Abstract
Streptococcus thermophilus is widely used for the manufacture of yoghurt and Swiss or Italian-type cheeses. These products have a market value of approximately $40 billion per year, making S. thermophilus a species that has major economic importance. Even though the fermentation properties of this bacterium have been gradually improved by classical methods, there is great potential for further improvement through genetic engineering. Due to the recent publication of three complete genome sequences, it is now possible to use a rational approach for designing S. thermophilus starter strains with improved properties. Progress in this field, however, is hampered by a lack of genetic tools. Therefore, we developed a system, based on natural transformation, which makes genetic manipulations in S. thermophilus easy, rapid, and highly efficient. The efficiency of this novel tool should make it possible to construct food-grade mutants of S. thermophilus, opening up exciting new possibilities that should benefit consumers as well as the dairy industry.
Bacteria that are competent for natural genetic transformation are able to take up naked DNA from the environment and incorporate it into their genomes by homologous recombination. Several streptococcal species belonging to the mitis, anginosus, and mutans phylogenetic groups have been shown to possess this property (4, 9, 17), but the phenomenon has never been demonstrated in most members of the genus Streptococcus. One of the best-studied naturally competent bacteria is Streptococcus pneumoniae. In this species and other streptococci shown to be naturally transformable, competence is not a constant property; rather, it is a transient state regulated by a quorum-sensing mechanism consisting of ComABCDE (4). comC encodes the precursor of a secreted peptide pheromone, the competence-stimulating peptide, which triggers development of the competent state when its external concentration in a pneumococcal culture reaches a critical threshold (7). The competence-stimulating peptide is secreted by ComAB (11) and acts through a two-component signal transduction pathway consisting of the histidine kinase ComD and the cognate response regulator ComE (4, 8, 21). The early genes are regulated by ComE, whereas the alternative sigma factor ComX is needed for expression of the late genes (5, 14, 22). Late genes share an 8-bp sequence in their promoter regions that is specifically recognized by a ComX-directed RNA polymerase holoenzyme (14). Circumstantial evidence indicates that ComX is encoded by one of the early genes and therefore depends on ComE for its expression (4). The 14 pneumococcal proteins known to be necessary for uptake of extracellular DNA and for subsequent incorporation of this DNA into the recipient's genome are all encoded by late genes (5, 22). Interestingly, recent genome sequencing has shown that the ComX regulon appears to be present in all streptococcal species (17). This finding suggests that most streptococci are naturally transformable provided that growth conditions promoting development of competence can be identified. Alternatively, the late genes of streptococcal species not known to be competent may have other functions or represent nonfunctional relicts inherited from a competent ancestor.
MATERIALS AND METHODS
Bacterial strains and growth media.
Streptococcus thermophilus strains LMG 18311 (= ATCC BAA-250) and LMD-9 (= ATCC BAA-491) were cultivated in Todd-Hewitt broth (Difco Laboratories) supplemented with 0.8% glucose (THG) or in Hogg-Jago glucose broth (HJG) consisting of 3% tryptone, 1% yeast extract, 0.2% beef extract, 0.5% KH2PO4, and 0.5% glucose. HJGL is Hogg-Jago glucose broth supplemented with 0.5% lactose, whereas HJGLS is HJGL supplemented with 0.4 M d-sorbitol. Agar plates were prepared by adding 1.5% (wt/vol) agar to the media.
Construction of plasmids.
The reporter plasmids pXP, pEAP, and pBP were constructed by fusing the promoters of comX, comEA (a late gene encoding part of the DNA uptake apparatus), and stbD (encoding a putative bacteriocin) to the firefly luciferase gene and ligating the resulting fragments into the pTRKH2 shuttle vector (20). Briefly, the luciferase gene was amplified in three separate PCRs using primer pairs LXP/LR (pXP), LCB/LR (pEAP), and LBP/LR (pBP) and a plasmid (pR424) carrying the luc gene as the template (3). Similarly, PCRs performed with primer pairs CXPF/CXPL, CBF/CBL, and BPF/BPL and genomic DNA from S. thermophilus LMG 18311 were used to amplify fragments corresponding to the comX (∼440-bp), comEA (∼210-bp), and stbD (∼250-bp) promoters, respectively. Then promoter and luc gene fragments with complementary overlapping ends were combined and amplified by PCRs using the appropriate external primers. Primer pairs CXPF/LR, CBF/LR, and BPF/LR were used to generate the XL, EAP, and BP fragments, respectively. Finally, the three fragments were cloned into the pCR 2.1-TOPO vector (Invitrogen), excised by XhoI and PstI, and ligated into the corresponding sites of the pTRKH2 vector. The resulting reporter plasmids, pXP, pEAP, and pBP, were electroporated into S. thermophilus LMG 18311 as described below, giving rise to the XP, EAP, and BP strains.
To construct the pXL plasmid, a DNA fragment containing the comX gene joined to the promoter of stbD was ligated into the pEAP plasmid (see above). The fragments corresponding to the bacteriocin promoter (PstbD) and the comX gene were amplified from S. thermophilus LMG 18311 DNA, using primers P1 and P2 and primers X1 and X2, respectively. The P2 and X1 primers contain NcoI sites at their 5′ ends coinciding with the start codon of the comX gene. Next, the PstbD and comX fragments were cloned separately into the pDrive vector (QIAGEN). The comX fragment was excised from pDrive by digestion with NcoI and XbaI and ligated into the corresponding sites of the pDrive vector carrying the stbD promoter fragment. Then the joined PstbD::comX fragment was excised from the pDrive vector by digestion with PstI and SacI and ligated into pTRKH2 precleaved with the same enzymes. Finally, the PstbD::comX fragment was excised from pTRKH2 by digestion with BamHI and EcoRV and ligated into pEAP precleaved with BamHI ans SmaI. The resulting construct, pXL, contained an expression module (PstbD::comX) and a reporter module (PcomEC::luc) inserted in opposite orientations. All PCRs described above were carried out with the Phusion high-fidelity DNA polymerase (Finnzymes). The sequences of the primers used are shown in Table 1.
TABLE 1.
Primers used in this study
| Primer | Sequence |
|---|---|
| CBF | 5′-AGTGTAACTGCAGAATACTTGCAGGTCTATCGATCGAT-3′ |
| CBL | 5′-TTTGGCGGATCTCATAAGGACCTCCTCATAAACCTATTC-3′ |
| CXPF | 5′-CGCTTTCTGCAGCTATCACTCTAATACAATCCTGTGGAA-3′ |
| CXPL | 5′-TTGGCGGATCTCATTGAACCTCCAATAATAAATATAAATTCTGT-3′ |
| BPF | 5′-GTAAATCTGCAGCTTCAAGGTCTAGTCCTCTCT-3′ |
| BPL | 5′-TTGGCGGATCTCATGGCAACTACCTCCTAAAATTTTTATC-3′ |
| LCB | 5′-TATGAGGAGGTCCTTATGAGATCCGCCAAAAACAT-3′ |
| LR | 5′-CATATGGCTCGAGTGCACTCTCAGTACAATCTGCTC-3′ |
| LXP | 5′-TATTGGAGGTTCAATGAGATCCGCCAAAAACATAAAGAAAGGC-3′ |
| LBP | 5′-TAGGAGGTAGTTGCCATGAGATCCGCCAAAAACATAAAGAAAGGC-3′ |
| P1 | 5′-GTTTGAGTTG CCATGGCAACTACCTCC-3′ |
| P2 | 5′-ATTAGGATCCTTCAAGGTCTAGTCCTCTCTTTTATGACG-3′ |
| X1 | 5′-ATTATCTAGACCAAGAATTACTGGAAACACAATAGAGG-3′ |
| X2 | 5′-GGAGGTTCCATGGAACAAGAAGTTTTTGTTAAGGC-3′ |
| EC1 | 5′-GAGGCATCATTGGAAGAATAGAGCAGC-3′ |
| EC2 | 5′-AAGCTTAAGATCTAGAGCTCGAGGATCAAAAACTAGAGAGAAGATTGCCGTCAG-3′ |
| EC3 | 5′-AGCATGCATATGCATCCGGAGTCCTAGCTTGTTTCAGTTTGTCTCAATG-3′ |
| EC4 | 5′-CCATCCCTTAAACCGAATGGCACC-3′ |
| Kana-F | 5′-ATCCTCGAGCTCTAGATCTTAAGCTT-3′ |
| Kana-R | 5′-ACTCCGGATGCATATGCATGCT-3′ |
Preparation of electrocompetent S. thermophilus LMG 18311 cells.
An overnight culture grown at 37°C was diluted 100-fold in preheated HJG (37°C) and incubated until the optical density at 660 nm (OD660) was 0.3. The culture (50 ml) was then diluted 1:1 in prewarmed HJG containing 20% glycine. After incubation at 37°C for 1 h, cells were harvested by centrifugation (4,000 × g for 10 min at 4°C) and washed twice with 1 volume of ice-cold electroporation buffer (5 mM KHPO4, 0.4 M d-sorbitol, 10% glycerol; pH 4.5). Finally, pelleted cells were resuspended in 4 ml ice-cold electroporation buffer, divided into aliquots, and frozen in an ethanol-dry ice bath. Electrocompetent S. thermophilus LMG 18311 cells were stored at −80°C.
Electroporation.
A Bio-Rad MicroPulser unit was used to transform S. thermophilus LMG 18311 cells by electroporation. Electrocompetent cells were thawed on ice, and an 80-μl cell suspension was mixed with 1 μg of recombinant pTRKH2 plasmid DNA. After incubation for 30 min on ice, the cells were transferred to an electroporation cuvette with a 0.1-cm gap between the electrodes. A single pulse of 1.6 kV lasting 2.5 ms was delivered. The electroporated cells were immediately resuspended in 1 ml of ice-cold HJGLS and incubated for 3 h at 37°C, before they were spread on HJGL plates containing 2 μg/ml erythromycin. Transformants were picked following 24 to 48 h of incubation at 37°C. Isolated clones were verified by PCR using primers M13F and M13R, which were complementary to sequences flanking the multiple cloning site of the pTRKH2 plasmid.
Luciferase reporter assay.
Detection of luciferase activity was performed essentially as previously described by Chastanet et al. (3). Strains were grown in THG to an OD550 of 0.4, aliquoted, and maintained as glycerol stocks at −80°C. Shortly before use, glycerol stocks were thawed and diluted 10 times in THG. For each test sample, 280 μl of diluted culture was mixed with 20 μl of firefly d-luciferin (10 mM solution in THG) and transferred into a 96-well Corning NBS plate with a clear bottom. If appropriate, the peptide pheromone known as Streptococcus thermophilus pheromone (STP) was added to a final concentration of 250 ng/ml immediately before the experiment was started. The plate was incubated at 37°C in an Anthos Lucy 1 luminometer for 7.5 h. The OD492 and luminescence were measured automatically by the luminometer at 10-min intervals.
Natural transformation of S. thermophilus LMG 18311.
S. thermophilus LMG 18311 cells harboring pXL were grown overnight at 37°C in Todd-Hewitt broth (Difco Laboratories) supplemented with 0.8% glucose and 2 μg/ml erythromycin. The next day the culture was diluted to obtain an OD550 of 0.5 in the same medium prewarmed to 37°C. Then 1 ml of the diluted culture was transferred to a 1.5-ml Eppendorf tube containing 250 ng STP, and the sample was placed in a water bath at 37°C. Two hours later transforming DNA was added. At this stage the OD550 of the culture did not exceed ∼0.3. The preparation was then incubated for an additional 2 h at 37°C. Finally, the sample was put on ice, serially diluted, and spread on HJGL agar plates containing the appropriate antibiotic (50 μg/ml streptomycin or 100 μg/ml kanamycin). To avoid loosing the pXL plasmid, 2 μg/ml erythromycin had to be added to the HJGL agar plates. Curing of the pXL plasmid was performed by cultivating transformants in antibiotic-free medium for about 100 generations.
Disruption of the comEC gene.
The comEC gene disruption cassette consists of a kanamycin resistance gene flanked by two 800- to 1,000-bp DNA fragments corresponding to the 5′ and 3′ regions of the comEC gene. In the first step for disruption of the comEC gene, the kanamycin resistance gene was amplified by PCR from the pFW13 vector (23), using primers Kana-F and Kana-R. In the second step, the 5′ and 3′ flanking fragments were generated in two separate PCRs with primer pairs EC1/EC2 and EC3/EC4 and genomic DNA from S. thermophilus LMG 18311 as the template. The EC2 and EC3 primers used to amplify the flanking sequences contained 22-bp extensions homologous to the 5′ and 3′ ends of the kanamycin gene, respectively. After agarose gel purification of all PCR fragments, the kanamycin resistance gene was first joined to the 5′ flanking fragment in a PCR performed with the two DNA fragments and the EC1 and Kana-R primers. In the same way, the kanamycin resistance gene was joined to the 3′ flanking fragment in a PCR performed with both fragments and the Kana-F and EC4 primers. Finally, the two combined fragments were joined in a PCR performed with the EC1 and EC4 primers. The resulting comEC gene disruption cassette was purified with a PCR purification kit obtained from QIAGEN and was used directly to transform competent S. thermophilus LMG 18311 cells carrying the pXL plasmid. In addition, the gene disruption cassette was cloned into the pCR 2.1-TOPO vector (Invitrogen) according to the manufacturer's instructions.
RESULTS
ComX is expressed in S. thermophilus during early logarithmic growth.
The recent sequencing of three S. thermophilus genomes opened the door to rational metabolic engineering of this important dairy species (2, 10, 25). However, in order to take full advantage of the available sequence information, better genetic tools have to be developed (26). Natural transformation is a highly efficient tool for genetic manipulation that has been used successfully with S. pneumoniae for more than 60 years. Drawing on our experience with S. pneumoniae, we therefore set out to determine whether S. thermophilus is a naturally transformable species. We chose to work on S. thermophilus LMG 18311, which was isolated from yoghurt produced in the United Kingdom in 1974, because close inspection of its genome sequence indicated that the ComX regulon is intact in this strain. Initially, experiments were carried out to establish if transformants could be obtained by adding homologous genomic DNA containing a streptomycin resistance marker to LMG 18311 cultures grown under various conditions. All results were negative, suggesting that ComX and/or the late genes are not expressed under the conditions used. To be able to monitor the activity of the comX promoter in a growing culture of LMG 18311 cells over time and under various conditions, we used the shuttle plasmid pTRKH2 (20) to construct a reporter plasmid, pXP, harboring a transcriptional fusion between the comX promoter and the firefly luciferase gene. The pXP plasmid was subsequently introduced into S. thermophilus LMG 18311 by electroporation, giving rise to the XP strain. Luciferase activity was monitored by growing cultures of the XP strain at 37°C in a Lucy 1 luminometer (Anthos) as described in Materials and Methods. Unexpectedly, we discovered that the comX promoter is active during early to approximately mid-logarithmic phase in XP cells grown in THG at 37°C. As the culture approached stationary phase, the activity of the comX promoter declined to zero (Fig. 1A).
FIG. 1.
Expression of the luc reporter gene (▴) during growth of S. thermophilus LMG 18311 from logarithmic to stationary phase (▪). (A) Expression of luciferase driven by the comX promoter. (B) Expression of luciferase driven by the comEC late gene promoter.
Low-level expression of late genes during early logarithmic phase.
Even though ComX is expressed in early logarithmic phase, we were not able to obtain transformants when cultures at this stage of growth were exposed to purified genomic DNA from a streptomycin-resistant mutant of strain LMG 18311. One possible explanation for this negative result is that there were undetected loss-of-function mutations in the transformation machinery. Alternatively, the level of ComX produced might have been too low to significantly activate expression of the late genes. To determine whether this could be the case, we constructed a plasmid similar to pXP, except that we exchanged the comX promoter with the promoter of the late gene comEA (stu1562). The resulting plasmid, pEAP, was electroporated into S. thermophilus LMG 18311, giving rise to the EAP strain. The activity of the comEA promoter was monitored by growing the EAP strain in the Lucy 1 luminometer exactly as described above for the XP strain. The data obtained revealed that there was low-level luciferase expression during early logarithmic phase, a pattern that roughly coincided with the activity of the comX promoter (Fig. 1A and B). From the reporter assay alone, it was not possible to determine whether the comEA promoter operated at a very low level in all bacteria in the culture or if it was highly expressed in just a tiny fraction of the cells. In any case, the results indicate that the level of ComX produced was too low to turn on the competent state in a significant fraction of the bacterial population.
Overexpression of ComX.
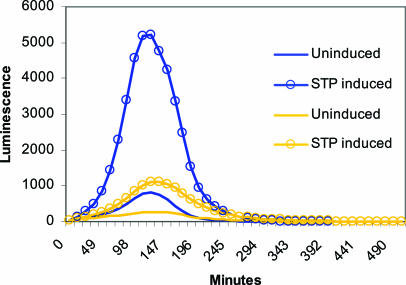
Morrison and coworkers have shown that in S. pneumoniae ectopic expression of ComX from a raffinose-inducible promoter in a comX mutant does not result in development of competence. However, when the comX gene was cloned into a multicopy plasmid and expressed from a nisin-inducible promoter, the number of transformants obtained was almost 10% of the wild-type number (15). In later studies these workers were able to show that a product of the early genes, termed ComW, is needed in addition to ComX for efficient induction of competence. Evidence indicates that ComW contributes to the stabilization of ComX against proteolysis and that it is also required for optimal functionality of the sigma factor (16, 24). We were not able to identify any homologue of ComW in S. thermophilus, but it is reasonable to assume that ComX is unstable also in this species. Judging from our results and the results of the studies carried out by Morrison and coworkers, it should be possible to induce competence in S. thermophilus if sufficiently high levels of ComX can be obtained. Using a strong constitutive promoter would probably not work, as constant high levels of ComX would interfere with the normal transcription pattern of the cell. Instead, a system that could provide high-level transient expression of ComX is needed. We recently developed an inducible expression system for S. thermophilus, and we decided to use this method for overexpression of ComX (1a). This method exploits a quorum-sensing system consisting of a peptide pheromone (STP) encoded by stbC (stu1688), its secretion apparatus (StbAB), its histidine kinase receptor (StbH) (stu1687), and the cognate response regulator StbR (stu1686). The STP secretion apparatus appears to be nonfunctional in S. thermophilus LMG 18311, due to a frameshift mutation in the stbB gene. Nevertheless, we found that synthetic STP (NH2-SGWMDYINGFLKGFGGQRTLPTKDYNIPQV-COOH), added to a culture of the LMG 18311 strain, activates transcription of the bacteriocin-like gene, stbD (stu1685), situated immediately upstream of stbR in the opposite transcriptional orientation. Consequently, by constructing transcriptional fusions between the stbD promoter and target genes, inducible expression of recombinant proteins can be obtained in S. thermophilus. To determine the efficiency of the expression system under various growth conditions, a luc reporter gene was placed behind the stbD promoter. The resulting reporter construct (pBP) was introduced into S. thermophilus LMG 18311 by electroporation, giving rise to the BP strain. Using this strain, we tested the STP-inducible expression system in different media and found that the level of luminescence obtained was by far the highest in THG (Fig. 2). This medium was therefore used for subsequent overexpression of ComX.
FIG. 2.
Effect of the growth medium on STP-induced overexpression of the luciferase reporter protein. Expression of the luc reporter gene was driven by the promoter of the putative bacteriocin gene, stbD. The S. thermophilus BP strain was grown in THG (blue lines) or HJGL (yellow lines) from logarithmic to stationary phase. The STP peptide pheromone was added at time zero.
Induction of the competent state in S. thermophilus.
To determine whether the new expression system could drive ComX production to the level required for activating transcription of the late genes, a new plasmid based on pEAP was constructed. This plasmid (pXL) was constructed by ligating a DNA fragment consisting of a transcriptional fusion between the stbD promoter and the comX gene into unique restriction sites of the pEAP plasmid. To avoid transcriptional readthrough, the expression and reporter modules of the pXL plasmid were inserted in opposite directions. The resulting construct was introduced into the LMG 18311 strain by electroporation. The effect of STP-induced overproduction of ComX on late gene expression was subsequently monitored by measuring light emission from XL cells with the Lucy 1 luminometer. The results showed that a culture of XL cells treated with 250 ng/ml of STP displayed an approximately 600-fold increase in luminescence compared to a corresponding culture of bacteria harboring the pEAP plasmid (Fig. 1B and 3). No effect on luciferase expression was seen when cultures of the EAP and XP strains were treated with the STP peptide pheromone, demonstrating that the dramatic increase in light production observed with the XL strain must have been due to STP-induced overexpression of ComX. Our results also revealed that ComX is expressed in uninduced XL cells due to a leaky stbD promoter. However, the peak luminescence of cultures treated with STP was about sevenfold higher than the luminescence of uninduced cultures (Fig. 3). By chance we discovered that in the absence of selection pressure the pXL plasmid is rapidly lost from its host. Presumably, the presence of ComX, expressed from the leaky stbD promoter, disturbs the normal functions of the bacterial cell.
FIG. 3.
Transcriptional activation of late competence genes by STP-induced overexpression of ComX in S. thermophilus LMG 18311 carrying the pXL plasmid. Plasmid pXL contains an expression cassette consisting of the STP-responsive stbD promoter transcriptionally fused to the comX gene. In addition, it contains a reporter cassette consisting of the comEC late gene promoter transcriptionally fused to the luc reporter gene. As ComX activates transcription from late gene promoters, this construct makes it possible to monitor expression of late competence genes in STP-induced (▴) and uninduced (▵) S. thermophilus cultures. ▪, growth curve for STP-induced culture; □, growth curve for uninduced culture. The STP peptide pheromone was added at time zero.
Having constructed an inducible ComX expression system that efficiently activated transcription from late gene promoters, we were anxious to find out whether the transformation machinery of S. thermophilus LMG 18311 was still functional. To our delight, 3 × 103 streptomycin-resistant CFU per ml of culture (standard error, ±0.9 × 103 CFU per ml; n = 4) were obtained when genomic DNA (1 μg/ml) that was isolated from a streptomycin-resistant LMG 18311 mutant (Strr through spontaneous conversion) was added to cultures of the XL strain pretreated with the STP pheromone (see Materials and Methods for experimental details). The total number of CFU in the culture was estimated in parallel and was determined to be 5 × 108 CFU per ml (standard error, ±1 × 108 CFU per ml; n = 4). To ensure that the observed acquisition of streptomycin resistance had taken place by natural transformation, we checked whether the process depended on a functional comEC gene. ComEC, also called CelB, has been shown to be essential for natural transformation in Bacillus subtilis, S. pneumoniae, and other competent bacteria and is believed to encode a transmembrane channel required for DNA internalization (1, 6). The gene encoding ComEC is located on the same transcription unit as comEA. To disrupt the comEC gene, we used PCR to generate a DNA fragment consisting of a kanamycin marker fused to ∼1,000-bp flanking regions amplified from the 5′ and 3′ halves of the comEC gene of S. thermophilus. This fragment was added to an STP-induced culture of the XL strain at a concentration of 1 μg/ml. After incubation for 2 h at 37°C, the bacteria were spread on agar plates containing 100 μg/ml of kanamycin and then incubated at 37°C for 18 to 24 h. We obtained 7 × 103 CFU per ml (standard error, ±1 × 103 CFU per ml; n = 4) on the agar plates containing kanamycin, and ∼5 × 108 CFU per ml on the control plates lacking the antibiotic. Correct integration of the gene disruption cassette into the comEC gene by double-crossover homologous recombination was verified by PCR in 10 randomly picked kanamycin-resistant colonies. Next, we tested the transformability of the XL ΔcomEC strain using genomic DNA from the streptomycin-resistant LMG 18311 mutant as a selectable marker. No transformants were obtained, demonstrating that the XL strain became nontransformable when the comEC gene was disrupted.
When transformation is performed with a linear DNA fragment, such as the comEC gene disruption cassette described above, the ends of the fragments may be attacked and shortened by nucleases, resulting in reduced transformation efficiency (13). In an attempt to further increase the transformation efficiency, we protected the ends of the comEC gene disruption cassette by cloning it into the pCR2.1-TOPO plasmid (Invitrogen). By using this strategy we obtained 3 × 106 kanamycin-resistant CFU per ml (standard error, ±0.4 × 106 CFU per ml; n = 4) when 3 μg/ml of plasmid DNA was added to STP-induced cultures of the XL strain. The total number of CFU per ml of competent culture was the same as before (∼5 × 108 CFU per ml). As described above, correct integration of the comEC gene disruption cassette was verified by PCR in 10 randomly picked colonies. These results showed that approximately 1% of the streptococcal chains present in the competent culture receiving 3 μg/ml of recombinant plasmid DNA gave rise to a colony when they were cultivated on agar plates containing kanamycin.
Unexpectedly, preliminary results indicated that S. thermophilus strain LMD-9 transformed very poorly using the method described above for strain LMG 18311. Therefore, more strains must be tested before it becomes clear whether the method described here works well with most S. thermophilus strains or whether we were just lucky to pick LMG 18311.
DISCUSSION
By using the highly efficient transformation procedure described above it should be possible to introduce mutations into the genome of S. thermophilus LMG 18311 without the use of a selectable antibiotic resistance marker. DNA fragments containing the desired insertion/deletion or point mutation(s) can be made by PCR or other molecular methods and cloned into a suitable plasmid. Following uptake of this construct by competent S. thermophilus LMG 18311 cells, targeted integration of the mutated region into the bacterial genome is mediated by ∼1,000-bp flanking regions through double-crossover homologous recombination. Due to the high transformation efficiency it should be relatively easy to identify transformants containing the sought-after genotype against the background of wild-type streptococci. Standard colony hybridization with a labeled oligonucleotide probe designed to specifically recognize mutants could be used for this purpose. Before plating, a mechanical blender (e.g., Ultra-Turrax model T25 [Ika Labotechnik, Staufen, Germany]) must be used to disrupt the long chains of S. thermophilus cells, as described previously (18). After identification of the desired mutant, it is easily cured of the unstable pXL helper plasmid by growth in the absence of erythromycin. S. thermophilus mutants made by using this technique fulfill the safety criteria described by Johansen (12) and should therefore attain “generally recognized as safe” status provided that DNA from organisms that are not generally recognized as safe is not introduced into the genetically engineered strain.
In the present work we showed that overexpression of ComX induces the competent state in S. thermophilus LMG 18311. An important question that remains to be answered, however, is how natural transformation is turned on spontaneously in this strain. Although it is possible that the mechanism controlling the development of competence has degenerated during adaptation to the dairy niche, it is more likely that spontaneous development of competence in S. thermophilus LMG 18311 requires special, as-yet-undiscovered growth conditions. The regulatory pathway controlling expression of the comX gene is not known, but our results show that this gene is actively transcribed during early logarithmic phase when the XP strain is grown in THG at 37°C. In spite of this, the levels of transcription of the late genes under these conditions stayed very low, indicating that the level of comX transcription was too low or that ComX was prevented from accumulating to levels required for late gene expression by a regulatory mechanism operating at the posttranscriptional level. It has been reported that the ClpP protease negatively controls ComX in S. pneumoniae (24) and Streptococcus pyogenes (19), and it is therefore likely that the same control mechanism operates in S. thermophilus. The fact that overexpression of ComX efficiently induces expression of the late genes suggests that a system that negatively controls the accumulation of ComX becomes saturated under these circumstances. In sum, preliminary data indicate that spontaneous induction of competence in S. thermophilus requires the joint action of at least two converging regulatory pathways. However, in contrast to other streptococci that have been shown to be competent for natural transformation, a quorum-sensing system of the ComCDE type does not seem to be involved.
Considering the high degree of degeneracy detected in the genome of S. thermophilus, it is remarkable that the genes involved in natural transformation have remained intact. Bolotin et al. (2) found that 10% of the genes in S. thermophilus strains LMG 18311 and CNRZ 1066 are nonfunctional pseudogenes, and they concluded that these strains have adapted to the dairy niche mainly through loss-of-function events. The intactness of the late competence genes in strain LMG 18311 indicates that even in a constant milk environment natural competence provides a selective advantage. Indeed, evidence of lateral gene transfer from other dairy bacteria to S. thermophilus LMG 18311 has been reported. A 17-kb mosaic region found in the pepD gene contains fragments with high homology to corresponding sequences in Lactobacillus bulgaricus and Lactococcus lactis, species that come into close contact with S. thermophilus during fermentation of yoghurt and cheeses, respectively (2). In light of the results presented here, it seems plausible that at least some of these gene transfer events have taken place by natural genetic transformation.
Acknowledgments
This work was supported by grants from the Research Council of Norway.
REFERENCES
- 1.Berge, M., M. Moscoso, M. Prudhomme, B. Martin, and J. P. Claverys. 2002. Uptake of transforming DNA in Gram-positive bacteria: a view from Streptococcus pneumoniae. Mol. Microbiol. 45:411-421. [DOI] [PubMed] [Google Scholar]
- 1a.Blomqvist, T., H. Steinmoen, L. S. Håvarstein. Pheromone-induced expression of recombinant proteins in Streptococcus thermophilus. Arch. Microbiol., in press. [DOI] [PubMed]
- 2.Bolotin, A., B. Quinquis, P. Renault, A. Sorokin, S. D. Ehrlich, S. Kulakauskas, A. Lapidus, E. Goltsman, M. Mazur, G. D. Pusch, M. Fonstein, R. Overbeek, N. Kyprides, B. Purnelle, D. Prozzi, K. Ngui, D. Masuy, F. Hancy, S. Burteau, M. Boutry, J. Delcour, A. Goffeau, and P. Hols. 2004. Complete sequence and comparative genome analysis of the dairy bacterium Streptococcus thermophilus. Nat. Biotechnol. 22:1554-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chastanet, A., M. Prudhomme, J. P. Claverys, and T. Msadek. 2001. Regulation of Streptococcus pneumoniae clp genes and their role in competence development and stress survival. J. Bacteriol. 183:7295-7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claverys, J. P., and L. S. Håvarstein. 2002. Extracellular-peptide control of competence for genetic transformation in Streptococcus pneumoniae. Front. Biosci. 7:d1798-1814. [DOI] [PubMed] [Google Scholar]
- 5.Dagkessamanskaia, A., M. Moscoso, V. Henard, S. Guiral, K. Overweg, M. Reuter, B. Martin, J. Wells, and J. P. Claverys. 2004. Interconnection of competence, stress and CiaR regulons in Streptococcus pneumoniae: competence triggers stationary phase autolysis of ciaR mutant cells. Mol. Microbiol. 51:1071-1086. [DOI] [PubMed] [Google Scholar]
- 6.Draskovic, I., and D. Dubnau. 2005. Biogenesis of a putative channel protein, ComEC, required for DNA uptake: membrane topology, oligomerization and formation of disulphide bonds. Mol. Microbiol. 55:881-896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Håvarstein, L. S., G. Coomaraswami, and D. A. Morrison. 1995. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc. Natl. Acad. Sci. USA 92:11140-11144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Håvarstein, L. S., P. Gaustad, I. F. Nes, and D. A. Morrison. 1996. Identification of the streptococcal competence-pheromone receptor. Mol. Microbiol. 21:863-869. [DOI] [PubMed] [Google Scholar]
- 9.Håvarstein, L. S., R. Hakenbeck, and P. Gaustad. 1997. Natural competence in the genus Streptococcus: evidence that streptococci can change pherotype by interspecies recombinational exchanges. J. Bacteriol. 179:6589-6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hols, P., F. Hancy, L. Fontaine, B. Grossiord, D. Prozzi, N. Leblond-Bourget, B. Decaris, A. Bolotin, C. Delorme, S. D. Ehrlich, E. Guédon, V. Monnet, P. Renault, and M. Kleerebezem. 2005. New insights in the molecular biology and physiology of Streptococcus thermophilus revealed by comparative genomics. FEMS Microbiol. Rev. 29:435-463. [DOI] [PubMed] [Google Scholar]
- 11.Hui, F. M., and D. A. Morrison. 1991. Genetic transformation in Streptococcus pneumoniae: nucleotide sequence analysis shows comA, a gene required for competence induction, to be a member of the bacterial ATP-dependent transport protein family. J. Bacteriol. 173:372-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johansen, E. 1999. Genetic engineering (b). Modification of bacteria, p. 917-921. In R. Robinson, C. Batt, and P. Patel (ed.), Encyclopedia of food microbiology. Academic Press, London, United Kingdom.
- 13.Lataste, H., J. P. Claverys, and A. M. Sicard. 1981. Relation between the transforming activity of a marker and its proximity to the end of the DNA particle. Mol. Gen. Genet. 183:199-201. [DOI] [PubMed] [Google Scholar]
- 14.Lee, M. S., and D. A. Morrison. 1999. Identification of a new regulator in Streptococcus pneumoniae linking quorum sensing to competence for genetic transformation. J. Bacteriol. 181:5004-5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo, P., H. Li, and D. A. Morrison. 2003. ComX is a unique link between multiple quorum sensing outputs and competence in Streptococcus pneumoniae. Mol. Microbiol. 50:623-633. [DOI] [PubMed] [Google Scholar]
- 16.Luo, P., H. Li, and D. A. Morrison. 2004. Identification of ComW as a new component in the regulation of genetic transformation in Streptococcus pneumoniae. Mol. Microbiol. 54:172-183. [DOI] [PubMed] [Google Scholar]
- 17.Martin, B., Y. Quentin, G. Fichant, and J. P. Claverys. 2006. Independent evolution of competence regulatory cascades in streptococci? Trends Microbiol. 14:339-345. [DOI] [PubMed] [Google Scholar]
- 18.Monnet, C., S. Pernoud, A. Sepulchre, C. Fremaux, and G. Corrieu. 2004. Selection and properties of Streptococcus thermophilus mutants deficient in urease. J. Dairy Sci. 87:1634-1640. [DOI] [PubMed] [Google Scholar]
- 19.Opdyke, J. A., J. R. Scott, and C. P. Moran, Jr. 2003. Expression of the secondary sigma factor σx in Streptococcus pyogenes is restricted at two levels. J. Bacteriol. 185:4291-4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Sullivan, D. J., and T. R. Klaenhammer. 1993. High-copy-number and low-copy-number Lactococcus shuttle cloning vectors with features for clone screening. Gene 137:227-231. [DOI] [PubMed] [Google Scholar]
- 21.Pestova, E. V., L. S. Håvarstein, and D. A. Morrison. 1996. Regulation of competence for genetic transformation in Streptococcus pneumoniae by an auto-induced peptide pheromone and a two-component regulatory system. Mol. Microbiol. 21:853-862. [DOI] [PubMed] [Google Scholar]
- 22.Peterson, S. N., C. K. Sung, R. Cline, B. V. Desai, E. C. Snesrud, P. Luo, J. Walling, H. Li, M. Mintz, G. Tsegaye, P. C. Burr, Y. Do, S. Ahn, J. Gilbert, R. D. Fleischmann, and D. A. Morrison. 2004. Identification of competence pheromone responsive genes in Streptococcus pneumoniae by use of DNA microarrays. Mol. Microbiol. 51:1051-1070. [DOI] [PubMed] [Google Scholar]
- 23.Podbielski, A., B. Spellerberg, M. Woischnik, B. Pohl, and R. Lütticken. 1996. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS). Gene 177:137-147. [DOI] [PubMed] [Google Scholar]
- 24.Sung, C. K., and D. A. Morrison. 2005. Two distinct functions of ComW in stabilization and activation of the alternative sigma factor ComX in Streptococcus pneumoniae. J. Bacteriol. 187:3052-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tettelin, H. 2004. Streptococcal genomes provide food for thought. Nat. Biotechnol. 22:1523-1524. [DOI] [PubMed] [Google Scholar]
- 26.Vadeboncoeur, C., and S. Moineau. 2004. The relevance of genetic analysis to dairy bacteria: building upon our heritage. Microb. Cell Fact. 3:15-18. [DOI] [PMC free article] [PubMed] [Google Scholar]