; ND, not determined.
TABLE 2.
16S rRNA gene sequence types obtained from the freshwater and brackish-water zones of the lagoon in March 2003a
| Accession no. (no. of nucleotides) | Origin | Closest relative (accession no.) | Similarity (%) | Phylogenetic affiliation and origin (if available) of closest relative |
|---|---|---|---|---|
| DQ435783 (1,445) | FW | Clone LiUU-9-115 (AY509418) | 94 | Uncultured freshwater Alphaproteobacteria (Acetobacteraceae) associated with a cyanobacterial bloom |
| DQ450165 (1,450) | FW | Clone FukuS110 (AJ289986) | >99 | Uncultured Alphaproteobacteria (Beijerinckiaceae) from lake plankton |
| DQ450166 (1,443) | FW | Clone B6 (AJ867909) | 95 | Uncultured Alphaproteobacteria (Rhizobiales) from a mountain lake |
| DQ450172 (1,491) | FW | Clone LiUU-5-340 (AY509446) | >99 | Uncultured Alphaproteobacteria (Herbaspirillum) associated with a cyanobacterial bloom |
| DQ435784 (1,489) | FW | Clone IRD18H05 (AY947979) | 95 | Uncultured Betaproteobacteria (Comamonadaceae) from a temperate river |
| DQ450167 (1,491) | FW | Clone DS160 (DQ234242) | 99 | Uncultured Betaproteobacteria (Polynucleobacter) from mangrove bacterioplankton |
| DQ450168 (1,498) | FW | Clone PRD01a011B (AF289159) | 97 | Uncultured freshwater Betaproteobacteria (Methylophilaceae) |
| DQ450169 (1,499) | FW | Clone IRD18C12 (AY947926) | >99 | Uncultured Betaproteobacteria (Rhodocyclaceae) from a temperate river |
| DQ450170 (1,489) | FW | Strain F1021 (AF236005) | >99 | Betaproteobacteria (Comamonadaceae) |
| DQ450171 (1,494) | FW | Clone PRD01b009B (AF289169) | 98 | Uncultured Betaproteobacteria (Comamonadaceae) from lakes and rivers |
| DQ450173 (1,492) | FW | Chitinibacter tainanensis strain BCRC (AY264287) | 98 | Betaproteobacteria (Chitinibacter) |
| DQ450175 (1,493) | FW | Clone BG.d11 (DQ228372) | 98 | Uncultured Betaproteobacteria (Rhodoferax) from a subglacial environment |
| DQ450176 (1,489) | FW | Clone EV818SWSAP26 (DQ337069) | 93 | Uncultured Betaproteobacteria (Burkholderiaceae) from subsurface waters of the Kalahari Shield |
| DQ450174 (770) | FW | Clone IRD18C04 (AY947919) | 93 | Uncultured Bacteroidetes (Flavobacteriales); environmental samples |
| DQ450177 (766) | FW | Planktothrix pseudagardhii strain T1-8-4 (AB045968) | 98 | Cyanobacteria; Oscillatoriales |
| DQ450178 (694) | FW | Planktothrix pseudagardhii strain T19-6′-6 (AB045965) | 94 | Cyanobacteria; Oscillatoriales |
| DQ450179 (699) | FW | Synechococcus sp. strain PS723 (AF216955) | 94 | Cyanobacteria; Chroococcales |
| DQ655709 (1,538) | FW | Stenotrophomonas maltophilia strain VUN10,003 (AF100733) | >99 | Gammaproteobacterial isolate from a polycyclic aromatic hydrocarbon-contaminated soil in Australia |
| DQ450180 (1,496) | FW | Clone WD2124 (AJ292676) | 92 | Uncultured Gammaproteobacteria from a polychlorinated biphenyl-polluted soil |
| DQ450181 (689) | FW | CHAB-III-7 (AJ240921) | 93 | Uncultured Gammaproteobacteria from marine plankton |
| DQ450182 (1,483) | FW | Clone PRD18H08 (AY948070) | 94 | Uncultured Bacteroidetes (Cryomorphaceae); temperate river |
| DQ450183 (699) | FW | Clone PRD18D12 (AY948032) | 94 | Uncultured Bacteroidetes (Crenotrichaceae); temperate river |
| DQ450184 (1,447) | BW | Prochlorothrix hollandica (AF132792) | 96 | Cyanobacteria; Prochlorales |
| DQ450185 (770) | BW | Clone MB11F01 (AY033309) | 93 | Uncultured Alphaproteobacteria (SAR11); coastal marine waters |
| DQ450186 (723) | BW | Clone AEGEAN_233 (AF406547) | 92 | Uncultured Alphaproteobacteria; marine water sample |
| DQ450187 (1,432) | BW | Clone SPOTSDEC01_5m17 (DQ009190) | >99 | Uncultured marine Alphaproteobacteria |
| DQ450188 (1,442) | BW | Clone Arctic97A-1 (AF353228) | 98 | Uncultured Alphaproteobacteria from the Arctic Ocean |
| DQ450191 (662) | BW | Clone HP1B39 (AF502216) | 92 | Uncultured Alphaproteobacteria (Rhodobacterales) from activated sludge |
| DQ450189 (1,495) | BW | Clone DS140 (DQ234222) | 98 | Uncultured Betaproteobacteria (Comamonadaceae) from mangrove sample |
| DQ450192 (757) | BW | Clone POCPN-5 (AB022337) | 93 | Oligotrophic Betaproteobacteria (Methylophilaceae) |
| DQ450194 (795) | BW | Clone S9JA-19 (AB154316) | 95 | Uncultured Cyanobacteria from an eutrophic lake |
| DQ450195 (655) | BW | Oscillatoria sp. strain Ant-SOS (AF263342) | 93 | Cyanobacteria (Oscillatoriales) from Antarctic samples |
| DQ450196 (673) | BW | Phormidium sp. strain MBIC10025 (AB183566) | 94 | Cyanobacteria (Oscillatoriales) |
| DQ450197 (801) | BW | Cylindrospermopsis raciborskii strain PMC98.14 (AJ582102) | 94 | Cyanobacteria (Nostocales) |
Alphaproteobacterial sequence types related to SAR11 and Sphingomonas are not listed because they are already depicted in Fig. 3. FW, freshwater; BW, brackish water.
Bacterial abundance and activity.
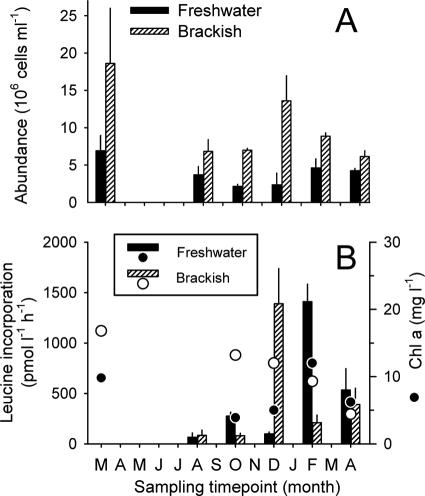
Numbers of bacterial cells were significantly higher in the brackish-water zone during the whole study period (Wilcoxon rank test; N = 36; P < 0.001) (Fig. 1A). In this area of the lagoon, two conspicuous peaks of bacterial abundance (1.8 × 107 ± 0.7 × 107 cells ml−1 and 1.3 × 107 ± 0.3 × 107 cells ml−1) were observed in March and December 2003, respectively. By contrast, the seasonal changes of bacterial cell numbers were less pronounced in the freshwater zone (range, 2.1 × 106 ± 0.3 × 106 cells ml−1 to 6.9 × 106 ± 2 × 106 cells ml−1) (Fig. 1A).
FIG. 1.
(A) Total prokaryotic cell numbers in the two zones of the lagoon. (B) Bulk leucine incorporation rates (bars) and concentrations of chlorophyll a (Chl a; circles). On two occasions, samples were lost: March 2003 (leucine incorporation) and August 2003 (chlorophyll a).
Bacterial activity was lowest in winter at both study sites (Fig. 1B). A remarkable single peak of activity (approximately 1,400 pmol liter−1 h−1) was found in the brackish-water zone in December. In the freshwater zone, a comparable maximum of bacterial activity occurred in February, a period of high water temperature, chlorophyll a concentrations, and conductivity (Table 1). Interestingly, the event of high bacterial production was matched by high bacterial cell numbers only in the brackish-water zone (Fig. 1A). Thus, the highest values for per-cell activity were found in the freshwater zone in February.
Microbial community composition.
On average, 66% ± 8% (mean ± 1 standard error) of DAPI-stained objects could be detected by CARD-FISH with probe EUB I-III in the freshwater zone, and 75% ± 8% were detected in the brackish-water zone. The sum of all cells that hybridized with the five probes for large phylogenetic lineages (Fig. 2) matched the fraction of all hybridized bacteria within the range of the counting error (92% ± 6% and 108% ± 8% in the freshwater and brackish-water zones, respectively). In general, members of the Alphaproteobacteria formed a significantly larger fraction of the bacterial assemblage in the brackish water than in the freshwater zone (Fig. 2), whereas few differences were found for Betaproteobacteria and the Cytophaga-Flavobacteria lineage of Bacteroidetes. With one noted exception (see below), Gammaproteobacteria were rare. Actinobacteria represented between 10 and 20% of total cells in the brackish-water zone during the winter and spring samplings (August and October).
FIG. 2.
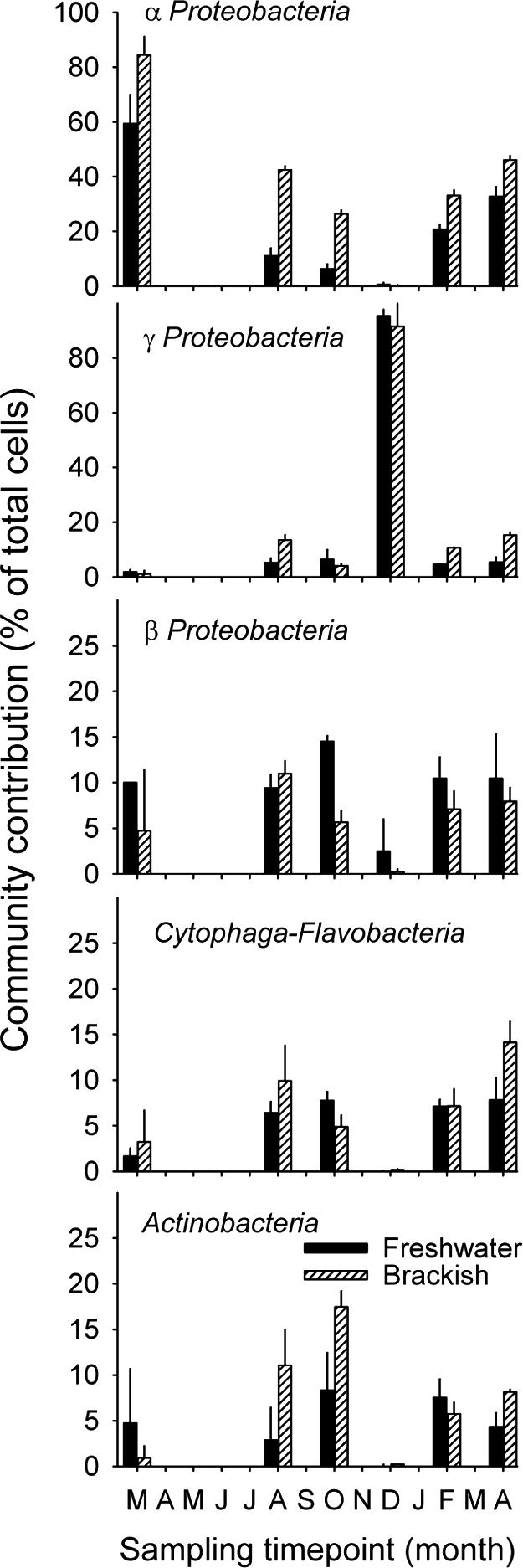
Relative abundances (percentages of total cell counts) of subphyla of Proteobacteria, of bacteria from the Cytophaga-Flavobacteria lineage, and of Actinobacteria in the two zones of the lagoon.
At two of the sampling time points (March and December 2003) the microbial assemblages in both zones of the lagoon were clearly dominated by members of either Alpha- or Gammaproteobacteria (Fig. 2). These bacteria were unusually numerous for a natural aquatic system (>107 cells ml−1 [Fig. 1A]), and they were conspicuously larger than typical pelagic bacteria. These findings inspired us to perform a more detailed analysis of the microbial community at the two time points and to focus our efforts on the identification of abundant members from these lineages.
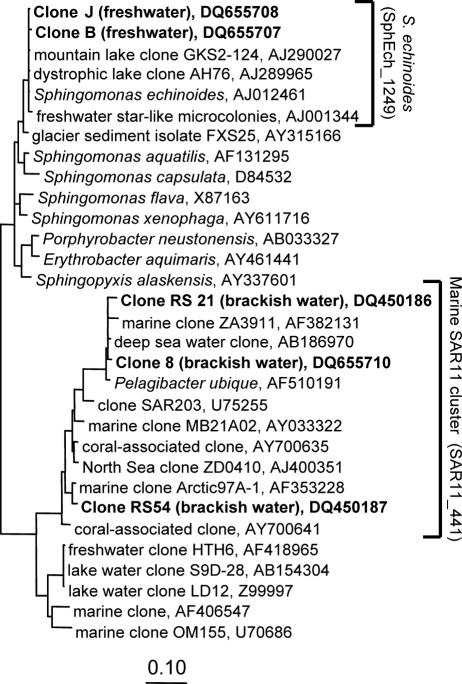
The analysis of the 16S rRNA gene clone library from March yielded sequence types related to typical freshwater and marine pelagic bacteria (Table 2). For example, the alphaproteobacterial sequence types from the freshwater zone were most closely related to sequences obtained from lakes during cyanobacterial blooms (AY509418) and from humic (AJ289986) and high mountain (AJ867909) lakes. However, the overall diversity in this library was low, and there were numerous identical sequence types (i.e., similarity > 99.8%) related to Sphingomonas echinoides (Alphaproteobacteria; 11 clones) (Fig. 3) and cyanobacteria (e.g., Prochlorothrix; 10 clones) (March). From the library of the December sample 12 clones were chosen randomly for initial sequencing. All 12 clones were identical and affiliated to the gammaproteobacterium Stenotrophomonas maltophilia.
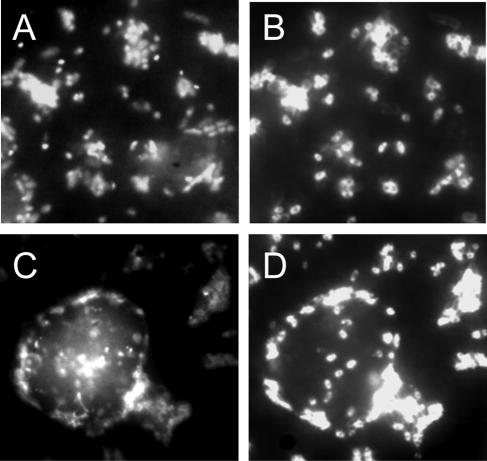
FIG. 4.
Photomicrographs of bacteria targeted by CARD-FISH with probe SteMal_439 in the brackish zone of the lagoon in December 2003. (Left) All bacteria (DAPI staining). (Right) Hybridized cells. The large object shown in panels C and D represents a heavily colonized phytoplankton cell.
Bacteria related to S. echinoides and S. maltophilia as targeted by probes SphEch_1249 and SteMal_439 were usually rare (<1%) in the microbial assemblages of both the freshwater and the brackish-water zones. However, virtually all Alphaproteobacteria (95% ± 3%) in the samples from March 2003 could be visualized by FISH with probe SphEch_1249, and all Gammaproteobacteria in December 2003 were targeted by probe SteMal_439 (Fig. 1 and 4) . Bacteria affiliated with S. maltophilia formed small aggregates and appeared to colonize algal cells (Fig. 4). The relative abundance of S. maltophilia rapidly declined over the 3-day period of repeated sampling that was performed at that time point, i.e., to <50% on day 2 and to <10% on day 3 (data not shown).
FIG. 3.
Phylogenetic affiliations of 16S rRNA gene sequences obtained from the Sphingomonas and SAR11 (“Candidatus Pelagibacter”) lineages (boldface). Only nearly complete sequences are depicted. The square brackets indicate the target ranges of the FISH probes SphEch_1249 and SAR11_441 (see reference 38). Bar, 10% estimated sequence divergence.
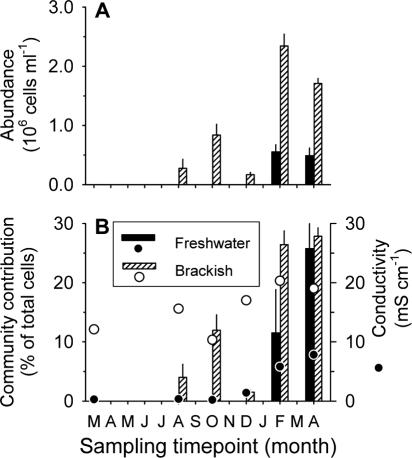
Two sequence types in the clone libraries from the brackish-water zone were affiliated with the SAR11 clade of Alphaproteobacteria (Fig. 3). Since Alphaproteobacteria formed a prominent fraction of total cells in the brackish-water zone at most of the sampling time points (Fig. 2), a probe for the SAR11 lineage (42) was chosen for further analysis of microbial community composition. Bacteria from the SAR 11 clade were common members of the bacterial assemblage in the brackish-water zone between August 2003 and April 2004 (Fig. 5), where they represented between 10% and 100% (mean, 59%) of all Alphaproteobacteria. A steep rise in the abundance of these bacteria was observed after the intrusion of marine water (February to April 2004). In the freshwater zone, bacteria related to SAR11 were only detected during February and April 2004, when the brackish water reached this area of the lagoon (as reflected by a rise in conductivity [Table 1 and Fig. 5B]).
FIG. 5.
Numbers of cells (A) and relative abundances (B) of bacteria from the marine SAR11 clade in the two zones of the lagoon. For comparison, panel B also depicts the conductivity data (circles) reported in Table 1.
DISCUSSION
Bacterial activity.
Coastal lagoons in densely populated areas are often severely affected by anthropogenic impact (21). Information about the dynamics and composition of pelagic microbial assemblages in lagoons typically originates from such heavily polluted systems (8, 35). Laguna de Rocha is increasingly threatened, too, by changes in land use and human settlements (14). However, the system has not been subjected to strong anthropogenic influence yet, and it thus may represent a good model for the natural state of coastal lagoons.
Bulk bacterial activity in the southern brackish-water zone of Laguna de Rocha was maximal in December 2003 (80.3 μg C liter−1 day−1) (Fig. 1B). Similarly high levels of bacterial activity were observed in the northern zone of the lagoon during the subsequent sampling period (February) (104.6 μg C liter−1 day−1), when this area was reached by a plume of brackish water from the south (Table 1). These values clearly exceed microbial production values reported from a Brazilian coastal lagoon (24). Comparable levels of bacterial activity were found in the Santa Rosa Sound (Florida) (66 μg C liter−1 day−1) (10), which is a strait that receives discharges from wastewater treatment plants, and in a eutrophic German lake (68.5 μg C liter−1 day−1) (24). Even higher bacterial production was described for an aquaculture area in Japan (172 μg C liter−1 day−1 on average) (53). In that system, microbial production was correlated with the concentration of dissolved organic nitrogen, suggesting that bacterial activity was supported by allochthonous organic matter from fish farming. Thus, the peaks in bacterial activity observed in our study (Fig. 1B) might indicate discrete inputs of water masses with increased concentrations of organic matter.
Although bacterial numbers were higher in the brackish southern part of the lagoon (Fig. 1A) and total bacterial production in this zone peaked in February 2004 (Fig. 1B), the cell-specific activity (i.e., [3H]leucine incorporation rate normalized to cell numbers) was on average three times higher in the freshwater zone (annual mean ± 1 standard deviation, 1.3 × 10−4 ± 1.1 × 10−4 fmol cell−1 h−1) than in the brackish-water zone (4.3 × 10−5 ± 4.0 × 10−5 fmol cell−1 h−1). This may be a consequence of better growth conditions. Conde et al. (12) have reported that annual DOC and nutrient concentrations are usually higher in the freshwater zone. Large areas of the lagoon, particularly in the northern zone, are covered by submerged marshes that are a major source of organic matter and inorganic nutrients (12). Moreover, the lagoon receives domestic waste from a small town (27,000 inhabitants) through its main tributary, Rocha River (13). On the other hand, estuarine bacteria may be less efficient than freshwater bacteria in using riverine DOM and thus exhibit lower growth efficiencies (36, 51). Physiological stress and growth reduction of both freshwater and marine microbes during periods of rapid horizontal mixing may be another cause of reduced cell-specific activity in the brackish-water zone (e.g., following a marine intrusion event) (45).
Microbial community composition. (i) Large phylogenetic lineages.
Members of all five phylogenetic groups studied were present in both zones of the lagoon (Fig. 2). The summed cell numbers of these groups formed the large majority (>90% [Fig. 1]) of all hybridized Bacteria, indicating a small role for other lineages that could potentially occur in pelagic habitats, such as freshwater Verrucomicrobia (62).
Alphaproteobacteria were significantly more abundant in brackish waters than in the freshwater zone, which agrees with patterns observed along salinity gradients in estuaries (9, 32). However, the Alphaproteobacteria also represented the largest single phylogenetic group in March 2003 and between February and April 2004 (Fig. 1). This is in contrast to reports from lakes (27), but it agrees with community patterns observed in some parts of the Hudson River in the United States (31). Indeed, considering the hydrodynamics in the freshwater part of the lagoon, the microbial assemblage more closely resembled a riverine community than a lake community. However, such a generalization appears premature, as, for example, the planktonic microbial assemblage in a eutrophic river was dominated by Betaproteobacteria (55).
No consistent differences between the two zones were observed for the other four phylogenetic lineages studied (Fig. 2). These findings confirm the wide distribution of the highly diversified aquatic Cytophaga-Flavobacteria across environments with variable salinities (32), whereas they somewhat contrast with current views about the typical habitats occupied by planktonic Betaproteobacteria and Actinobacteria (9, 27, 32). It is, however, conceivable that some representatives from the latter two lineages might specifically inhabit brackish-water habitats. Crump et al. (16) obtained two sequence types from the Parker River estuary that were related to a betaproteobacterium from freshwater sediments and to an actinobacterium from a salt marsh.
(ii) Bacterial activity and community composition.
Analysis at the level of large phylogenetic entities revealed that the brackish and freshwater zones strikingly differed in community diversity at the respective time points of maximal bacterial production (December 2003 and February 2004) (Fig. 1B and 2). While the microbial assemblage of the lagoon was completely dominated by members of the Gammaproteobacteria in December 2003, representatives of all five phylogenetic lineages were abundant at the next sampling (Fig. 2). In addition, a more detailed molecular analysis revealed that virtually all pelagic Gammaproteobacteria in December had a single 16S rRNA genotype related to Stenotrophomonas maltophilia (Table 2 and Fig. 4). Moreover, although microbial production in the two zones was extremely different at the two time points (Fig. 1B), microbial community composition at the level of large phylogenetic groups was rather similar (Fig. 2). This counterintuitive observation suggests that the microbial assemblages from one zone of the lagoon were passively transported to the other zone (in December, from north to south; in February, from south to north), where they encountered substantially better growth conditions. In order to test such a hypothesis it would be necessary to more closely monitor horizontal transport processes within the lagoon (50) and to investigate the activities of different microbial taxa.
(iii) SAR11 bacteria.
Members of the SAR11/“Candidatus Pelagibacter” lineage (Fig. 3) are common in the planktonic microbial assemblages of marine environments (42), whereas they are considered to be rare or absent in brackish-water or freshwater environments. For example, SAR11 bacteria were abundant at the marine end of the Delaware Estuary but not at the river mouth (32). In our 16S rRNA gene clone library from the brackish-water zone, we found sequence types that were closely affiliated to “Candidatus Pelagibacter ubique,” but none from the closely related freshwater LD12 clade (62) (Fig. 3). Moreover, our FISH data show that marine SAR11 bacteria could persist in high numbers at low salinities for extended periods of time (Fig. 5). Members of the marine SAR11 clade (as detected by probe SAR11-441) represented 10 to 25% of all cells in the northern freshwater zone of the lagoon (Fig. 5B) on the last two sampling dates, when conductivity ranged between 5 and 10 mS cm−1 (∼3 to 7 practical salinity units). Unfortunately, we did not determine whether SAR11 bacteria were active or growing in the lagoon. Considering the development of conductivity in the northern zone (Table 1 and Fig. 5B) and the substantially higher cell numbers of SAR11 bacteria in the south (Fig. 5A), it is possible that these bacteria were introduced during the sporadic intrusions of marine water, as has been shown for other communities in coastal lagoons (7), and then passively distributed throughout the lagoon by wind-driven horizontal mixing. However, Elifantz et al. reported the assimilation of extracellular polymeric substances by SAR11 bacteria even at salinities as low as 13 practical salinity units (∼20 mS cm−1) (23). Moreover, members of this lineage reached abundances of 2.3 × 106 cells ml−1 in the southern brackish zone of the lagoon in February 2004 (Fig. 5), which was as high as the total number of bacterial cells in the coastal waters immediately outside the sand bar at that time point (data not shown). This suggests that bacteria from the marine SAR11 clade might have been active and growing members of the microbial assemblages in the southern zone of the lagoon. In addition, it is conceivable that SAR11 bacteria persisted because they were consumed at a lower rate than other bacteria by phagotrophic protists (48), for example, as a consequence of their small cell size.
(iv) Dominance of single microbial genotypes.
Among the most interesting findings of our study were two pronounced blooms of heterotrophic bacteria that were affiliated to single 16S rRNA genotypes (>99.8% sequence similarity) (Fig. 3 and Table 2). In March 2003, the majority of bacteria in the lagoon (but especially in the brackish zone) were Alphaproteobacteria (Fig. 2) that were identified as Sphingomonas echinoides by 16S rRNA gene sequence analysis (Fig. 3) and CARD-FISH with a specific oligonucleotide probe. This species was also present, albeit at very low numbers (<1% total DAPI counts), in the microbial assemblages at the other sampling time points. The conspicuous dominance of S. echinoides coincided with a pronounced bloom of cyanobacteria that were affiliated to either Pseudanabaena (microscopic identification) or the morphologically similar Prochlorothrix (16S rRNA gene sequence data). The genus Sphingomonas is highly diversified (Fig. 3) and includes culturable representatives from many habitats, including the freshwater and marine plankton (26, 54). The occurrence of Sphingomonas spp. during cyanobacterial blooms has been reported previously (22). Some strains from this genus are even capable of degrading the cyanobacterial toxin microcystin and using it as a carbon source (46).
During the second summer, we observed an even more striking dominance of another single bacterial genotype affiliated to Gammaproteobacteria (Fig. 1 and 4 and Table 2), which showed more than 99.5% similarity with Stenotrophomonas maltophilia. This species is affiliated to the Xanthomonadaceae and has been obtained from lake plankton (20), sewage (58), sediments (5), and soil (18). In addition, it has been described as a growth-promoting or symbiotic agent in the rhizosphere of crops (30) and as a biocontrol agent against phytopathogenic fungi (61). S. maltophilia has also been isolated from a wide range of nosocomial sources (57), and it is increasingly recognized as an opportunistic human pathogen in hospitals (19).
Our data suggest that Laguna de Rocha provided adequate conditions for the mass development of this bacterium in December 2003. If extrapolated from the two sampling sites to the whole lagoon, a total population of >1020 cells of S. maltophilia (concentration [1010 cells liter−1] × average depth [0.6 m] × area [72 km2]) would be expected, representing >90% of the pelagic microbial assemblage (Fig. 1 and 4). Moreover, it is likely that these bacteria were highly active in the brackish zone of the lagoon, as reflected by the elevated leucine incorporation rates at that time point (Fig. 1B). Since the high bulk bacterial activity during the S. maltophilia dominance coincided with a peak in ammonium concentration (Table 1), we speculate that the dominance and activity of S. maltophilia in the southern zone of the lagoon could also be related to this nutrient. This may be particularly relevant for the management and land use of catchments of lagoons with comparable characteristics. It is conceivable that high input of nitrogen compounds (such as commercial fertilizers commonly used in agriculture) in combination with increased influx of organic carbon by terrestrial runoff might induce the development of large populations of potentially pathogenic bacteria. On the other hand, the rapid decline of S. maltophilia during a period of only 3 days points to a remarkable resilience of microbial assemblages in the lagoons against invasions by allochthonous bacteria, potentially mediated by protistan grazing or viral lysis (48, 60).
Acknowledgments
We thank Clemente Olivera, Lorena Rodríguez, and Gissell Lacerot for helping during samplings and Rudolf Amann for continuous support.
This study was supported by grant IFS A-2917/2, by a grant from the German Academic Exchange Program awarded to C.P., and by the Max Planck Society. This work was carried out in partial fulfillment of the requirements of C.P. for the doctoral degree from the University of Uruguay.
REFERENCES
- 1.Abreu, P. C., C. Odebrecht, and A. Gonzalez. 1994. Particulate and dissolved phytoplankton production of the Patos Lagoon Estuary, southern Brazil: comparison of methods and influencing factors. J. Plankton Res. 16:737-753. [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.APHA. 1995. Standard methods for the examination of water and wastewater. American Public Health Association, Washington, D.C.
- 4.Ashelford, K. E., N. A. Chuzhanova, J. C. Fry, A. J. Jones, and A. J. Weightman. 2005. At least 1 in 20 16S rRNA sequence records currently held in public repositories is estimated to contain substantial anomalies. Appl. Environ. Microbiol. 71:7724-7736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aznar, R., E. Alcaide, and E. Garay. 1992. Numerical taxonomy of pseudomonads isolated from water, sediment and eels. Syst. Appl. Microbiol. 14:235-246. [Google Scholar]
- 6.Beardsley, C., J. Pernthaler, W. Wosniok, and R. Amann. 2003. Are readily cultured bacteria in coastal North Sea waters suppressed by selective grazing mortality? Appl. Environ. Microbiol. 69:2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bell, K. N. I., P. D. Cowley, and A. K. Whitfield. 2001. Seasonality in frequency of marine access to an intermittently open estuary: implications for recruitment strategies. Estuar. Coast. Shelf Sci. 52:327-337. [Google Scholar]
- 8.Benlloch, S., F. Rodriguez-Valera, and A. J. Martinez-Murcia. 1995. Bacterial diversity in two coastal lagoons deduced from 16s rDNA PCR amplification and partial sequencing. FEMS Microbiol. Ecol. 18:267-280. [Google Scholar]
- 9.Bouvier, T. C., and P. A. del Giorgio. 2002. Compositional changes in free-living bacterial communities along a salinity gradient in two temperate estuaries. Limnol. Oceanogr. 47:453-470. [Google Scholar]
- 10.Coffin, R. B., and J. P. Connolly. 1997. Bacteria and heterotrophic microflagellate production in the Santa Rosa Sound, Florida. Hydrobiologia 353:53-61. [Google Scholar]
- 11.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33:D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conde, D., L. Aubriot, and R. Sommaruga. 2000. Changes in UV penetration associated with marine intrusions and freshwater discharge in a shallow coastal lagoon of the Southern Atlantic Ocean. Mar. Ecol. Prog. Ser. 207:19-31. [Google Scholar]
- 13.Conde, D., S. Bonilla, L. Aubriot, R. de Leon, and W. Pintos. 1999. Comparison of the areal amount of chlorophyll a of planktonic and attached microalgae in a shallow coastal lagoon. Hydrobiologia 409:285-291. [Google Scholar]
- 14.Conde, D., and R. Sommaruga. 1999. A review of the state of limnology in Uruguay, p. 1-31. In J. Gopal and R. Wetzel (ed.), Limnology in developing countries. SIL/LDC, Stuttgart, Germany.
- 15.Covert, J. S., and M. A. Moran. 2001. Molecular characterization of estuarine bacterial communities that use high- and low-molecular weight fractions of dissolved organic carbon. Aquat. Microb. Ecol. 25:127-139. [Google Scholar]
- 16.Crump, B. C., C. S. Hopkinson, M. L. Sogin, and J. E. Hobbie. 2004. Microbial biogeography along an estuarine salinity gradient: combined influences of bacterial growth and residence time. Appl. Environ. Microbiol. 70:1494-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daims, H., A. Bruhl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 18.Debette, J., and R. Blondeau. 1977. Caractérization de bactéries telluriques assimilables á Pseudomonas maltophilia. Can. J. Microbiol. 23:1123-1127. [PubMed] [Google Scholar]
- 19.Denton, M., and K. G. Kerr. 1998. Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia. Clin. Microbiol. Rev. 11:57-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Wever, A., K. Muylaert, K. Van der Gucht, S. Pirlot, C. Cocquyt, J. P. Descy, P. D. Plisnier, and W. Vyverman. 2005. Bacterial community composition in Lake Tanganyika: vertical and horizontal heterogeneity. Appl. Environ. Microbiol. 71:5029-5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dos Santos Furtado, A. L., P. Casper, and F. de Assis Estevesqq. 2002. Methanogenesis in an impacted and two dystrophic coastal lagoons (Macaé, Brazil). Braz. Arch. Biol. Technol. 45:195-202. [Google Scholar]
- 22.Eiler, A., and S. Bertilsson. 2004. Composition of freshwater bacterial communities associated with cyanobacterial blooms in four Swedish lakes. Environ. Microbiol. 6:1228-1243. [DOI] [PubMed] [Google Scholar]
- 23.Elifantz, H., R. R. Malmstrom, M. T. Cottrell, and D. L. Kirchman. 2005. Assimilation of polysaccharides and glucose by major bacterial groups in the Delaware Estuary. Appl. Environ. Microbiol. 71:7799-7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furtado, A. L. S., P. Casper, and F. A. Esteves. 2001. Bacterioplankton abundance, biomass and production in a Brazilian coastal lagoon and in two German lakes. An. Acad. Bras. Cienc. 73:39-49. [DOI] [PubMed] [Google Scholar]
- 25.Geiss, U., I. Bergmann, M. Blank, R. Schumann, M. Hagemann, and A. Schoor. 2003. Detection of Prochlorothrix in brackish waters by specific amplification of pcb genes. Appl. Environ. Microbiol. 69:6243-6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gich, F., K. Schubert, A. Bruns, H. Hoffelner, and J. Overmann. 2005. Specific detection, isolation, and characterization of selected, previously uncultured members of the freshwater bacterioplankton community. Appl. Environ. Microbiol. 71:5908-5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glöckner, F. O., B. M. Fuchs, and R. Amann. 1999. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl. Environ. Microbiol. 65:3721-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grossart, H.-P., and M. Simon. 1993. Limnetic macroscopic organic aggregates (lake snow): occurrence, characteristics, and microbial dynamics in Lake Constance. Limnol. Oceanogr. 38:532-546. [Google Scholar]
- 29.Jeffrey, S. W., and G. F. Humphrey. 1975. New spectrophotometric equations for determining chlorophylls a, b, c1 and c2 in higher plants, algae and natural phytoplankton. Biochem. Physiol. Pflanz. 167:191-194. [Google Scholar]
- 30.Juhnke, M. E., D. E. Mathre, and D. C. Sands. 1987. Identification and characterization of rhizosphere-competent bacteria of wheat. Appl. Environ. Microbiol. 53:2793-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirchman, D. L., A. I. Dittel, S. E. G. Findlay, and D. Fischer. 2004. Changes in bacterial activity and community structure in response to dissolved organic matter in the Hudson River, New York. Aquat. Microb. Ecol. 35:243-257. [Google Scholar]
- 32.Kirchman, D. L., A. I. Dittel, R. R. Malmstrom, and M. T. Cottrell. 2005. Biogeography of major bacterial groups in the Delaware Estuary. Limnol. Oceanogr. 50:1697-1706. [Google Scholar]
- 33.Kirchman, D. L., and H. W. Ducklow. 1993. Estimating conversion factors for the thymidine and leucine methods for measuring bacterial production, p. 513-517. In P. F. Kemp, B. F. Sherr, E. B. Sherr, and J. J. Cole (ed.), Handbook of methods in aquatic microbial ecology. Lewis Publishers, Boca Raton, Fla.
- 34.Kirchman, D. L., L. Y. Yu, B. M. Fuchs, and R. Amann. 2001. Structure of bacterial communities in aquatic systems as revealed by filter PCR. Aquat. Microb. Ecol. 26:13-22. [Google Scholar]
- 35.LaMontagne, M. G., and P. A. Holden. 2003. Comparison of free-living and particle-associated bacterial communities in a coastal lagoon. Microb. Ecol. 46:228-237. [DOI] [PubMed] [Google Scholar]
- 36.Langenheder, S., V. Kisand, J. Wikner, and L. J. Tranvik. 2003. Salinity as a structuring factor for the composition and performance of bacterioplankton degrading riverine DOC. FEMS Microbiol. Ecol. 45:189-202. [DOI] [PubMed] [Google Scholar]
- 37.Lebaron, P., P. Servais, M. Troussellier, and C. Courties. 2001. Microbial community dynamics in Mediterranean nutrient-enriched seawater. FEMS Microbiol. Ecol. 34:255-266. [DOI] [PubMed] [Google Scholar]
- 38.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Forster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. Konig, T. Liss, R. Lussmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K.-H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum Cytophaga-Flavobacter-Bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 40.Manz, W., R. Amann, W. Ludwig, M. Wagner, and K.-H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 41.Miller, S. D., S. H. D. Haddock, C. D. Elvidge, and T. F. Lee. 2005. Detection of a bioluminescent milky sea from space. Proc. Natl. Acad. Sci. USA 102:14181-14184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morris, R. M., M. S. Rappe, S. A. Connon, K. L. Vergin, W. A. Siebold, C. A. Carlson, and S. J. Giovannoni. 2002. SAR11 clade dominates ocean surface bacterioplankton communities. Nature 420:806-810. [DOI] [PubMed] [Google Scholar]
- 43.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oliveira, A. M., and B. Kjerfve. 1993. Environmental responses of a tropical coastal lagoon system to hydrological variability—Mundau-Manguaba, Brazil. Estuar. Coast. Shelf Sci. 37:575-591. [Google Scholar]
- 45.Painchaud, J., J. C. Therriault, and L. Legendre. 1995. Assessment of salinity-related mortality of freshwater bacteria in the Saint Lawrence Estuary. Appl. Environ. Microbiol. 61:205-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park, H. D., Y. Sasaki, T. Maruyama, E. Yanagisawa, A. Hiraishi, and K. Kato. 2001. Degradation of the cyanobacterial hepatotoxin microcystin by a new bacterium isolated from a hypertrophic lake. Environ. Toxicol. 16:337-343. [DOI] [PubMed] [Google Scholar]
- 47.Pernthaler, A., J. Pernthaler, and R. Amann. 2004. Sensitive multicolour fluorescence in situ hybridization for the identification of environmental organisms, p. 711-726. In G. A. Kowalchuk, F. J. De Bruijn, I. M. Head, A. D. L. Akkermans, and J. D. van Elsas (ed.), Molecular microbial ecology manual, 2nd ed. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 48.Pernthaler, J. 2005. Predation on procaryotes in the water column and its ecological implications. Nat. Rev. Microbiol. 3:537-546. [DOI] [PubMed] [Google Scholar]
- 49.Pernthaler, J., A. Pernthaler, and R. Amann. 2003. Automated enumeration of groups of marine picoplankton after fluorescence in situ hybridization. Appl. Environ. Microbiol. 69:2631-2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ranasinghe, R., and C. Pattiaratchi. 1999. Circulation and mixing characteristics of a seasonally open tidal inlet: a field study. Mar. Freshw. Res. 50:281-290. [Google Scholar]
- 51.Raymond, P. A., and J. E. Bauer. 2000. Bacterial consumption of DOC during transport through a temperate estuary. Aquat. Microb. Ecol. 22:1-12. [Google Scholar]
- 52.Roller, C., M. Wagner, R. Amann, W. Ludwig, and K.-H. Schleifer. 1994. In situ probing of gram-positive bacteria with high DNA G+C content using 23S rRNA-targeted oligonucleotides. Microbiology 140:2849-2858. [DOI] [PubMed] [Google Scholar]
- 53.Sakami, T., K. Abo, K. Takayanagi, and S. Toda. 2003. Effects of water mass exchange on bacterial communities in an aquaculture area during summer. Estuar. Coast. Shelf Sci. 56:111-118. [Google Scholar]
- 54.Schut, F., J. C. Gottschal, and R. A. Prins. 1997. Isolation and characterization of the marine ultrabacterium Sphingomonas sp. strain RB2256. FEMS Microbiol. Rev. 20:363-369. [Google Scholar]
- 55.Simek, K., J. Armengol, M. Comerma, J. C. Garcia, P. Kojecka, J. Nedoma, and J. Hejzlar. 2001. Changes in the epilimnetic bacterial community composition, production, and protist-induced mortality along the longitudinal axis of a highly eutrophic reservoir. Microb. Ecol. 42:359-371. [DOI] [PubMed] [Google Scholar]
- 56.Šimek, K., K. Horòák, J. Jezbera, M. Mašín, J. Nedoma, J. M. Gasol, and M. Schauer. 2005. Influence of top-down and bottom-up manipulations on the R-BT065 subcluster of β-proteobacteria, an abundant group in bacterioplankton of a freshwater reservoir. Appl. Environ. Microbiol. 71:2381-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Swings, J., P. De Vos, M. Van den Mooter, and J. De Ley. 1983. Transfer of Pseudomonas maltophilia Hugh 1981 to the genus Xanthomonas as Xanthomonas maltophilia (Hugh 1981) comb. nov. Int. J. Syst. Bacteriol. 33:409-413. [Google Scholar]
- 58.Wallace, W. H., J. F. Rice, D. C. White, and G. S. Sayler. 1994. Distribution of alginate genes in bacterial isolates from corroded metal surfaces. Microb. Ecol. 27:213-223. [DOI] [PubMed] [Google Scholar]
- 59.Warnecke, F., J. Pernthaler, and R. Amann. 2004. Actinobacterial 16S rRNA genes from freshwater habitats cluster in four distinct lineages. Environ. Microbiol. 6:242-253. [DOI] [PubMed] [Google Scholar]
- 60.Weinbauer, M. G. 2004. Ecology of prokaryotic viruses. FEMS Microbiol. Rev. 28:127-181. [DOI] [PubMed] [Google Scholar]
- 61.Zhang, Z., G. Y. Yuen, G. Sarath, and A. R. Penheiter. 2001. Chitinases from the plant disease biocontrol agent, Stenotrophomonas maltophilia C3. Phytopathology 91:204-211. [DOI] [PubMed] [Google Scholar]
- 62.Zwart, G., B. C. Crump, M. Agterveld, F. Hagen, and S. K. Han. 2002. Typical freshwater bacteria: an analysis of available 16S rRNA gene sequences from plankton of lakes and rivers. Aquat. Microb. Ecol. 28:141-155. [Google Scholar]