Abstract
Heterotrophic flagellates are key components of all ecosystems. Understanding the patterns of biodiversity of these organisms is thus particularly important. Here we analyzed the intraspecific diversity of 10 morphospecies of heterotrophic flagellates comprising representatives of the Apusozoa (2 morphospecies) and Kinetoplastea (8 morphospecies), all belonging to the most common flagellates with worldwide distribution. Most morphospecies showed a mixing of lineages isolated from diverse habitats, indicating that some lineages of these morphospecies had been able to colonize different habitats several times. Furthermore, our results revealed remarkable levels of genetic divergence within most of the morphospecies studied, underlining the difficulty of correctly determining species by means of morphology alone. Many cryptic or pseudocryptic species seem to occur. Our results revealed clear divergence between marine and freshwater lineages of the morphospecies Ancyromonas sigmoides, showing that freshwater lineages have not been able to colonize marine environments and marine lineages have not been able to colonize freshwater environments for a long time.
Application of the concept of a morphological species in eukaryotic microbiology has promoted the view that only a limited number of species with cosmopolitan distribution exist (12). The introduction of molecular tools into microbial research has started to change this view (34) and has shown that the fine structure of phylogenetic trees can yield information about the ecology of eukaryotic microbial populations (31).
For bacteria, tolerance to high salinity has been recognized as an important physiological property that can be used to define a phylogenetic lineage (18, 29). Moreover, evidence is increasing that microbial communities differ significantly in marine and freshwater environments (15). Environmental factors thus influence the composition and species distribution of microbial communities and can be major forces of diversification (19, 21).
It can be expected that lineages of microbial eukaryotes (protists) that have evolved in distinct directions are also present in marine and freshwater environments. This conclusion is supported by the fact that the specific tolerance to NaCl concentrations of several strains of the cosmopolitan flagellate Neobodo designis might be due to genetic adaptation (10, 22). Koch and Ekelund (22) showed that most strains of N. designis isolated from freshwater were not able to grow if the salinity was greater than 15 practical salinity units. A study of strains of the flagellate Oxyrrhis marina showed that different salinity preferences and levels of salinity tolerance could be associated with different types of habitats (26). Von der Heyden et al. (37) reported that there is clear divergence between marine and freshwater lineages of the flagellate Goniomonas.
Should we therefore not expect to find genetic divergence between marine and freshwater strains in other morphospecies? Cosmopolitan morphospecies are interesting candidates to answer this question. In this analysis we focused on the intraspecific divergence of the small rRNA gene of the morphospecies Ancyromonas sigmoides, Bodo saltans, Neobodo designis, and Rhynchomonas nasuta, four species regarded as cosmopolitan and belonging to the 20 most commonly reported morphospecies of heterotrophic flagellates (30). Furthermore, we sequenced members of the morphospecies Apusomonas proboscidea, Neobodo curvifilus, Neobodo saliens, Parabodo caudatus, and Procryptobia sorokini. The strains used were isolated from marine and freshwater sites, as well as from soil and groundwater.
MATERIALS AND METHODS
Heterotrophic flagellate strain cultivation and DNA sequencing.
The clonal cultures used and the sampling locations are shown in Table 1. Deep-sea strains were obtained with a multiple corer system. Sediment was taken from undisturbed cores with a sterile syringe. Freshwater samples were obtained with a water sampler. Soil samples were taken with a sterile syringe. All samples were placed into 50-ml tissue culture flasks (Sarstedt) and cultivated in autoclaved artificial seawater (containing [per liter] 28.15 g NaCl, 0.67 g KCl, 5.51 g MgCl2 · 6H2O, 1.45 g CaCl2 · 2H2O, 6.92 g MgSO4 · 7H2O, 0.1 g KNO3, and 0.01 g K2HPO4 · 3H2O) (marine isolates) or with autoclaved WC medium (17) (other isolates) together with sterile wheat grain as food for the bacteria. Clonal strains were established from enrichment cultures by isolating single cells by micromanipulation and were kept in culture until DNA extraction. Morphological identification of each strain was based on light microscopy. All strains are referred to below by their GenBank accession numbers.
TABLE 1.
Sample locations with descriptions of the habitat type and the salinity for all species sequenced and GenBank accession numbers
| Species | GenBank accession no. | Sample location | Salinity (PSU) |
|---|---|---|---|
| Ancyromonas sigmoides | DQ207563 | River Rhine at Cologne (Germany) | 0 |
| Ancyromonas sigmoides | DQ207564 | River Rhine at Cologne (Germany) | 0 |
| Ancyromonas sigmoides | DQ207565 | Eutrophic pond with macrophytes, Zoological Institute, University of Cologne (Germany) | 0 |
| Ancyromonas sigmoides | DQ207566 | River Rhine at Cologne (Germany) | 0 |
| Apusomonas proboscidea | DQ207567 | Dystrophic pond in Borok near Yaroslavl (Russia) | 0 |
| Apusomonas proboscidea | DQ207568 | Groundwater from a depth of 18 m, Bornheim near Cologne (Germany) | 0 |
| “Bodo curvifilus” | DQ207577 | Mesotrophic Lake Schoehsee, Ploen (Germany) | 0 |
| Bodo saltans | DQ207569 | Dystrophic pond in Borok near Yaroslavl (Russia) | 0 |
| Bodo saltans | DQ207570 | Sewage plant Stammheim in Cologne (Germany) | 0 |
| Bodo saltans | DQ207571 | Black Sea near Gelendchik (Russia) | 17 |
| Bodo saltans | DQ207572 | Eutrophic pond with macrophytes, Zoological Institute, University of Cologne (Germany) | 0 |
| Bodo saltans | DQ207573 | River Rhine at Cologne (Germany) | 0 |
| Bodo saltans | DQ207574 | Eutrophic pond in Randwick, Sydney (Australia) | 0 |
| Bodo saltans | DQ207575 | River Rhine at Cologne (Germany) | 0 |
| Dimastigella mimosa | DQ207576 | Dystrophic pond in Borok near Yaroslavl (Russia) | 0 |
| Neobodo designis | DQ207578 | Littoral of the Baltic Sea at the island Hiddensee (Germany) | 8 |
| Neobodo designis | DQ207579 | River Rhine at Cologne (Germany) | 0 |
| Neobodo designis | DQ207580 | Surface of ice floe, 79°N, 2°W, North Atlantic Ocean | 1 |
| Neobodo designis | DQ207581 | Surface of ice floe, 79°N, 12°W, North Atlantic Ocean | 1 |
| Neobodo designis | DQ207582 | River Rhine at Cologne (Germany) | 0 |
| Neobodo designis | DQ207583 | River Rhine at Cologne (Germany) | 0 |
| Neobodo designis | DQ207584 | Garden soil, Zoological Institute, University of Cologne (Germany) | 0 |
| Neobodo designis | DQ207585 | Eutrophic pond with macrophytes, Zoological Institute, University of Cologne (Germany) | 0 |
| Neobodo designis | DQ207586 | Sandy soil, Bienen near Rees (Germany) | 0 |
| Neobodo designis | DQ207587 | Garden soil, Grietherbusch near Rees (Germany) | 0 |
| Neobodo designis | DQ207588 | Clay soil, Millinger Waard near Nijmegen (The Netherlands) | 0 |
| Neobodo saliens | DQ207589 | Littoral of the Baltic Sea at the island Hiddensee (Germany) | 8 |
| Parabodo caudatus | DQ207590 | River Rhine at Cologne (Germany) | 0 |
| Parabodo caudatus | DQ207591 | Mesotrophic Lake Schoehsee, Ploen (Germany) | 0 |
| Procryptobia sorokini | DQ207592 | Littoral of the White Sea near Kartesh, Karelia (Russia) | 32 |
| Procryptobia sorokini | DQ207593 | Surface of an ice floe, 79°N, 2°W, North Atlantic Ocean | 1 |
| Rhynchobodo sp. | DQ207594 | Garden soil, Zoological Institute, University of Cologne, (Germany) | 0 |
| Rhynchomonas nasuta | DQ207595 | Dystrophic pond in Borok near Yaroslavl (Russia) | 0 |
| Rhynchomonas nasuta | DQ207596 | Deep-sea sediment from a depth of 5,392 m in the Angola Basin, 18°25.3′S, 4°44.0′E, South Atlantic Ocean | 37 |
| Rhynchomonas nasuta | DQ207597 | Deep-sea sediment from a depth of 5,415m in the Angola Basin, 17°11.6′S, 4°45.9′E, South Atlantic Ocean | 37 |
| Rhynchomonas nasuta | DQ207598 | River Rhine at Cologne (Germany) | 0 |
| Rhynchomonas nasuta | DQ207599 | Deep-sea sediment from a depth of 5,423m in the Angola Basin, 19°06.9′S, 3°52.0′E, South Atlantic Ocean | 37 |
The cultured isolates were grown to high densities (104 cells · ml−1) and harvested by centrifugation in 50-ml tubes (Sarstedt) for 30 min at 4,000 × g and 4°C. Each supernatant was discarded, and the cells were resuspended in 50 μl 1× Tris-EDTA buffer and transferred into PCR tubes. The collected cells were lysed, and the genomic DNA was isolated using a cetyltrimethylammonium extraction method with phenol and chloroform as previously described (4). Amplification of the small-subunit (SSU) rRNA gene by PCR was performed in a 50-μl reaction mixture containing each primer at a concentration of 0.1 μM, each deoxynucleoside triphosphate at a concentration of 200 μM, up to 100 ng genomic DNA, 2 mM MgCl2, 1× reaction buffer, and 2.5 U AmpliTaq DNA polymerase (Applied Biosystems). General eukaryotic PCR primers were used for amplification of Apusozoa (primers 18Sfor [AAC CTG GTT GAT CCT GCC AGT] and 18Srev [TGA TCC TTC CGC AGG TTC ACC TAC]). Specific PCR primers were used for amplification of Kinetoplastea (primers 18Sfor-Bodo [CTG GTT GAT TCT GCC AGT AGT] and 18Srev-Bodo [TGA TCC AGC TGC AGG TCC ACC]). Each PCR was started with an initial denaturation step at 98°C for 3 min, after which the polymerase was added; this was followed by 35 cycles of 98°C for 15 s, 56°C for 30 s, 53°C for 30 s and 72°C for 2 min. PCR products were purified with the Rapid PCR purification system (Marligen Biosciences). Cycle sequencing was performed with a BigDye terminator cycle sequencing kit (version 3.1; Applied Biosystems), and the sequence was determined with an ABI 3100 automated sequencer. For all these steps we used the manufacturers' specifications. All internal sequencing primers have been described by Scheckenbach et al. (33).
Phylogenetic analysis.
The sequence fragments determined were checked and assembled manually. The sequences determined were aligned with sequences retrieved from GenBank using the ClustalX multiple alignment program (version 1.83; IGBMC, Strasbourg, France) (ftp://ftp-igbmc.u-strasbg.fr/pub/ClustalX). Uncorrected genetic p-distances were calculated using MEGA (version 3) (http://www.megasoftware.net). Phylogenetic analyses were carried out by using a distance method (minimum evolution) and the maximum likelihood method. The nucleotide substitution models which best fit the data set were estimated with J. A. A. Nylander's MrAIC (version 1.4; Department of Systematic Zoology, Uppsala University, Sweden) (http://www.ebc.uu.se/systzoo/staff/nylander.html), evaluating 56 different models. Minimum evolution phylogenetic trees were calculated with FastME (version 1.9; LIRMM, Montpellier, France) (http://atgc.lirmm.fr/fastme) and maximum likelihood phylogenetic trees were calculated with Phyml (version 2.4.4; LIRMM, Montpellier, France) (http://atgc.lirmm.fr/phyml) using all aligned sites. TrN + Γ + I distances for minimum evolution analysis of Apusozoa and GTR + Γ distances for Kinetoplastea, both using substitution rates estimated by Phyml, were calculated with Tree-Puzzle (version 5.2) (http://www.tree-puzzle.de). Parametric bootstrapping with 100 resamplings was performed using Seqboot, and consensus trees (50% majority rule) were constructed using Consense; both Seqboot and Consense were obtained from J. Felsenstein's PHYLIP package (version 3.63; Department of Genetics, University of Washington) (http://evolution.genetics.washington.edu/phylip.html).
RESULTS AND DISCUSSION
Marine and freshwater lineages of A. sigmoides.
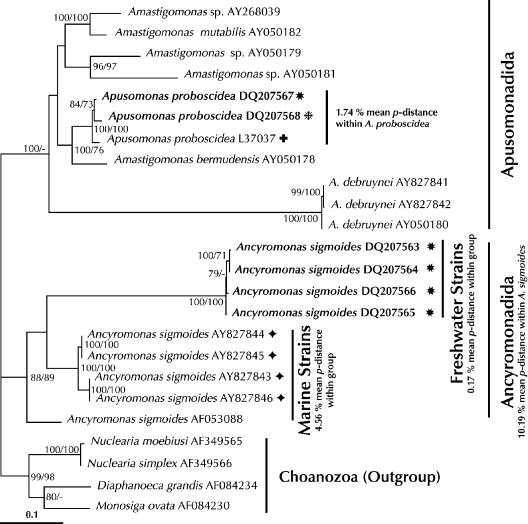
There is complete phylogenetic separation between the marine and the freshwater lineages of the morphospecies Ancyromonas sigmoides (Fig. 1). The freshwater lineages are ancestrally related but phylogenetically distinct from the marine lineages, and there is a high level of genetic divergence between these types of lineages (mean p-distance between the five marine strains and the freshwater strains, 16.16%; mean p-distance within the freshwater cluster, 0.17%). The freshwater cluster is stable as determined by all phylogenetic methods used and is clearly separated from the marine lineages by bootstrap support values of 100% and long evolutionary branches.
FIG. 1.
Maximum likelihood tree of the phylum Apusozoa obtained by using all aligned positions. The tree was rooted using Choanozoa as the outgroup. The numbers at the nodes are bootstrap support percentages calculated from 100 replicates for maximum likelihood (left) and minimum evolution (right) analyses. Values less than 70% are omitted or are indicated by a hyphen. The model for nucleotide substitution chosen was TrN + Γ + I. GenBank accession numbers are indicated. Strains sequenced in this study are indicated by boldface type. Scale bar = 0.1 substitution per site. Freshwater and marine clades of A. sigmoides are labeled. Mean p-distances for the species studied (Ancyromonas and Apusomonas) and for the major clades within A. sigmoides are shown. Sample locations for the morphospecies strains studied are indicated as follows: diamonds, marine; stars, freshwater; plus sign, soil; and asterisk, groundwater.
Regarding the high level of divergence between the phylogenetically coherent marine and freshwater lineages of A. sigmoides, it is obvious that populations of one habitat have not colonized the other habitat for a long time. The dispersal rates for this morphospecies should be as high as those for other microbial species, and one could expect to find marine lineages in freshwater environments and freshwater lineages in marine environments (12, 16). Since this is apparently not the case, the observed pattern must be the result of different ecophysiological traits (13). The ecological differences between marine and freshwater habitats are therefore sufficient barriers for dispersal of at least some lineages of A. sigmoides, as previously reported for Goniomonas (37). The phylogeny of protistan morphospecies can therefore serve as an indicator of ecophysiological differentiation below the morphospecies level. However, it is not possible to deduce from the phylogeny which abiotic or biotic factors are shared by lineages with similar rRNA genes. Considering salinity tolerance the main factor for the distribution of Ancyromonas might be wrong since another flagellated morphospecies, Goniomonas pacifica, which has been found only in marine environments, was able to tolerate salt concentrations ranging from 35 PSU down to 0 PSU (1).
Successive occurrence between environments.
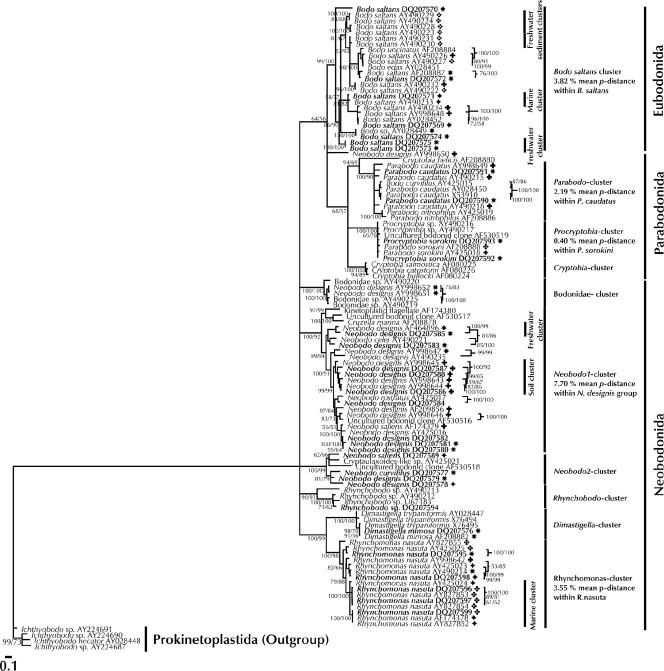
The morphospecies B. saltans, N. designis, P. caudatus, P. sorokini, and R. nasuta contain strains that cluster among other strains isolated from different habitats (Fig. 2). The marine strains of B. saltans (DQ207571 and AY490233) cluster with freshwater, freshwater sediment, and soil isolates. Freshwater, freshwater sediment, and soil isolates of the same morphospecies cluster together. Soil isolates of P. caudatus and N. designis cluster together with freshwater isolates of these morphospecies. Marine isolates of P. sorokini and R. nasuta cluster together with freshwater isolates, and one soil strain of R. nasuta (AY998642) clusters between marine and freshwater strains.
FIG. 2.
Consensus tree (50% majority rule) of the class Kinetoplastea obtained by using all aligned positions. The tree was rooted using Prokinetoplastida as the outgroup. The model for nucleotide substitution chosen was GTR + Γ. Scale bar = 0.1 substitution per site. Some clades are labeled according to their sample locations. Mean p-distances for all of the mono- and paraphyletic species studied and for the major clade of N. designis are shown. Sample locations for the morphospecies strains studied are indicated as follows: large diamonds, marine; stars, freshwater; plus signs, soil; cloverleaves, marine sediment; and clusters of four small diamonds, freshwater sediment. For further details see the legend to Fig. 1.
Strains of these morphospecies obviously do not cluster together according to their habitats. The succession of strains according to their sample locations alternates in the phylogenetic trees within one morphospecies. The taxa mentioned above have obviously been able to move across ecosystem boundaries and have even been able to colonize several habitats multiple times. Tolerance to high salinity has been demonstrated for a marine strain of N. designis (AY998646), as well as one soil strain of the same morphospecies (AY998645), which could survive and grow at salinities ranging from 0 PSU to 45 PSU (22). Furthermore, this is in accordance with previous findings (1) which showed that marine strains of B. saltans and N. designis could have been transferred slowly and gradually into an environment with a salt concentration of 0 PSU and that a freshwater strain of B. saltans could have been transferred into an environment with a salt concentration of 35 PSU.
Identical genotypes of the same morphospecies isolated from different habitats.
R. nasuta DQ207599 from sediments of the Angola Basin and sequence AF174378 from a biovent serpulid zone of the East Pacific Rise are identical (p-distance, 0.00%) at the SSU rRNA gene level. A previous study described one identical pair of A. sigmoides strains from different sample locations (AY827843 from brackish waters of the Baltic Sea and AY827846 from the deep-sea environment of the North Atlantic Ocean) (33).
Some taxa are obviously able to move across ecosystem boundaries on a global scale. However, this might be the result of our inability to recognize the actual nature of their microhabitats and their ecological niches, as well as the insufficient resolution of the SSU rRNA genes at or below the species level (2). For bacteria it is known that even phylogenetically identical strains can represent different ecotypes (20, 25). Protists with identical SSU rRNA genes can exhibit a high level of genetic divergence if more variable markers, such as the internal transcribed spacer, are used (36) and can have different ecophysiological characteristics, as has been shown for the heterotrophic dinoflagellate Oxyrrhis marina (26) and the prasinophycean Ostreococcus (31). Thus, phylogenetic markers and especially slowly evolving markers, such as the SSU rRNA genes, might not be sufficient to detect ecophysiological differences between populations of the same morphospecies.
Strains isolated from the same habitat clustering together.
Within some morphospecies, clusters of similar or identical strains from the same habitat type were present (Fig. 2). With the exception of the B. saltans freshwater clade, which was composed of two strains isolated from the River Rhine in Cologne (DQ207573 and DQ207575), all strains of these clusters were isolated from geographically different locations and all were phylogenetically distinct from other strains of the corresponding morphospecies. There are several such environment-specific clades of freshwater and marine isolates within B. saltans. A freshwater cluster and a soil cluster exist in N. designis, and a large cluster of exclusively marine isolates is present within R. nasuta.
It is impossible to decide with the current data set whether these clusters are simply the result of undersampling or might remain after further sampling of the morphospecies. A previous phylogenetic study (38) revealed a major exclusively marine clade within the morphospecies N. designis. Our data, however, show that two sequences of freshwater isolates of N. designis (DQ207581 and DQ207582) cluster in the presumably “marine” clade (data not shown). Interestingly, with the exception of P. sorokini and R. nasuta, the rRNA gene divergence was generally higher between marine and freshwater habitats than, e.g., between freshwater and soil habitats. Even though one has to be very careful when detecting biogeographical patterns within protistan morphospecies, one might at least expect ecophysiologically coherent phylogenetic clusters to be detected within these morphospecies by further sampling.
High rRNA gene divergence.
The rRNA gene divergence, expressed as p-distance, was generally very high within most morphospecies studied (Fig. 1 and 2). In addition, the pairwise p-distances were 14.30% for N. curvifilus and 11.81% for N. saliens. The mean p-distance for N. designis was 9.64%. These are very high values considering that p-distances are not corrected for multiple base changes at individual nucleotide positions.
With the exception of A. sigmoides, A. proboscidea, P. sorokini, and R. nasuta, all morphospecies sequenced in this study were para- or even polyphyletic. Before this study, 3 of 15 kinetoplastid morphospecies (B. saltans, N. designis, and P. caudatus) were considered to be para- or polyphyletic. The present findings revealed that two additional morphospecies (N. curvifilus and N. saliens) are para- or polyphyletic (Fig. 2). One has to keep in mind that irrespective of the species concept applied, every species must at least fulfill the criteria of monophyly.
Although it is well known that species identification based on light microscopy might lead to lumping together of different species (30), light microscopy is widely accepted for species identification. Most studies which have led to a cosmopolitan view of microbial species distribution have been based on morphospecies identified by light microscopy. In view of the present high intraspecific rRNA gene diversity, one might ask to what degree the conclusions drawn by some authors (11, 12, 30) regarding protist biogeography can be generalized (6, 14, 23, 27).
It has been argued that rRNA gene sequence divergence is selectively neutral and that genotypic patterns based on ribosomal genes do not reflect phenotypic patterns (11). Given the relationship between phenotypic evolution within morphospecies and the genetic divergence of ribosomal genes demonstrated by several studies (5, 7, 9, 24, 28, 32), high genetic divergence of slowly evolving housekeeping genes, such as the rRNA gene, may reflect phenotypic variation. It is therefore plausible to expect phenotypically distinct lineages within the morphospecies studied.
Conclusion.
We showed that the habitat selects very particular phylogenetic lineages of some protists, such as A. sigmoides. Ecologically important patterns can obviously be found below the morphospecies level. This points to the conclusions that the morphospecies concept does not reflect protist biodiversity and that the species problem of protists must receive further attention (34). Our study therefore supports the idea that integration of molecular techniques into protist taxonomy (8, 35) will be crucial for future ecological studies (3).
Acknowledgments
This study was financially supported by grant Ar 288/5 from the German Research Foundation to H.A. Additional support came from the Russian Foundation for Basic Research to A.P.M. (grant 05-04-48180).
We thank the crew of the R/V “Meteor” 48/1 expedition and the R/V “Polarstern” 18/1 expedition. Special thanks for cooperation on the vessels go to K. Hausmann and M. Türkay. The A. proboscidea sequence (DQ207568) was kindly provided by N. Loquay and R. Bieg. For technical support we are indebted to C. Barth, R. Bieg, and B. Gräfe.
REFERENCES
- 1.Arndt, H., D. Dietrich, B. Auer, E. J. Cleven, T. Gräfenhan, M. Weitere, and A. P. Mylnikov. 2000. Functional diversity of heterotrophic flagellates in aquatic ecosystems, p. 240-268. In B. S. C. Leadbeater and J. C. Green (ed.), The flagellates. The Systematic Association special volume 59. Unity, diversity and evolution. Taylor & Francis Ltd., London, United Kingdom.
- 2.Avise, J. C. 2004. Molecular markers, natural history, and evolution, 2nd ed. Sinauer Associates, Sunderland, MA.
- 3.Caron, D. A., P. D. Countway, and M. V. Brown. 2004. The growing contribution of molecular biology and immunology to protistan ecology: molecular signatures as ecological tools. J. Eukaryot. Microbiol. 51:38-48. [DOI] [PubMed] [Google Scholar]
- 4.Clark, C. G. 1992. DNA purification from polysaccharide-rich cells, p. D3.1-D3.2. In J. J. Lee and A. T. Soldo (ed.), Protocols in protozoology. Allen Press, Lawrence, KS.
- 5.Coleman, A. W. 2001. Biogeography and speciation in the Pandorina/Volvulina (Chlorophyte) superclade. J. Phycol. 37:836-851. [Google Scholar]
- 6.Coleman, A. W. 2002. Microbial eukaryote species. Science 297:337. [DOI] [PubMed] [Google Scholar]
- 7.Darling, K. F., M. Kucera, C. J. Pudsey, and C. M. Wade. 2004. Molecular evidence links cryptic diversification in polar planktonic protists to Quaternary climate dynamics. Proc. Natl. Acad. Sci. USA 101:7657-7662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dayrat, B. 2005. Towards integrative taxonomy. Biol. J. Linn. Soc. 85:407-415. [Google Scholar]
- 9.De Vargas, C., R. Norris, L. Zannetti, S. W. Gibb, and J. Pawlowski. 1999. Molecular evidence of cryptic speciation in planktonic foraminifers and their relation to oceanic provinces. Proc. Natl. Acad. Sci. USA 96:2864-2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ekelund, F. 2002. Tolerance of soil flagellates to increased NaCl levels J. Eukaryot. Microbiol. 49:324-328. [DOI] [PubMed] [Google Scholar]
- 11.Fenchel, T. 2005. Cosmopolitan microbes and their “cryptic” species. Aquat. Microb. Ecol. 41:49-54. [Google Scholar]
- 12.Finlay, B. J. 2002. Global dispersal of free-living microbial eukaryote species. Science 296:1061-1063. [DOI] [PubMed] [Google Scholar]
- 13.Finlay, B. J. 2004. Protist taxonomy: an ecological perspective. Philos. Trans. R. Soc. Lond. Ser. B 359:599-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foissner, W. 1999. Protist diversity: estimates of the near-imponderable. Protist 150:363-368. [DOI] [PubMed] [Google Scholar]
- 15.Glöckner, F. O., E. Zaichikov, N. Belkova, L. Denissova, J. Pernthaler, A. Pernthaler, and R. Amann. 2000. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria. Appl. Environ. Microbiol. 66:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffin, D. W., C. A. Kellogg, V. H. Garrison, and E. A. Shinn. 2002. The global transport of dust—an intercontinental river of dust, microorganisms and toxic chemicals flows through the Earth's atmosphere. Am. Sci. 90:398. [Google Scholar]
- 17.Guillard, R., and C. Lorenzen. 1972. Yellow-green algae with chlorophyllide. J. Phycol. 8:10-14. [Google Scholar]
- 18.Hiraishi, A., and Y. Ueda. 1994. Intrageneric structure of the genus Rhodobacter sulfidophilus and related marine species to the genus Rhodovulvum gen. nov. Int. J. Syst. Bacteriol. 44:15-23. [Google Scholar]
- 19.Horner-Devine, M. C., K. M. Carney, and B. J. M. Bohannan. 2003. An ecological perspective on bacterial biodiversity. Proc. R. Soc. Lond. B 271:113-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaspers, E., and J. Overmann. 2004. Ecological significance of microdiversity: identical 16S rRNA gene sequences can be found in bacteria with highly divergent genomes and ecophysiologies. Appl. Environ. Microbiol. 70:4831-4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kassen, R., and P. B. Rainey. 2004. The ecology and genetics of microbial diversity. Annu. Rev. Microbiol. 58:207-231. [DOI] [PubMed] [Google Scholar]
- 22.Koch, T. A., and F. Ekelund. 2005. Strains of the heterotrophic flagellate Bodo designis from different environments vary considerably with respect to salinity preference and SSU rRNA gene composition. Protist 156:97-112. [DOI] [PubMed] [Google Scholar]
- 23.Lachance, M. A. 2004. Here and there or everywhere? BioScience 54:884. [Google Scholar]
- 24.LaJeunesse, T. C. 2001. Investigating the biodiversity, ecology, and phylogeny of endosymbiotic dinoflagellates in the genus Symbiodinium using the ITS region: in search of a “species” level marker. J. Phycol. 37:866-880. [Google Scholar]
- 25.Lebuhn, M., W. Achouak, M. Schloter, O. Berge, H. Meier, M. Barakat, A. Hartmann, and T. Heulin. 2000. Taxonomic characterization of Ochrobactrum sp. isolates from soil samples and wheat roots, and description of Ochrobactrum tritici sp. nov. and Ochrobactrum grignonense sp. nov. Int. J. Syst. Evol. Microbiol. 50:2207-2223. [DOI] [PubMed] [Google Scholar]
- 26.Lowe, C. D., A. Day, S. J. Kemp, and D. J. S. Montagnes. 2005. There are high levels of functional and genetic diversity in Oxyrrhis marina. J. Eukaryot. Microbiol. 52:250-257. [DOI] [PubMed] [Google Scholar]
- 27.Nanney, D. L. 2004. No trivial pursuit. BioScience 54:720-721. [Google Scholar]
- 28.Norris, R. D., and C. de Vargas. 2000. Evolution all at sea. Nature 405:23-24. [DOI] [PubMed] [Google Scholar]
- 29.Nübel, U., F. Garcia-Pichel, and G. Muyzer. 2000. The halotolerance and phylogeny of cyanobacteria with tightly coiled trichomes (Spirulina Turpin) and the description of Halospirulina tapeticola gen. nov., sp. nov. Int. J. Syst. Bacteriol. 50:1265-1277. [DOI] [PubMed] [Google Scholar]
- 30.Patterson, D. J., and W. Lee. 2000. Geographic distribution and diversity of free-living heterotrophic flagellates, p. 269-287. In B. S. C. Leadbeater and J. C. Green (ed.), The flagellates. The Systematic Association special volume 59. Unity, diversity and evolution. Taylor & Francis Ltd., London, United Kingdom.
- 31.Rodríguez, F., E. Derelle, L. Guillou, F. Le Gall, D. Vaulot, and H. Moreau. 2005. Ecotype diversity in the marine picoeukaryote Ostreococcus (Chlorophyta, Prasinophyceae). Environ. Microbiol. 7:853-859. [DOI] [PubMed] [Google Scholar]
- 32.Sáez, A. G., I. Probert, M. Geisen, P. Quinn, J. R. Young, and L. K. Medlin. 2003. Pseudo-cryptic speciation in coccolithophores. Proc. Natl. Acad. Sci. USA 100:7163-7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scheckenbach, F., C. Wylezich, M. Weitere, K. Hausmann, and H. Arndt. 2005. Molecular identity of strains of heterotrophic flagellates isolated from surface waters and deep-sea sediments of the South Atlantic based on SSU rDNA. Aquat. Microb. Ecol. 38:239-247. [Google Scholar]
- 34.Schlegel, M., and R. Meisterfeld. 2003. The species problem in Protozoa revisited. Eur. J. Protistol. 39:349-355. [Google Scholar]
- 35.Tautz, D., P. Arctander, A. Minelli, R. H. Thomas, and A. P. Vogler. 2003. A plea for DNA taxonomy. Trends Ecol. Evol. 18:70-74. [Google Scholar]
- 36.Tsuchiya, M., H. Kitazato, and J. Pawlowski. 2003. Analysis of internal transcribed spacer of ribosomal DNA reveals cryptic speciation in Planoglabratella opercularis. J. Foramin. Res. 33:285-293. [Google Scholar]
- 37.Von der Heyden, S., E. E. Chao, and T. Cavalier-Smith. 2004. Genetic diversity of goniomonads: an ancient divergence between marine and freshwater species. Eur. J. Phycol. 39:343-350. [Google Scholar]
- 38.Von der Heyden, S., and T. Cavalier-Smith. 2005. Culturing and environmental DNA sequencing uncover hidden kinetoplastid biodiversity and a major marine clade within ancestrally freshwater Neobodo designis. Int. J. Syst. Evol. Microbiol. 55:2605-2621. [DOI] [PubMed] [Google Scholar]