Abstract
Nonsteroidal anti-inflammatory drugs (NSAIDs) are commonly prescribed for a variety of inflammatory conditions; however, the benefits of this class of drugs are accompanied by deleterious side effects, most commonly gastric irritation and ulceration. NSAID-induced ulceration is thought to be exacerbated by intestinal microbiota, but previous studies have not identified specific microbes that contribute to these adverse effects. In this study, we conducted a culture-independent analysis of ∼1,400 bacterial small-subunit rRNA genes associated with the small intestines and mesenteric lymph nodes of rats treated with the NSAID indomethacin. This is the first molecular analysis of the microbiota of the rat small intestine. A comparison of clone libraries and species-specific quantitative PCR results from rats treated with indomethacin and untreated rats revealed that organisms closely related to Enterococcus faecalis were heavily enriched in the small intestine and mesenteric lymph nodes of the treated rats. These data suggest that treatment of NSAID-induced ulceration may be facilitated by addressing the microbiological imbalances.
Despite being one of the most frequently used classes of drugs, nonsteroidal anti-inflammatory drugs (NSAIDs) have adverse effects that result in substantial morbidity and mortality (3, 11, 46). Toxicities to the gastrointestinal, renal, and cardiovascular systems are documented for a variety of prescribed and over-the-counter (e.g., aspirin, ibuprofen, and naproxen) NSAIDs (14, 15, 36). Dyspepsia secondary to gastroduodenal mucosal injury is the most commonly cited side effect of prolonged ingestion of NSAIDs. Additionally, epidemiological and experimental studies have demonstrated that NSAIDs can cause ulceration of the small intestine (12). Overall, severe gastrointestinal complications from NSAIDs, including peptic ulceration, perforation, and hemorrhage, require over 100,000 hospitalizations annually in the United States (46).
NSAID-induced toxicity arises from a combination of localized and systemic effects. NSAIDs are thought to mediate systemic effects that result in deleterious changes in mucus hydrophobicity and epithelial pH, erosion of the gastroduodenal mucosa, and increases in intestinal permeability (44, 46). The gastric mucosa therefore becomes susceptible to bacterial invasion and the activities of endotoxins. The deleterious effects are exacerbated by prolonged drug exposure due to enterohepatic circulation and redelivery of NSAIDs to the small intestine. Furthermore, bacterial infiltration of the intestinal wall activates the local inflammatory response. Current understanding links systemic damage to small-intestine microvascular permeability that allows intestinal flora, or their products, to access other host organs. Translocation is demonstrated in the loading of the mesenteric lymph nodes (MLNs) and other organs by intestinal flora and upregulation of inflammatory factors in the liver, lung, kidney, and spleen (7, 17, 47).
There is strong evidence for microbial involvement in ulceration, although little is known about the specific causative microorganisms. For example, germfree rats injected with the NSAID indomethacin do not develop symptoms of inflammation and ulceration (32). Indomethacin is a commonly prescribed NSAID and is often used as a model of such agents. Furthermore, when indomethacin is coadministered with broad-range antibiotics or antimicrobials, experimental animals do not develop gastric ulcers (7, 8, 17, 47). Investigations of microbial influences on ulceration have been limited to culture-based studies, which indicate an expansion of gram-negative bacteria within the intestine following challenge with indomethacin. Similarly, culture-based studies have also shown increased microbial loads in the MLNs, but the results have not identified the organisms (47). Thus, the specific microbes involved in NSAID-induced insult (e.g., inflammation and ulceration) are poorly understood.
Characterization of the microbial response to NSAIDs has been hampered by the traditional requirement for culture to detect and identify microbes because most natural microbes are not cultured using standard methods (13, 27, 29, 45). Through the use of rRNA gene-based techniques, however, it has become possible to identify the intestinal microbiota molecularly, independently of culture (9). With these techniques, microbial rRNA genes are obtained, for instance, by PCR and cloning, and then sequenced to identify the corresponding organisms. The results are interpreted using a phylogenetic framework. Contextualization of a sequence, by phylogenetic comparison to sequences of known organisms, allows inference of some information related to metabolic properties and possible pathogenicity. Previous rRNA gene-based studies of the gut ecosystems in mammals have dramatically expanded our understanding of the microbial ecology of the intestine (2, 6, 19, 20, 26, 29, 40). The gastrointestinal tract contains many microbes, often mutualistic, that participate in nutrient absorption, digestion, stimulation of host mucosal immunity, and myriad other roles (1, 37). Understanding the distribution and balance of different microbial groups is one key to understanding the intestinal functions; conversely, understanding disequilibrium in the system may be a key to understanding NSAID ulceration, as well as other inflammatory bowel diseases (22). In order to identify microbes potentially involved in NSAID effects, we have conducted an rRNA-based analysis of the impact of administration of indomethacin on the microbiota of the gastrointestinal tract and regional lymph nodes in a rat model system.
MATERIALS AND METHODS
Experimental animals.
A total of 16 female Lewis rats weighing approximately 200 g were randomly assigned to control (6 individuals) and treatment (10 individuals) groups. The experimental rats received two subcutaneous injections of indomethacin (7.5 mg/kg of body weight), 24 h apart, in a solution of 5% NaHCO3. The control rats received subcutaneous injections of 5% NaHCO3. The rats were sacrificed 48 h after the first injection; 10 cm of the affected small intestine, as well as the mesenteric lymph nodes, was removed. Specimens to be used in rRNA gene analyses were snap frozen. Specimens for histology were fixed in formalin.
Histopathology.
A 10-cm section of the jejunum in the area at risk for indomethacin-induced inflammation and ulceration was evaluated microscopically for lesions. Five equally spaced cross sections of formalin-fixed intestine were processed for paraffin embedding, sectioned, stained with hematoxylyn and eosin, and scored according to the following criteria: minimal, less than 10% of the section area was affected by inflammation and mucosal necrosis; mild, 11 to 25% of the section area was affected; moderate, 26 to 50% of the section area was affected; severe, greater than 50% of section area was affected by inflammation and mucosal necrosis. Bacterial colonization was scored on “−” to “+++” scale, with − indicating no colonization, + indicating minor colonization, ++ indicating moderate colonization, and +++ assigned to sections with marked diffuse colonization in necrotic tissue of lesion areas.
Preparation of sample DNA.
For extraction of bulk nucleic acids from rat tissues, lymph nodes and small-intestine samples were washed extensively in TE (10 mM Tris [pH 8.0], 1 mM EDTA) containing 0.15 M NaCl in order to minimize contamination from the small-intestine lumenal contents. Approximately 150 to 250 mg of small-intestine tissue or ∼10 to 50 mg of MLN tissue samples was minced with a sterile razor blade and placed in sterile microcentrifuge tubes that contained 500 μl of buffer A (200 mM NaCl, 200 mM Tris-Cl [pH 8.0], 20 mM EDTA), 220 μl of 20% sodium dodecyl sulfate, 250 μl of phenol, and 250 μl of chloroform. A total of 0.5 g of 0.1-mm zirconia/silica beads was added to the suspension, and samples were mechanically disrupted in a Mini Beadbeater-8 (Biospec Products, Bartlesville, OK) at the highest setting for 2 minutes. The samples then were centrifuged (16,100 × g) for 5 minutes, and the aqueous layer was reextracted two more times with an equal volume of phenol-chloroform and once with an equal volume of chloroform.
DNAs were precipitated by the addition of NaCl (to 0.280 M) and 2.5 volumes of 100% ethanol, followed by centrifugation (16,100 × g; 30 min). The pellets were washed with 200 μl of 70% ethanol and centrifugation (16,100 × g; 10 min), dried in air, and then resuspended in 150 μl of sterile TE. DNA concentrations were assessed using the Quant-iT PicoGreen dsDNA Reagent kit according to the manufacturer's suggestions (Molecular Probes, Eugene, OR) and a Turner Quantech Digital Filter Flourometer (Barnstead Thermolyne Corporation, Dubuque, IA).
PCR, cloning, sequencing, and analysis.
Bacterial small-subunit (SSU) rRNA genes were amplified by PCR from specimen DNA using the bacterium-specific primers 8F (5′-AGAGTTTGATCCTGGCTCAG) and 805R (5′-GACTACCAGGGTATCTAAT) (18). Thirty-microliter PCR mixtures contained 1× PCR buffer (Bioline USA Inc., Randolph, MA), 2.5 mM MgCl2, deoxynucleoside triphosphates (0.21 mM each), 25 ng of each primer, 1 unit of Taq DNA polymerase (Bioline USA Inc., Randolph, MA), and ∼2 mg (2 μl) of genomic DNA lysate.
The PCR mixtures were amplified using touchdown cycling (5). Genomic DNAs were first denatured at 94°C for 2 min. The first 20 cycles consisted of a 30-second denaturation step at 94°C and a 30-second annealing step, initially 65°C and decreased 1°C every cycle. Annealing was followed by a 1-minute extension step at 72°C. Cycles 21 through 35 had denaturation and elongation steps that were identical to those in the first 20 cycles; however, the 30-second annealing step remained constant at 45°C. After all cycling was complete, the reaction mixtures were incubated at 72°C for an additional 20 min in order to tail the PCR products with polyadenylate.
The PCR products were resolved and visualized by electrophoresis through 1% agarose Tris-borate-EDTA gels stained with ethidium bromide (Gel Logic 200; Eastman Kodak Company, Rochester, NY); all control reactions that lacked DNA were negative. PCR products of sufficient intensity and appropriate length were excised from the gel and purified using QIAquick gel purification columns (QIAGEN, Valencia, CA). The purified PCR products were then ligated into the pCR4-TOPO TA vector (Invitrogen, Carlsbad, CA) and transformed into One Shot TOP 10 competent cells (Invitrogen, Carlsbad, CA), following the manufacturer's instructions. Ideally, clone libraries from each sample consisted of 96 randomly chosen transformants, but in several libraries, poor cloning efficiency limited the total number of clones that could be screened (see Table 2). Transformants were grown in 96-well culture plates overnight at 37°C with vigorous shaking; each well contained 1.5 ml of 2× YT medium (24).
TABLE 2.
Ranked abundances of phylogenetic clusters encountered in control and indomethacin-injected rats
| Cluster representativeb | Accession no.c | Abundancea
|
Total no. of clonesd | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control small intestines
|
Control MLNs
|
Indomethacin-injected small intestines
|
Indomethacin-injected MLNs
|
||||||||||||||||||||||
| BR01 | BR02 | BR05 | BR06 | BR13 | BR14 | BR01 | BR02 | BR06 | BR03 | BR04 | BR08 | BR11 | BR12 | BR15 | BR16 | BR03 | BR04 | BR10 | BR11 | BR15 | BR16 | ||||
| Firmicutes | |||||||||||||||||||||||||
| SFB | X87244 | 64 | 20 | 39 | 95 | 3 | 157 | ||||||||||||||||||
| Bacillus licheniformis | X68416 | 20 | 17 | 3 | 29 | 31 | 24 | 22 | 4 | 11 | 19 | 28 | 24 | 1 | 152 | ||||||||||
| Lactobacillus murinus | AJ308393 | 16 | 18 | 2 | 68 | 61 | 5 | 51 | 5 | 143 | |||||||||||||||
| Enterococcus faecalis | AF515223 | 43 | 12 | 1 | 40 | 1 | 49 | 4 | 40 | 122 | |||||||||||||||
| Clostridium difficile | AJ306761 | 4 | 6 | 44 | 51 | 94 | |||||||||||||||||||
| Lactobacillus intestinalis | AJ306299 | 25 | 18 | 37 | |||||||||||||||||||||
| Lactobacillus reuteri | AJ308392 | 26 | 4 | 6 | 29 | ||||||||||||||||||||
| Lactobacillus acidophilus | AF371477 | 1 | 24 | 1 | 1 | 24 | |||||||||||||||||||
| Lactobacillus sp. | AF243163 | 20 | 16 | ||||||||||||||||||||||
| Streptococcus sp. | AF385545 | 4 | 2 | 1 | 4 | ||||||||||||||||||||
| Enterococcus faecalis | AF515223 | 2 | 1 | 3 | |||||||||||||||||||||
| Proteobacteria | |||||||||||||||||||||||||
| γ Group | |||||||||||||||||||||||||
| Escherichia coli | AE000345 | 27 | 35 | 36 | 4 | 36 | 41 | 33 | 51 | 52 | 16 | 16 | 56 | 2 | 1 | 47 | 60 | 43 | 33 | 57 | 396 | ||||
| Stenotrophomonas maltophilia | AB021406 | 6 | 4 | 2 | 2 | 2 | 6 | 3 | 3 | 17 | |||||||||||||||
| Acinetobacter johnsonii | AF467299 | 4 | 2 | 4 | 1 | 3 | 3 | 11 | |||||||||||||||||
| Pseudomonas stutzeri | D85998 | 2 | 1 | 2 | 1 | 3 | 2 | 8 | |||||||||||||||||
| β Group | |||||||||||||||||||||||||
| Aquabacterium commune | AY135860 | 7 | 6 | 3 | 2 | 10 | 19 | 3 | 32 | ||||||||||||||||
| Burkholderiales species | AY053477 | 2 | 5 | 1 | 3 | 2 | 8 | 6 | 8 | 26 | |||||||||||||||
| Achromobacter xylosoxidans | AJ306836 | 6 | 4 | 10 | 18 | ||||||||||||||||||||
| Oxalobacteraceae species | AJ252575 | 11 | 6 | 9 | |||||||||||||||||||||
| Delftia acidovorans | AB074256 | 4 | 5 | 6 | |||||||||||||||||||||
| Curvibacter lanceolatus | AJ387862 | 7 | 2 | 5 | |||||||||||||||||||||
| Comamonadaceae | AF445700 | 4 | 5 | 5 | |||||||||||||||||||||
| Rubrivivax gelatinosus | AF293003 | 2 | 2 | 1 | 4 | ||||||||||||||||||||
| Herbaspirillum autotrophicum | AY133107 | 2 | 4 | 4 | |||||||||||||||||||||
| Variovorax paradoxus | AJ292635 | 1 | 2 | 2 | |||||||||||||||||||||
| Burkholderia caryophylli | AF052387 | 1 | 3 | 2 | |||||||||||||||||||||
| α Group | |||||||||||||||||||||||||
| Bradyrhizobium japonicum | AF417546 | 3 | 1 | 3 | 1 | 5 | 8 | 12 | |||||||||||||||||
| Agrobacterium tumefaciens | AF531767 | 4 | 4 | ||||||||||||||||||||||
| Phyllobacterium myrsinacearum | D12789 | 2 | 3 | 3 | |||||||||||||||||||||
| Methylobacterium | AB015566 | 2 | 1 | 2 | 3 | ||||||||||||||||||||
| Sphingobium yanoikuyae | AB091683 | 2 | 1 | 3 | |||||||||||||||||||||
| Sphingomonas taejonensis | AB022601 | 3 | 3 | ||||||||||||||||||||||
| Bosea thiooxidans | AF531764 | 3 | 3 | ||||||||||||||||||||||
| Bacteroidetes |
|
||||||||||||||||||||||||
| Flavobacterium columnare | AF493636 | 2 | 7 | 3 | 6 | 2 | 8 | 16 | 3 | 35 | |||||||||||||||
| Saprospiraceae | AY053480 | 3 | 1 | 4 | |||||||||||||||||||||
| Flavobacterium indologenes | AF217562 | 4 | 3 | 4 | |||||||||||||||||||||
| Sphingobacterium heparinum | AJ252710 | 7 | 3 | ||||||||||||||||||||||
| Saprospiraceae | AF368190 | 1 | 1 | 2 | |||||||||||||||||||||
| Actinobacteria | |||||||||||||||||||||||||
| Dietzia maris | AB010904 | 7 | 3 | ||||||||||||||||||||||
| Brachybacterium alimentarium | X91031 | 2 | 1 | 2 | |||||||||||||||||||||
| Othere | 13 | 7 | 3 | 6 | 0 | 2 | 4 | 0 | 0 | 9 | 2 | 0 | 1 | 0 | 0 | 2 | 3 | 13 | 0 | 15 | 3 | 8 | 53 | ||
| Total no. of clonesf | 45 | 45 | 66 | 80 | 85 | 89 | 95 | 94 | 42 | 45 | 42 | 25 | 89 | 43 | 82 | 89 | 79 | 47 | 88 | 39 | 63 | 91 | 1,468 | ||
Abundances are expressed as percentages of the total number of clones from a particular library rounded to the nearest integer.
Relatedness cluster representatives were identified by proximity to experimental sequences in a phylogenetic tree constructed by parsimony insertion in the program ARB. Cultivars with the highest average percent sequence identity were chosen as representatives of their respective clusters. In some cases, clusters were less than 97% identical to known organisms; in these cases, broader taxonomic identifiers are given.
Closest relative of relatedness cluster as identified by BlastN search. The best BLAST hits do not necessarily correspond to the cluster representative.
The total number of clones for each relatedness cluster. Each relatedness cluster is comprised of sequences that share at least 97% sequence similarity.
All sequences that are present in only one library and for which there are two or fewer representatives.
The total number of clones for each library.
Restriction fragment length polymorphism (RFLP) analysis was carried out in 96-well plates. Twenty microliters of culture from each transformant was heated at 95°C in 20 μl of 10 mM Tris (pH 8.0), the precipitates were pelleted by centrifugation (4,000 × g; 10 min), and 1.5 μl of the supernatants was used as a template in PCRs, as previously described (4). The PCR products were analyzed by agarose gel electrophoresis to verify the presence of inserts, and the products of PCRs were digested with MspI and HinPI (2 U) at 37°C for 3 h (4). The products of the RFLP digestion were separated on a 4% Gene-Pure HiRes Agarose gel (ISC BioExpress, Kaysville, UT). Digitized images of the RFLP gels were compared manually (Gel Logic 200; Eastman Kodak Company, Rochester, NY). At least two representative clones were sequenced for RFLP types that occurred multiple times within a particular clone library.
Selected clones were sequenced on a Licor automated DNA sequencer using the ThermoSequenase cycle-sequencing kit (USB Corporation, Cleveland, OH) according to the manufacturer's protocol. Inserts were sequenced in both directions, and sequences were obtained for both strands. All sequence and associated data were managed using the software package XplorSeq (D. N. Frank, unpublished data). Sequences were identified initially by BlastN searches (National Center for Biotechnology Information [NCBI], Bethesda, MD) using the client application BlastCl3 (NCBI, Bethesda, MD). All sequences that were longer than 500 nucleotides and that had BLAST bit scores to other SSU rRNA genes greater than 500 were used in further analyses. The sequences were aligned to an existing database of aligned SSU rRNA genes (23) using the ARB software package (http://www.carb-home.de). Aligned sequences were clustered at the 97% identity level using the program SortX (D. N. Frank, unpublished data); sequence relatedness clusters were generated by mean-linkage clustering of sequences base on uncorrected pairwise sequence similarities. Phylogenetic analyses were carried out using ARB. Estimates of sample coverage (Good's coverage) and species richness (SChao1) (see below) were calculated for each of the 22 tissue specimens using the software packages EstimateS (R. K. Colwell, EstimateS: statistical estimation of a species richness and shared species from samples, version 7.5 [http//purl.oclc.org/estimates]) and XplorSeq (Frank, unpublished). The data set for these calculations included the number of representatives of each observed RFLP type. Phylotypes were resampled 100 times (28).
Q-PCR.
Segmented filamentous bacteria (SFB) and Enterococcus faecalis group-specific primers were designed using the primer design function in the ARB software package. For SFB assays, the primers SFB1F (5′-AGGAGGAGTCTGCGGCAC) and SFB2R (5′-CCTTCCTCTTCCCTGCT) were used. For the E. faecalis assays, the primers EFAEC1F (5′-GCATAAGAGTGAAAGGCG) and EFAEC2R (5′-TAGATACCGTCAGGGGAC) were used. In order to verify the specificity of the assays, clone libraries from 11 of the quantitative-PCR (Q-PCR) products were prepared, and 192 clones were sequenced from the 11 Q-PCRs following the methods described above.
Q-PCR mixtures contained 25 ng of each primer, 11 μl of 2× Sybr Green JumpStart Taq ReadyMix (Sigma, St. Louis, MO), 11 μl of H2O, and between 50 and 350 ng of bulk genomic DNA. SFB Q-PCR mixtures were initially denatured at 94°C for 5 minutes. Next, for 45 cycles, the reaction mixtures were denatured at 94°C for 30 seconds, annealed at 62°C for 30 seconds, elongated at 72°C for 30 seconds, and heated to 83°C for 5 seconds, and then a fluorescence reading was taken. The E. faecalis assay cycling parameters were identical to the SFB parameters with the exceptions that the annealing temperature for the E. faecalis Q-PCR was 58°C and the PCR ran for 50 cycles instead of 45.
Plasmid quantitation standards for both Q-PCR assays were a representative clone of each specific group, purified using QIA Miniprep columns according to the manufacturer's protocol (QIAGEN, Valencia, CA) and quantified by fluorimetry as described above. A 10-fold dilution series, ranging from 1 × 108 copies to 1 copy, was made by serial dilution in H2O.
Q-PCRs used 10-fold dilutions of bulk genomic DNA from rat specimens with a DNA Engine Opticon System (MJ Research, Incline Village, NE). For each Q-PCR assay, PCR was carried out on the plasmid dilution series to generate a standard curve. Three replicate PCRs were performed for each sample. Quantifications of template concentrations were made by linear extrapolation of baseline-subtracted data from the plasmid dilution series standard curves. Because Q-PCR data were measured in triplicate for each sample, a multivariate analysis of variance (MANOVA) was performed to determine whether E. faecalis and/or SFB populations differed significantly between control and experimental groups. Statistical analyses used the R software package (31). A plot of the residuals obtained in the initial MANOVA versus the expected values for a normal distribution revealed a nonlinear relationship, suggesting that the variance of the data was not normally distributed. A logarithmic transformation of the Q-PCR data produced a high correlation between residuals and expected values (r > 0.97), which indicated that error variance in this model was normally distributed. The reported P values, which test the null hypotheses of equivalent mean Q-PCR results between control and treatment groups, were therefore produced by MANOVA of transformed data sets.
Nucleotide sequence accession numbers.
The nucleotide sequences of the SSU rRNA genes described in this paper have been deposited in the GenBank database (NCBI, Bethesda, MD) with accession numbers DQ856621 to DQ857242.
RESULTS
Two injections of indomethacin (7.5 mg/kg) produced microscopic alterations of intestinal inflammation and ulceration ranging from minimal to severe in 10 rats. Six additional rats served as uninjected controls, and no abnormalities were observed in these animals (Table 1). Semiquantitative microscopic assessments of the bacterial load (the number of bacterial colonies per field) in mucosal lesions were made during histopathology evaluation for rats injected with indomethacin (Table 1).
TABLE 1.
Pathology overview for control and treatment groups
| Subject | Groupa | Ileum/jejunumb | MLNb | Pathology | Colonizationc |
|---|---|---|---|---|---|
| BR01 | − | + | + | Normal | ND |
| BR02 | − | + | + | Normal | ND |
| BR05 | − | + | − | Normal | ND |
| BR06 | − | + | + | Normal | ND |
| BR13 | − | + | − | Normal | ND |
| BR14 | − | + | − | Normal | ND |
| BR03 | + | + | + | Severe | +++ |
| BR04 | + | + | + | Mild | − |
| BR07 | + | − | − | Severe | +++ |
| BR08 | + | + | − | Mild | + |
| BR09 | + | − | − | Severe | ++ |
| BR10 | + | − | + | Moderate | ++ |
| BR11 | + | + | + | Mild | +++ |
| BR12 | + | + | − | Severe | ++ |
| BR15 | + | + | + | Mild | ND |
| BR16 | + | + | + | Moderate | ND |
Indicates control group (−) or indomethacin-injected group (+).
Indicates whether rRNA PCR was successful (+) or unsuccessful (−) for a particular sample.
ND, no data; −, no bacterial colonization; +, minor colonization; ++, moderate colonization; +++, marked necrosis.
DNAs were prepared from tissues as described in Materials and Methods, and bacterial SSU rRNA genes were successfully amplified from 13/16 gastrointestinal DNAs and 9/16 MLN DNAs. PCR products were not obtained from some samples, which were not considered further. rRNA clone libraries were constructed from PCR products and screened by RFLP assay (see Materials and Methods) in order to identify and enumerate related clone types. Multiple sequences were determined for each unique RFLP type. Sequences with either BLAST bit scores of less than 500 or lengths of less than 500 nucleotides were not analyzed further. A total of 622 sequences from the 1,468 clones analyzed were included in the final data set; 570 of the 622 sequences (∼91%) were at least 97% identical to those of previously sequenced rRNA genes. This corresponds approximately to species level identification (39).
rRNA gene sequences from different samples seldom are exactly identical but generally belong to relatedness clusters (6, 10, 21, 38, 43). In order to assess the diversity and novelty of microorganisms present in the gastrointestinal-tract and MLN samples, sequences were clustered into relatedness groups with pairwise sequence identities of ≥97%, a “species level” bin. With this constraint, 84 such relatedness groups were identified. Table 2 summarizes the identification of these relatedness clusters in individual rat specimens and the closest GenBank BLAST hit for the particular cluster.
To determine the extent to which the clone libraries adequately represented the microorganisms present in each specimen, the distributions of sequences in each library (corrected for RFLP occurrence) were analyzed by Good's coverage statistic, an estimator of the completeness of sampling (16). By this statistic, the libraries were sampled, on average, with 90% coverage (range, 69% to 95% coverage). An estimate of species richness, the SChao1 statistic (16), indicated that a relatively low number of species (mean, 23; range, 4.5 to 55.5) were present in each clone library.
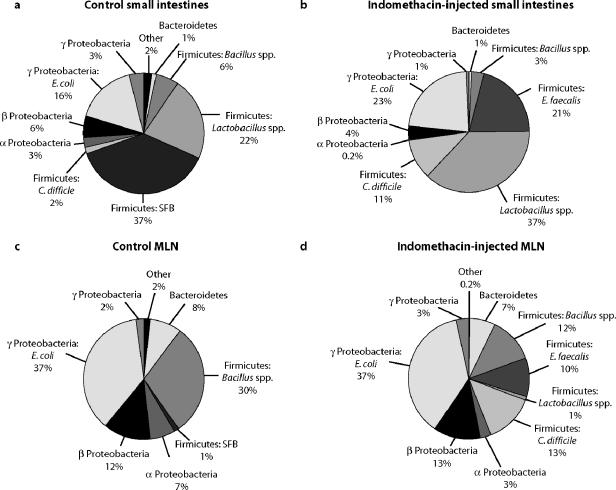
Molecular studies of large-bowel ecosystems have revealed relatively low diversity at the phylum level (1, 6, 20), and we found the same in the rat small intestine. The rRNA sequences obtained in this study of the small intestine belonged to organisms from only 4 of the >75 major bacterial phyla (phylogenetic divisions) (21, 30, 35): actinobacteria, firmicutes, bacteriodetes, and proteobacteria. Figure 1a to d summarizes the findings for each of the tissue groups. Sequences from the small-intestine mucosa of both indomethacin-injected and control rats belonged predominantly to the firmicutes (low-G+C gram-positive bacteria). In control rats, the firmicute sequences primarily consisted of SFB, Lactobacillus spp., and Bacillus spp., whereas in the indomethacin-injected rats, the majority of firmicute sequences were Lactobacillus spp., E. faecalis spp., and Clostridium difficile spp. (Fig. 1). In both control and experimental samples, proteobacterial (enterobacterial) sequence types were the second most prevalent (Fig. 1). Most of the enterobacterial sequences could be further classified as Escherichia coli-type sequences and did not differ significantly between the control and experimental-group samples (Table 2).
FIG. 1.
SSU clone distribution in control and experimental rat small intestines and mesenteric lymph nodes. Sequences are classified by phylum. In some cases, further taxonomic classification is given, depending on the diversity of sequences in a particular wedge. Percentages were calculated from pooled samples for each group. (a) Control rat small-intestine clone libraries. (b) Indomethacin-injected rat small-intestine clone libraries. (c) Control rat MLN clone libraries. (d) Indomethacin-injected rat MLN clone libraries.
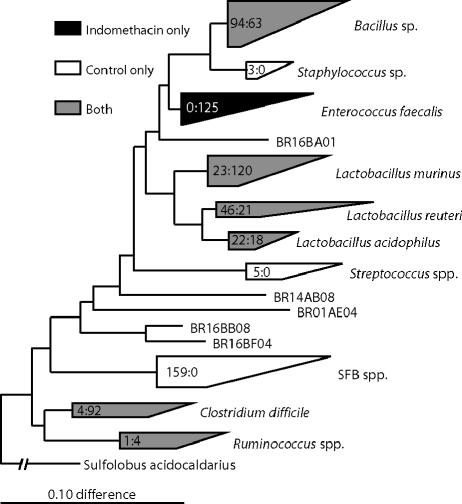
Although the microbial makeup of individual samples varied in detail, the dominant firmicute phylotypes taken together differed significantly between control and indomethacin-injected rats. This is summarized in the phylogenetic diagram in Fig. 2. Control rat libraries contained abundant unique sequences that were not observed in experimental samples. Staphylococcus spp., Streptococcus spp., and SFBs, all firmicutes, were present in the controls but were not detected in rats injected with indomethacin. The SFB sequences, particularly, comprised a large component of the control rat libraries but were not detected among analyzed sequences from experimental rats.
FIG. 2.
A parsimony insertion phylogenetic tree exported from ARB representing the prevalences of different firmicute lineages in different samples. The wedges are classified by a cultured representative and in some cases by broader taxonomic identifiers. The numbers inside the wedges correspond to the numbers of clones in the control samples, followed by the numbers of clones in the experimental samples. Gray wedges represent lineages that are found in both experimental and control samples. Black wedges represent phylogenetic groups found only in indomethacin-injected samples, and white wedges correspond to families of organisms found only in control samples.
Specific firmicute sequences distinct from those in control animals were prevalent in rats injected with indomethacin (Fig. 2). Notably, E. faecalis-like sequences were conspicuous in the experimental rats but completely absent in control rat libraries. Six of the seven small-intestine samples from the rats injected with indomethacin contained representative E. faecalis sequences. Lactobacillus murinus-like organisms also appeared to bloom in indomethacin-injected specimens. Additionally, C. difficile-like sequences were present in high abundance in both the mucosa and MLN of experimental rat 16; however, the C. difficile group sequences were present only in low numbers in two other samples, including one control sample (Fig. 2).
Although the distribution of firmicute sequences differed between treatment groups, many were represented in both the experimental and control rats (Fig. 2). The Bacillus sp., Lactobacillus sp. (with the exception of the L. murinus relatedness cluster), and Ruminococcus sp. groups all contained roughly equal numbers of representatives from both control and experimental samples.
In contrast to the small-intestine clone libraries, the MLN libraries were dominated by Enterobacteriales sequences (Fig. 1) with the firmicutes as the second most abundant group of bacteria. Enterococcus faecalis and Clostridium difficile relatives were most prominently different between experimental and control mesenteric lymph node libraries. Otherwise, the sequences in MLN libraries were similar in composition and abundance between experimental and control samples.
rRNA library analysis indicated that indomethacin injection induced a reduction in SFB sequences relative to those of other organisms and the expansion of E. faecalis sequences. The apparent loss of SFB sequences could be due either to an actual loss or to a dilution of the SFB sequences by a bloom of other organisms, including E. faecalis. Consequently, we used Q-PCR to estimate the absolute numbers of SFB sequences compared to E. faecalis sequences in the tissue DNAs. The Q-PCR assays were designed to amplify species-specific fragments of the rRNA genes from the respective organisms (see Materials and Methods). The specificities of primer sets were verified by cloning and sequencing PCR products from multiple amplification reactions; only clones that were closely related to SFB or E. faecalis sequences were recovered. The results of Q-PCR are summarized in Table 3 and demonstrate that SFB sequences were detectable in 9/12 specimens; furthermore, the abundance of SFBs was not affected by injection with indomethacin (P = 0.70 for MANOVA of control and treatment groups) (Table 3). All four small-intestine specimens in which SFB rRNA gene sequences were observed in intestinal clone libraries were judged to be SFB positive by Q-PCR.
TABLE 3.
Summary of species-specific Q-PCR data for control and indomethacin-injected rats
| Label | Groupa | Pathology | SFB
|
E. faecalis
|
||
|---|---|---|---|---|---|---|
| No. of moleculesb | No. of clonesc | No. of molecules | No. of clones | |||
| BR1A | − | Normal | 2,254 | 62 | 5,398 | 0 |
| BR2A | − | Normal | 0 | 0 | 11,722 | 0 |
| BR5A | − | Normal | 363 | 19 | 7,797 | 0 |
| BR6A | − | Normal | 3,838 | 37 | 6,191 | 0 |
| BR13A | − | Normal | 985 | 91 | 2,531 | 0 |
| BR14A | − | Normal | 52 | 0 | 1,632 | 0 |
| BR3A | + | Severe | 223 | 0 | 5 × 107 | 0 |
| BR4A | + | Mild | 8,621 | 0 | 1 × 108 | 41 |
| BR11A | + | Mild | 28 | 0 | 1 × 106 | 1 |
| BR12A | + | Severe | 0 | 0 | 6 × 106 | 38 |
| BR15A | + | Mild | 0 | 0 | 4 × 105 | 1 |
| BR16A | + | Moderate | 95 | 0 | 2 × 108 | 47 |
−, control group; +, indomethacin-injected group.
The number of molecules normalized to the amount of input DNA.
The number of clones normalized to 100.
Q-PCR results indicated that E. faecalis-like organisms were present in all samples (Table 3). In contrast to SFB sequences, however, E. faecalis sequences were significantly enriched (typically >1,000-fold; P = 0.0028 for MANOVA of control and treatment groups) in samples from rats injected with indomethacin compared to control samples (Table 3). Thus, the SFB do not vanish; rather, their relative abundance is diminished by blooms of other organisms, notably the E. faecalis relatedness cluster.
DISCUSSION
Previous studies of the effects of NSAIDs on the gastrointestinal tract have indicated that the microbiota of the small intestine may be involved in ulceration, but these studies did not identify specific bacteria, nor did they elucidate the impact of NSAIDs on gastrointestinal-tract microbial diversity. In order to gain insight into the effects of indomethacin on the microbial ecology of the small intestine, we used a culture-independent approach based on phylogenetic analysis of bacterial rRNA gene sequences. In the study, we identified >1,400 rRNA genes from 16 rats. The vast majority of clones (∼91%) were related by at least 97% sequence identity, the species level, to other sequences in the GenBank database. Sampling coverage statistics (Good's coverage [16]) indicated that we sampled, on average, 90% of the species level phylotypes in the DNA libraries; in other words, in all cases, we identified by rRNA sequence criteria the majority of “species” present in the samples.
The rat clone libraries were comprised primarily of sequences of the kinds of organisms commonly found in the mammalian intestinal tract. The sequences were confined to four of the nine bacterial phyla previously found in the human intestine and feces (20). Similar to previous studies of rodent and human intestines, firmicute sequences were dominant in the clone libraries (2, 6, 20, 26). Bacteroidetes and Clostridium spp. sequences were relatively rare in these clone libraries from the small intestine. In contrast, molecular studies of the rat and mouse large intestine found bacteroidetes to comprise a major portion of the total number of clones identified (2, 20). These differences most likely can be attributed to the samples analyzed, small-intestinal mucosa and lymph nodes in the present case compared to colon and fecal samples in the other studies. Bacteroidetes and clostridia are enriched in feces compared to the ileal mucosa in gnotobiotic mice cultured with Shaedler flora (34).
The clone libraries collectively indicated that indomethacin injection can produce two major effects on the gastrointestinal microbiota. First, E. faecalis group organisms (as well as C. difficile and L. murinus to a lesser extent) were greatly enriched in multiple examples of the treatment group relative to controls. Second, SFB sequences were prevalent in control libraries but were not detected among analyzed clones from the treatment group libraries. These results suggested either loss of SFB or their overgrowth by E. faecalis and other organisms. To test these two hypotheses, we used Q-PCR to estimate the numbers of SFB and E. faecalis rRNA genes in the sample sets. The Q-PCR results indicated that SFB sequences did not differ significantly in numbers between control and experimental samples but that E. faecalis sequences were significantly enriched in the indomethacin-treated specimens. The lack of SFB sequences among analyzed clones from indomethacin-injected rats thus was most likely due to overgrowth of other organisms that diluted the SFB sequences in clone libraries.
Previous culture-based studies noted the importance of bacterial expansion coincident with the onset of NSAID-induced inflammation and ulceration (17, 47). In addition to the significant increase in E. faecalis-like organisms identified in our studies, other bacterial groups probably also expand upon administration of indomethacin, since the relative proportions of many rRNA genes were similar in control and experimental animals. Although the significance of the expansion of general intestinal flora is unclear, the conspicuous bloom of enterococci indicated by the clone library and Q-PCR results may be particularly important. E. faecalis is known to induce colitis in rodent experimental models, and recent studies have demonstrated that this group of bacteria is capable of maintaining constitutive expression of proinflammatory genes (33). Expansion of the enterococcal populations as a consequence of indomethacin injection may, therefore, participate in induction or exacerbation of ulceration. In rat 16, in addition to the bloom of E. faecalis-like organisms, C. difficile-like organisms were enriched. Such organisms are also known to cause health-related complications, such as diarrhea, in humans (42).
Overall, this study provides evidence from clone libraries, as well as Q-PCR data, that injection with indomethacin causes an overgrowth of specific bacterial groups, notably members of the E. faecalis relatedness cluster. The bloom of some groups of microbes resulted in relative diminution, as a percentage of the total bacterial load, of other microorganisms, such as the SFB. The SFB apparently adhere to mononuclear cells, epithelial cells overlaying the Peyer's patches in the ileum (25). This colonization is correlated with immunoglobulin A production, which implicates the SFB as stimulators of mucosal immunity (41). The SFB may, therefore, perform a protective function for the gastrointestinal tract that is lost or overwhelmed upon indomethacin-induced expansion of other bacteria, such as E. faecalis. The overgrowth by other bacteria of groups such as the SFB suggests a microbiological imbalance and perhaps reflects a failure of mucosal immunity to check the growth of microbes that exacerbate NSAID-induced ulceration. Thus, one key to a better understanding and therapy of NSAID gastritis may lie in addressing the microbiological imbalances that result from NSAID ingestion. Finally, although differences in microbial populations were evident between treatment and control groups, no correlation was observed between the extent of NSAID-induced pathology and the resultant intestinal microbiota. Clearly, other factors are also involved in determining the severity of the host response to indomethacin treatment.
Acknowledgments
We thank Catherine Lozupone and members of the Pace laboratory for their helpful comments and discussion.
This work was supported in part by an NIH grant to N.R.P.
REFERENCES
- 1.Backhed, F., R. E. Ley, J. L. Sonnenburg, D. A. Peterson, and J. I. Gordon. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915-1920. [DOI] [PubMed] [Google Scholar]
- 2.Brooks, S. P., M. McAllister, M. Sandoz, and M. L. Kalmokoff. 2003. Culture-independent phylogenetic analysis of the faecal flora of the rat. Can. J. Microbiol. 49:589-601. [DOI] [PubMed] [Google Scholar]
- 3.Caldwell, B., S. Aldington, M. Weatherall, P. Shirtcliffe, and R. Beasley. 2006. Risk of cardiovascular events and celecoxib: a systematic review and meta-analysis. J. R. Soc. Med. 99:132-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Don, R. H., P. T. Cox, B. J. Wainwright, K. Baker, and J. S. Mattick. 1991. ‘Touchdown’ PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 19:4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eckburg, P. B., E. M. Bik, C. N. Bernstein, E. Purdom, L. Dethlefsen, M. Sargent, S. R. Gill, K. E. Nelson, and D. A. Relman. 2005. Diversity of the human intestinal microbial flora. Science 308:1635-1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evans, S. M., and B. J. Whittle. 2003. Role of bacteria and inducible nitric oxide synthase activity in the systemic inflammatory microvascular response provoked by indomethacin in the rat. Eur. J. Pharmacol. 461:63-71. [DOI] [PubMed] [Google Scholar]
- 8.Fang, W. F., A. Broughton, and E. D. Jacobson. 1977. Indomethacin-induced intestinal inflammation. Am. J. Dig. Dis. 22:749-760. [DOI] [PubMed] [Google Scholar]
- 9.Frank, D. N., and N. R. Pace. 2001. Molecular-phylogenetic analyses of human gastrointestinal microbiota. Curr. Opin. Gastroenterol. 17:52-57. [DOI] [PubMed] [Google Scholar]
- 10.Frank, D. N., G. B. Spiegelman, W. Davis, E. Wagner, E. Lyons, and N. R. Pace. 2003. Culture-independent molecular analysis of microbial constituents of the healthy human outer ear. J. Clin. Microbiol. 41:295-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham, D. J., D. Campen, R. Hui, M. Spence, C. Cheetham, G. Levy, S. Shoor, and W. A. Ray. 2005. Risk of acute myocardial infarction and sudden cardiac death in patients treated with cyclo-oxygenase 2 selective and non-selective non-steroidal anti-inflammatory drugs: nested case-control study. Lancet 365:475-481. [DOI] [PubMed] [Google Scholar]
- 12.Graham, D. Y., A. R. Opekun, F. F. Willingham, and W. A. Qureshi. 2005. Visible small-intestinal mucosal injury in chronic NSAID users. Clin. Gastroenterol. Hepatol. 3:55-59. [DOI] [PubMed] [Google Scholar]
- 13.Harmsen, H. J., G. R. Gibson, P. Elfferich, G. C. Raangs, A. C. Wildeboer-Veloo, A. Argaiz, M. B. Roberfroid, and G. W. Welling. 2000. Comparison of viable cell counts and fluorescence in situ hybridization using specific rRNA-based probes for the quantification of human fecal bacteria. FEMS Microbiol. Lett. 183:125-129. [DOI] [PubMed] [Google Scholar]
- 14.Hippisley-Cox, J., and C. Coupland. 2005. Risk of myocardial infarction in patients taking cyclo-oxygenase-2 inhibitors or conventional non-steroidal anti-inflammatory drugs: population based nested case-control analysis. BMJ 330:1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hippisley-Cox, J., C. Coupland, and R. Logan. 2005. Risk of adverse gastrointestinal outcomes in patients taking cyclo-oxygenase-2 inhibitors or conventional non-steroidal anti-inflammatory drugs: population based nested case-control analysis. BMJ 331:1310-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kemp, P. F., and J. Y. Aller. 2004. Bacterial diversity in aquatic and other environments: what 16S rDNA libraries can tell us. FEMS Microbiol. Ecol. 47:161-177. [DOI] [PubMed] [Google Scholar]
- 17.Kent, T. H., R. M. Cardelli, and F. W. Stamler. 1969. Small intestinal ulcers and intestinal flora in rats given indomethacin. Am. J. Pathol. 54:237-249. [PMC free article] [PubMed] [Google Scholar]
- 18.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In S. A. M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, N.Y.
- 19.Leser, T. D., J. Z. Amenuvor, T. K. Jensen, R. H. Lindecrona, M. Boye, and K. Moller. 2002. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68:673-690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ley, R. E., F. Backhed, P. Turnbaugh, C. A. Lozupone, R. D. Knight, and J. I. Gordon. 2005. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 102:11070-11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ley, R. E., J. K. Harris, J. Wilcox, J. R. Spear, S. R. Miller, B. M. Bebout, J. A. Maresca, D. A. Bryant, M. L. Sogin, and N. R. Pace. 2006. Unexpected diversity and complexity of the Guerrero Negro hypersaline microbial mat. Appl. Environ. Microbiol. 72:3685-3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahida, Y. R., and V. E. Rolfe. 2004. Host-bacterial interactions in inflammatory bowel disease. Clin. Sci. (London) 107:331-341. [DOI] [PubMed] [Google Scholar]
- 23.Maidak, B. L., J. R. Cole, C. T. Parker, Jr., G. M. Garrity, N. Larsen, B. Li, T. G. Lilburn, M. J. McCaughey, G. J. Olsen, R. Overbeek, S. Pramanik, T. M. Schmidt, J. M. Tiedje, and C. R. Woese. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Meyerholz, D. K., T. J. Stabel, and N. F. Cheville. 2002. Segmented filamentous bacteria interact with intraepithelial mononuclear cells. Infect. Immun. 70:3277-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ott, S. J., M. Musfeldt, D. F. Wenderoth, J. Hampe, O. Brant, U. R. Folsch, K. N. Timmis, and S. Schreiber. 2004. Reduction in diversity of the colonic mucosa associated bacterial microflora in patients with active inflammatory bowel disease. Gut 53:685-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pace, N. R. 1997. A molecular view of microbial diversity and the biosphere. Science 276:734-740. [DOI] [PubMed] [Google Scholar]
- 28.Papineau, D., J. J. Walker, S. J. Mojzsis, and N. R. Pace. 2005. Composition and structure of microbial communities from stromatolites of Hamelin Pool in Shark Bay, Western Australia. Appl. Environ. Microbiol. 71:4822-4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pryde, S. E., A. J. Richardson, C. S. Stewart, and H. J. Flint. 1999. Molecular analysis of the microbial diversity present in the colonic wall, colonic lumen, and cecal lumen of a pig. Appl. Environ. Microbiol. 65:5372-5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rappe, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 31.R Development Core Team. 2005. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria.
- 32.Robert, A., and T. Asano. 1977. Resistance of germfree rats to indomethacin-induced intestinal lesions. Prostaglandins 14:333-341. [DOI] [PubMed] [Google Scholar]
- 33.Ruiz, P. A., A. Shkoda, S. C. Kim, R. B. Sartor, and D. Haller. 2005. IL-10 gene-deficient mice lack TGF-beta/Smad signaling and fail to inhibit proinflammatory gene expression in intestinal epithelial cells after the colonization with colitogenic Enterococcus faecalis. J. Immunol. 174:2990-2999. [DOI] [PubMed] [Google Scholar]
- 34.Sarma-Rupavtarm, R. B., Z. Ge, D. B. Schauer, J. G. Fox, and M. F. Polz. 2004. Spatial distribution and stability of the eight microbial species of the altered schaedler flora in the mouse gastrointestinal tract. Appl. Environ. Microbiol. 70:2791-2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schloss, P. D., and J. Handelsman. 2004. Status of the microbial census. Microbiol. Mol. Biol. Rev. 68:686-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Silverstein, F. E., G. Faich, J. L. Goldstein, L. S. Simon, T. Pincus, A. Whelton, R. Makuch, G. Eisen, N. M. Agrawal, W. F. Stenson, A. M. Burr, W. W. Zhao, J. D. Kent, J. B. Lefkowith, K. M. Verburg, and G. S. Geis. 2000. Gastrointestinal toxicity with celecoxib vs nonsteroidal anti-inflammatory drugs for osteoarthritis and rheumatoid arthritis: the CLASS study: a randomized controlled trial. Celecoxib Long-term Arthritis Safety Study. JAMA 284:1247-1255. [DOI] [PubMed] [Google Scholar]
- 37.Sonnenburg, J. L., L. T. Angenent, and J. I. Gordon. 2004. Getting a grip on things: how do communities of bacterial symbionts become established in our intestine? Nat. Immunol. 5:569-573. [DOI] [PubMed] [Google Scholar]
- 38.Spear, J. R., J. J. Walker, T. M. McCollom, and N. R. Pace. 2005. Hydrogen and bioenergetics in the Yellowstone geothermal ecosystem. Proc. Natl. Acad. Sci. USA 102:2555-2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 40.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Umesaki, Y., H. Setoyama, S. Matsumoto, A. Imaoka, and K. Itoh. 1999. Differential roles of segmented filamentous bacteria and clostridia in development of the intestinal immune system. Infect. Immun. 67:3504-3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Voth, D. E., and J. D. Ballard. 2005. Clostridium difficile toxins: mechanism of action and role in disease. Clin. Microbiol. Rev. 18:247-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walker, J. J., J. R. Spear, and N. R. Pace. 2005. Geobiology of a microbial endolithic community in the Yellowstone geothermal environment. Nature 434:1011-1014. [DOI] [PubMed] [Google Scholar]
- 44.Whittle, B. J. 2004. Mechanisms underlying intestinal injury induced by anti-inflammatory COX inhibitors. Eur. J. Pharmacol. 500:427-439. [DOI] [PubMed] [Google Scholar]
- 45.Wilson, K. H., and R. B. Blitchington. 1996. Human colonic biota studied by ribosomal DNA sequence analysis. Appl. Environ. Microbiol. 62:2273-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wolfe, M. M., D. R. Lichtenstein, and G. Singh. 1999. Gastrointestinal toxicity of nonsteroidal antiinflammatory drugs. N. Engl. J. Med. 340:1888-1899. [DOI] [PubMed] [Google Scholar]
- 47.Yamada, T., E. Deitch, R. D. Specian, M. A. Perry, R. B. Sartor, and M. B. Grisham. 1993. Mechanisms of acute and chronic intestinal inflammation induced by indomethacin. Inflammation 17:641-662. [DOI] [PubMed] [Google Scholar]