Abstract
Bacterial virulence is an integrative process that may involve quorum sensing. In this work, we compared by global expression profiling the wild-type entomopathogenic Photorhabdus luminescens subsp. laumondii TT01 to a luxS-deficient mutant unable to synthesize the type 2 quorum-sensing inducer AI-2. AI-2 was shown to regulate more than 300 targets involved in most compartments and metabolic pathways of the cell. AI-2 is located high in the hierarchy, as it controls the expression of several transcriptional regulators. The regulatory effect of AI-2 appeared to be dose dependent. The luxS-deficient strain exhibited decreased biofilm formation and increased type IV/V pilus-dependent twitching motility. AI-2 activated its own synthesis and transport. It also modulated bioluminescence by regulating the synthesis of spermidine. AI-2 was further shown to increase oxidative stress resistance, which is necessary to overcome part of the innate immune response of the host insect involving reactive oxygen species. Finally, we showed that the luxS-deficient strain had attenuated virulence against the lepidopteran Spodoptera littoralis. We concluded that AI-2 is involved mainly in early steps of insect invasion in P. luminescens.
Interest in the insect pathogen Photorhabdus luminescens, a gram-negative luminescent gammaproteobacterium, has increased greatly since the genome sequence of P. luminescens subsp. laumondii TT01 was deciphered (15). The organism has two host-associated stages in its life cycle: the symbiotic stage, in which bacteria colonize the intestinal tract of a nematode belonging to the Heterorhabditis genus, and a second pathogenic stage, in which the bacterium-nematode couple invades and kills a wide variety of insect larvae. The antimicrobial response of insects comprises both cellular and humoral reactions. Plasmatocytes, the major hemolymph cells in Drosophila, show strong phagocytic activity (33). They also trigger a first line of defense comprising highly toxic molecules: nitric oxide (NO), the superoxide ion and hydrogen peroxide (H2O2), and even peroxynitrite (ONOO−), a highly reactive species that is rapidly generated by the reaction between NO and the superoxide ion (21). In addition to these reactive oxygen species (ROS), the insect host defense also includes melanization. The humoral reaction is induced by the microbial challenge to antimicrobial peptide genes in the body fat, followed by the secretion of the peptides into the hemolymph (33). The processes by which P. luminescens circumvents these host defenses are not well characterized. Nevertheless, recent work has shown that the type III secretion system plays a key role in preventing phagocytosis by insect macrophage cells during insect colonization by P. luminescens (5). By contrast, the way by which P. luminescens kills the insect is better known. As the bacteria multiply, they produce many toxins, such as the Mcf (Makes Caterpillars Floppy) toxin, a virulence factor critical for pathogenesis (11); the PirA (PLU4093) and PirB (PLU4092) toxins, which can kill larvae belonging to two insect orders (15, 74); and four forms of a toxin complex, Tca, Tcb, Tcc, and Tcd, which are highly toxic to Manduca sexta (4, 36). The bacteria also produce various antibiotics, which are presumed to inhibit the growth of competing microorganisms in the insect cadaver (12, 15, 76) and to enhance conditions for nematode reproduction by providing nutrients and growth factors (22). After several rounds of reproduction, a new generation of infective juvenile nematodes finally reacquires the bacteria and leaves the insect carcass (3).
Upon regurgitation of the few cells contained in the nematode intestine, the bacteria then rapidly multiply to reach high concentrations within the insect cadaver (10). In bacteria, one or several quorum-sensing processes are generally induced at this step. Two quorum-sensing-related genes, mtnN (pfs, plu0906) and luxS (plu1253), are present in P. luminescens (15). These genes code for, respectively, 5′-methylthioadenosine/S-adenosylhomocysteine (AdoHcy) nucleosidase (SwissProt nomenclature) (58) and the autoinducer 2 (AI-2) synthesis protein. The function of the P. luminescens LuxS protein was recently characterized, leading to the identification of a first target, the cpm operon. This operon is responsible for producing a carbapenem-like antibiotic that is repressed in the presence of AI-2 (12).
We wondered whether AI-2 regulates other processes in this insect pathogen, such as virulence, and whether the role of AI-2 is similar to that in other bacteria. Thus, we carried out a global expression profiling study with a luxS-deficient strain of P. luminescens to identify the pathways involved by analyzing the transcriptome and proteome under various experimental conditions. We followed this with complementary in vivo experiments to verify the most prominent regulatory pathways determined from the expression profiling. As the luxS strain cannot synthesize AI-2, this allowed us to characterize the processes involving AI-2.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study were Photorhabdus luminescens TT01 (wild type) (53) and PL2102 (TT01 luxS::Cm) (12). Strains were routinely grown at 30°C in Schneider medium (BioWhittaker) containing 10 μM sodium borate. All experiments were performed in accordance with the European requirements concerning the contained use of genetically modified organisms of group I (agreement no. 2736 CAII).
DNA arrays.
Fragments of 4,225 genes of the genome of P. luminescens TT01 were amplified from 100 ng of chromosomal DNA, using the Dinazyme kit (Finzyme) or Yield Ace kit (Stratagene). Specific primers were designed using CAAT-Box software (23) and Primer3 software; 18- to 25-base-long primers (melting temperatures of between 51.0 and 62.0°C) were chosen to amplify a 450- to 550-bp fragment specific for each coding sequence. The specificity of the PCR products was tested against the complete genome sequence by excluding nonspecific PCR products shorter than 3,000 base pairs. All PCR products were verified on agarose gels. For DNA array preparation, nylon membranes (Genetix) were soaked in 10 mM Tris-1 mM EDTA, pH 7.6. PCR products were printed using a QPix robot (Genetix). Immediately following spot deposition, the membranes were denatured for 15 min in 0.5 M NaOH-1.5 M NaCl-0.1% sodium dodecyl sulfate and then neutralized for 15 min in 0.5 M Tris HCl (pH 7.5)-1.5 M NaCl. Finally, the membranes were dried on Whatman paper and fixed with UV light at 0.51 J/cm2.
Transcriptome experiments.
Total RNA from three independent cultures of each strain grown to optical densities at 600 nm (OD600) of 2.5 (mid-exponential growth phase) and 7.5 (end of exponential growth phase) was purified as previously described (12). The direct effect of AI-2 was studied by adding in vitro-synthesized AI-2 to 5 ml of a strain PL2102 (luxS) culture 2 h before sampling. LsrB, a known target of AI-2 in Salmonella enterica serovar Typhimurium (68), was used to optimize the quantity and timing of AI-2 addition by measuring the increase of lsrB expression by reverse transcription-PCR (RT-PCR) at the end of the exponential growth phase. Two cDNA targets were synthesized from 10 μg of each RNA preparation: after heating at 80°C for 5 min without enzyme and radioactivity, followed by slow cooling, cDNAs were synthesized for 2 h at 42°C with AMV reverse transcriptase (Roche); a mixture of specific primers; dATP, dGTP, and dTTP at 125 μM; and a dCTP mix (2.5 μM dCTP and 100 μCi [α-33P]dCTP [2,000 to 3,000 Ci mmol−1]). The remaining RNA was subjected to alkaline lysis as previously described (48), and cDNAs were purified through a G-25 Sephadex column (Roche). Prehybridization and hybridization were carried out as previously described (29). Hybridization was carried out with six different arrays, which were used once for each culture condition and quantified using ArrayVision software (Imaging Research). Two distinct statistical methods were used for data analysis: (i) the Wilcoxon signed rank test, a nonparametric statistical method contained in the Statview 5.0.1 package, as previously described (29), and (ii) a Student t test with Welch correction and a one-way analysis of variance using BioConductor packages from the statistical environment for data analysis R, after log transformation and global mean normalization of the data. The number of false-positive genes was limited in these results by correcting the obtained P values with different multiple-test procedures (16, 63). We selected differentially expressed genes having a P value of ≤0.05 in at least one of the analysis procedures and a ratio between the normalized data for the wild-type and mutant strains of ≥2. The functions of the genes of interest were extracted from the PhotoList database (http://genolist.pasteur.fr/PhotoList/) (15). Genes with unknown function in PhotoList were further characterized using BLAST and CD searches on updated data at http://www.ncbi.nlm.nih.gov/BLAST/, with COG search at http://www.ncbi.nlm.nih.gov/COG/old/xognitor.html and with membrane domain search at http://www.cbs.dtu.dk/services/TMHMM-2.0/.
Overexpression and purification of LuxS and Pfs/MtnN.
Photorhabdus luminescens luxS and pfs genes were amplified and inserted into the NdeI and XhoI sites of the pET-22b vector (Novagen), giving plasmids pDIA610 and pDIA611, respectively. Recombinant proteins were overproduced by introducing the plasmids into strain BL21(DE3) in the presence of plasmid pDIA17, which synthesizes the lacI repressor (44). The strains, grown in Hyper Broth (Athena Enzyme Systems), were induced at an OD600 of 3 with 3 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 2 h. After centrifugation, bacterial pellets were disrupted with a Fastprep apparatus (BIO 101) in buffer A (20 mM sodium phosphate [pH 7.2] and 200 mM NaCl). Cell debris was removed by centrifugation for 20 min at 7,500 × g, and supernatants were loaded onto an NiSO4 chelation column. After washing with buffer A containing 5 mM imidazole, proteins were eluted with an imidazole gradient in buffer A. The buffer was changed to 50 mM sodium phosphate buffer (pH 7.2) using a PD 10 desalting column (Pharmacia). Proteins were then concentrated with an Amicon Ultra-4 filter (limited 10,000-molecular-weight cutoff) (Millipore).
In vitro production of AI-2.
AI-2 was produced by incubation with 1 mM S-adenosylhomocysteine and 1 mg/ml of the purified P. luminescens proteins Pfs/MtnN and LuxS for 2 h at 30°C in 50 mM sodium phosphate buffer at pH 8 (54). The AI-2 concentration was estimated using Ellman's assay to quantify homocysteine, assuming that AI-2 and homocysteine are equimolar in the reaction (54); the assay was carried out using an aliquot of the reaction mixture filtered through an Amicon Ultra-4 filter (limited 10,000-molecular-weight cutoff) (Millipore).
Two-dimensional gel electrophoresis, MALDI-TOF mass spectrometry, and database searches.
Gel electrophoresis was performed on proteins extracted from disrupted cells grown to mid-exponential growth phase (OD600 of 2.5) from two cultures in 50 ml Schneider medium plus 10 μM sodium borate. The proteins were separated on three two-dimensional sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels for each extract, silver stained, scanned, and analyzed as previously described (13). Proteins were identified by peptide mass fingerprinting with a Voyager DE-STR matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectrometer (MS) (Applied Biosystems, Framingham, MA). Spots of interest were excised, and the Investigator Progest system (Genomic Solutions) was used for in-gel digestion of proteins with modified porcine trypsin (Promega). The Investigator ProMS system (Genomic Solutions) was used for peptide purification with Zip-Tip C18 (Millipore) and for loading the sample with the matrix (alpha-cyano-4-hydroxycinnamic Acid) on the MALDI sample plate. We used the P. luminescens database with the MS-Fit3.2 part of the Protein Prospector package (University of California Mass Spectrometry Facility, San Francisco) to identify the proteins. Search parameters were set as previously described (13).
Polyamine dansylation procedure.
Three independent cultures for each strain were grown to an OD600 of 7, washed three times with 50 mM sodium phosphate buffer at pH 7, and then centrifuged. The bacterial proteins were precipitated by adding 1 ml of ice-cold 0.2 N perchloric acid to the cell pellet, disrupted using a Fastprep apparatus (BIO 101), and centrifuged at 10,000 × g for 10 min. Perchloric supernatants and protein precipitates were stored at −80°C until analyzed (within 1 month). Dansylation was carried out as previously described (57) using 1,10-diamino-dodecane (DAD) as an internal standard. Aliquots (200 μl) of the perchloric supernatants were reacted with four volumes of dansyl chloride in acetone (5 mg/ml) in the presence of solid sodium carbonate. Excess dansyl chloride was removed by reaction with proline, and acetone was removed by evaporation. Dansylated polyamines were extracted using 2 ml of cyclohexane. The cyclohexane extract containing the dansyl derivatives was evaporated to dryness and the residue taken up in 200 μl acetonitrile.
LC-MS analysis of the polyamine content.
The liquid chromatography-MS (LC-MS) analysis of the polyamine content was as previously described (24). The LC-MS, which was from Agilent Technologies, was supplied with the Chem Station 1100 software (Wilmington, DE). Dansylated polyamines were analyzed by flow injection analysis, omitting separation with a LC column. Samples were directly injected from an HP1100 series autosampler into a flowing stream of a mixture of water and acetonitrile (1:9, vol/vol) at a flow rate of 0.5 ml/min. Selected ion monitoring mode data mass were obtained with an atmospheric pressure chemical ionization source. Selective parent ions [(M + H)+] were detected in the selected ion monitoring mode at m/z 555.2, 845.3, 1135.4, and 639.3 for putrescine, spermidine, spermine, and the internal standard DAD, respectively. The area under the ionic peak, determined for each selective ion, was corrected from the ionic intensity of the internal standard DAD, and polyamine concentrations were deduced from calibration curves. The statistical significance of the data was determined using a Mann-Whitney test with a P value of <0.05.
Real-time quantitative RT-PCR.
After heating 5 μg of total RNA at 90°C for 2 min without enzyme, followed by slow cooling, cDNAs were synthesized for 2 h at 42°C with AMV reverse transcriptase (Roche), a mixture of hexanucleotides, and 1.33 mM deoxynucleoside triphosphates. Real-time quantitative PCRs were performed twice in a 20-μl reaction volume containing several cDNA dilutions (100 ng to 0.1 ng), SYBR PCR master mix (Applied Biosystems), and 400 nM of gene-specific primers (designed using the Primer3 software). Amplification and detection were performed as previously described (42). The quantity of cDNA for each experimental gene was normalized to the quantity of udp cDNA in each sample, because udp was considered (and confirmed by DNA array analysis) to represent a stably expressed housekeeping gene. To check whether contaminating chromosomal DNA was present, each sample was tested in control reactions without reverse transcriptase. The relative change in gene expression was recorded as the ratio of normalized target concentrations (ΔΔct) (37).
Bioluminescence assay.
Luminescence was measured by quantifying light production over 2 seconds in 10 μl of culture, using a Lumat LB9507 apparatus (Berthold). Experiments were performed twice.
Bacterial growth in the presence of H2O2.
An overnight culture was washed in fresh Schneider medium and diluted to an OD600 of 0.15. When the OD600 reached 0.5, the culture was separated into three equal parts and exposed to 0, 0.5, and 1 mM H2O2, and the absorbance at 600 nm was monitored. Experiments were performed twice.
Motility assay. (i) Swimming.
The experiment was carried out twice on semisolid medium (1% tryptone, 0.2% NaCl, and 0.3% agar) by spotting 5 μl of Schneider medium cultures grown at an OD600 of 2. The diameters of the halos were measured after 20 h of growth at 30°C.
(ii) Slide culture twitching assay.
The slide culture twitching assay was adapted from that described by Rashid and Kornberg (50). Strains were point inoculated with a toothpick onto the surface of a slab of agar medium (1% tryptone, 0.5% yeast extract, 0.2% NaCl, and 1% agar) placed on a microscope slide. The inoculated agar medium was then covered with a glass coverslip and incubated for 16 h at 30°C. Colony expansion zones were visualized by light microscopy.
Biofilm assay.
Four independent cultures were assayed. After growth in Schneider medium, cultures were diluted to an OD600 of 0.6 into fresh medium and incubated for 4 days at 30°C before being stained with crystal violet. After resuspension in ethanol, staining was measured at 570 nm as previously described (30).
Electron microscopy.
The strains of interest were grown at 30°C with mild agitation (90 rpm) to preserve appendages, and a flagellated strain was used as the positive control. For negative staining, bacteria were placed on 200-mesh copper grids, fixed with 2% glutaraldehyde (Sigma) in 1× phosphate-buffered saline, and stained with 2% uranyl acetate. Samples were examined at 80 kV with a transmission electron microscope (model 1200 EX; Jeol Ltd.) and a charge-coupled-device camera (model Megaview; Eloise, Ltd.).
In vivo pathogenicity assays.
Insect lethality analysis was performed as previously described (14).
RESULTS
Comparative analysis of expression profiling of wild-type and luxS-deficient strains at the end of the exponential growth phase.
Wild-type P. luminescens (TT01) and its luxS-deficient counterpart (PL2102) (12) were used to characterize the AI-2 targets. As AI-2 is involved in quorum sensing, the effect of disrupting the luxS gene was first studied at the end of the exponential growth phase in Schneider medium supplemented with 10 μM sodium borate (AI-2 is described as a boron-associated molecule [9] [see discussion]). In the luxS-deficient strain, some intermediary metabolism genes could be indirectly induced or repressed due to the accumulation of AdoHcy, a molecule that is potentially toxic for the cell through the inhibition of S-adenosylmethionine (AdoMet)-dependent methyltransferases (52). To identify authentic AI-2-regulated genes in intermediary metabolism, we purified total RNA from cultures of strain PL2102 cells supplemented with 4.25 μM AI-2 synthesized in vitro (concentration measured according to the Ellman assay [see Materials and Methods]) 2 hours before sampling, in parallel with experiments involving RNAs from the wild-type and nonsupplemented luxS-deficient strain cultures.
Transcriptome experiments were performed with two cDNA preparations synthesized from each RNA preparation and six arrays, each used once for each strain and/or culture condition. After statistical analysis of the hybridization signal intensity, we identified 221 genes having expression levels in the luxS-deficient strain different from those in the wild-type strain (see Table S1 in the supplemental material). Among these, the expression levels of 108 were partially or completely restored (decrease of ≥20% of difference level of expression) to wild-type levels after the addition of AI-2 to the growth medium. Table 1 lists those genes we chose for further studies.
TABLE 1.
Genes involved in specific processes and differentially expressed in wild-type (TT01) and luxS-deficient (PL2102) P. luminescens strains
| Process | plu no. | Gene | Wild-type/luxS expression ratio at:
|
Function | |
|---|---|---|---|---|---|
| Mid-exponential phase | End of exponential phase | ||||
| AI-2- and polyamine-related synthesis | plu0394a | argD | 4.70 | Acetylornithine delta-aminotransferase | |
| plu0842a | speD | 0.41 | S-Adenosylmethionine decarboxylase | ||
| plu0843a | speE | 0.21 | Spermidine synthase | ||
| plu1307a | gltI | 2.18 | Glutamate/aspartate ABC transport system permease | ||
| plu2942a | 6.03 | 58% similarity with DNA-methyltransferase MunI, AdoMet dependent | |||
| plu3141a | lsrK | 2.24 | AI-2 kinase | ||
| plu3146a | lsrB | 8.17 | AI-2 binding protein of ABC transport system | ||
| plu3147a | lsrF | 2.58 | AI-2-processing aldolase | ||
| plu3148a | lsrG | 3.20 | AI-2-processing protein | ||
| plu4397a | metA | 11.50 | Homoserine O-succinyltransferase | ||
| plu4420a | metE | 24.13 | 5-Methyltetrahydropteroyltriglutamate-homocysteine methyltransferase | ||
| plu4566a | 2.48 | N-Dimethylarginine dimethylaminohydrolase family protein | |||
| plu4567a | 2.52 | Argininosuccinate synthase domain (arginine biosynthesis) | |||
| plu4568a | 2.14 | Putative cystathionine beta-synthase | |||
| plu4743a | argB | 2.81 | Cetylglutamate kinase (arginine biosynthesis) | ||
| plu4756a | metB | 13.06 | Cystathionine gamma-synthase | ||
| Oxidative stress | plu0842a | speD | 0.41 | S-Adenosylmethionine decarboxylase | |
| resistance | plu0843a | speE | 0.21 | Spermidine synthase | |
| plu2027a | uvrC | 7.69 | Excinuclease ABC subunit C | ||
| plu2182 | idgB | 4.85 | Indigoidine synthesis protein | ||
| plu2496a | adhE | 2.74 | Aldehyde-alcohol dehydrogenase | ||
| plu2557a | msrB | 10.91 | Peptide methionine sulfoxide reductase B | ||
| plu3281a | iscA | 3.89 | Iron binding protein IscA (iron-sulfur cluster) | ||
| plu3282 | iscU | 2.02 | Involved in formation of [Fe-S] clusters | ||
| plu3343a | rseC | 3.20 | Sigma-E factor regulatory protein | ||
| plu4728a | bfr | 0.34 | Bacterioferritin (cytochrome B-1, cytochrome B-557) | ||
| Motility and biofilm related | |||||
| Flagellum regulon | plu1848a | flhC | 0.39 | Flagellum biosynthesis transcription activator | |
| plu1912a | flgN | 0.36 | Flagellum synthesis protein | ||
| plu1945 | fliI | 0.22 | Flagellum-specific ATP synthase | ||
| plu4688 | hdfR | 0.49 | H-NS-dependent flhDC regulator | ||
| Pili and fimbriae | plu0417 | 0.05 | Putative fimbrial minor pilin subunit precursor | ||
| plu0769a | mrfA | 2.13 | 3.59 | Major fimbrial subunit polypeptide | |
| plu0782 | 0.36 | Putative fimbrial chaperone protein | |||
| plu0783 | 0.09 | 48% similarity with chaperone protein FimC precursor (E. coli O6) | |||
| plu1053 | pilP | 0.04 | Pilus assembly protein | ||
| plu1056 | pilS | 0.21 | Type IV prepilin | ||
| plu1057 | pilU | 0.27 | Prepilin peptidase | ||
| plu1058 | pilV | 0.20 | Type V prepilin | ||
| plu1734 | 0.50 | Type V prepilin-like | |||
| Other | plu1855a | cheR | 0.36 | Chemotaxis protein methyltransferase | |
| plu4793a | cpxP | 0.47 | Periplasmic protein precursor | ||
| plu4795a | cpxA | 5.52 | Sensory kinase in two-component regulatory system with CpxR | ||
| Virulence related | plu0635a | 0.15 | Hemolysin secretion protein D (HlyD) family secretion protein | ||
| plu0769a | mrfA | 2.13 | 3.59 | Major fimbrial subunit polypeptide | |
| plu0884a | 0.32 | 2.70 | Putative killer protein of pyocin S3 | ||
| plu0887a | 2.81 | 70% similarity with C-terminal region of klebicin B pyocin S2 and the killer protein of pyocin S1 | |||
| plu0888a | 2.52 | 50% similarity with colicin immunity protein operon | |||
| plu0962a | tcdA1 | 2.39 | Insecticidal toxin complex protein TcdA1 (toxin A) | ||
| plu1115 | 0.40 | Syringomycin synthetase involved in antibiotic biosynthesis | |||
| plu1356 | 0.22 | 64% similarity with hemolysin/hemagglutinin-like protein HecA precursor (Erwinia chrysanthemi) | |||
| plu1665 | 2.73 | Orf15 (Photorhabdus W14), C terminus similar to phage tail sheath protein | |||
| plu1666 | 2.54 | Orf14 (Photorhabdus W14), C terminus similar to Phage tail sheath protein FI | |||
| plu1706a | 0.35 | 77% similarity with Orf15 (Photorhabdus W14), C terminus similar to phage tail sheath protein | |||
| plu1878 | 0.24 | Antibiotic synthetase | |||
| plu1893 | 0.47 | Colicin immunity protein | |||
| plu1894 | 0.40 | S-type pyocin | |||
| plu2279 | ccdB | 0.33 | Cytotoxic protein CcdB | ||
| plu2393 | 4.32 | Phage baseplate assembly protein W | |||
| plu2753 | 0.23 | 84% similarity with hypothetical phage protein (Yersinia pestis) | |||
| plu2875 | 0.21 | 60% similarity with hypothetical protein (bacteriophage Aaφ23) | |||
| plu2902 | 0.30 | Bacteriophage protein | |||
| plu2905a | 2.84 | Bacteriophage lambda endopeptidase | |||
| plu2959 | 0.40 | Putative tail fiber protein | |||
| plu3026a | 10.84 | 59% similarity with putative bacteriophage protein STY2015 (S. enterica serovar Typhimurium) | |||
| plu3028a | 11.28 | 68% similarity with putative bacteriophage protein STY2017 (S. enterica serovar Typhimurium) | |||
| plu3029 | 0.43 | 73% similarity with putative bacteriophage protein STY2019 (S. enterica serovar Typhimurium) | |||
| plu3030a | 7.62 | 61% similarity with putative bacteriophage protein STY2023 (S. enterica serovar Typhimurium) | |||
| plu3031a | 11.28 | 57% similarity with putative bacteriophage protein STY2024 (S. enterica serovar Typhimurium) | |||
| plu3035a | 16.28 | 78% similarity with putative bacteriophage protein STY2028 (S. enterica serovar Typhimurium) | |||
| plu3036a | 14.03 | 69% similarity with putative bacteriophage protein STY2029 (S. enterica serovar Typhimurium) | |||
| plu3037a | 12.58 | 69% similarity with putative bacteriophage protein STY2030 (S. enterica serovar Typhimurium) | |||
| plu3123a | 0.48 | Syringopeptin family antibiotic synthetase | |||
| plu3126 | rtxD | 0.34 | Toxin ABC transporter | ||
| plu3127 | rtxB | 0.21 | Toxin ABC transporter | ||
| plu3128 | mcf2 | 0.43 | Toxin protein (Mcf-like protein) | ||
| plu3238 | 0.04 | Toxin motif IcmF in C-terminal region | |||
| plu3385 | 0.34 | 52% similarity with hypothetical protein ORF45 (Burkholderia cepacia phage Bcep781) | |||
| plu3428a | 5.06 | Phage protein U | |||
| plu3429a | 8.43 | 54% similarity with protein gp44 (Burkholderia cenocepacia phage BcepMu) | |||
| plu3460a | 3.14 | Mu-like prophage protein gp16 | |||
| plu3477 | 11.40 | 66% similarity with protein encoded by cryptic prophage CP-933P | |||
| plu3780 | sctE | 0.15 | Type III secretion component protein | ||
| plu3781a | sctF | 7.71 | Type III secretion component protein | ||
| plu4167 | tccC1 | 0.22 | Insecticidal toxin complex protein TccC1 | ||
| plu4780 | 0.38 | 75% similarity with gpFI (bacteriophage Wφ) | |||
| plu4781 | 0.49 | Phage-related protein | |||
The expression level was partially or completely restored to the wild-type level after the addition of AI-2 to the growth medium (see the text).
Interestingly, the presence of AI-2 activated the expression of four genes (plu3141, -3146, -3147, and -3148) that were very similar to and had the same genomic organization as the lsr genes, which are essential for AI-2 transport and processing in Salmonella enterica serovar Typhimurium (67, 68).
Among the regulated genes, one-third were involved in cell envelope-related processes. The synthesis of outer membrane-associated proteins (e.g., flagella, pili, and fimbriae) was also luxS sensitive. Almost all the relevant genes were repressed, with a noteworthy exception being that expression of the gene mrfA, which encodes a major fimbrial subunit, was activated. Moreover, the expression of many transporters was affected, with most being repressed in the presence of AI-2 [e.g., the iron(III) dicitrate and thiamine transport systems] and some being activated (such as a glutamate transporter [gltI] and a nucleoside transporter [plu0519]).
Seven transcriptional regulators were regulated in the presence of AI-2: two flagellum regulator genes, flhC and hdfR, as well as two flagellar genes, flgN and fliI, were repressed; however, most of the regulators were not functionally characterized. Several affected genes were involved in protecting or repairing nucleic acids: the radA and vgrG families of genes were repressed, whereas uvrC, plu2942, plu2935, and rseC were activated. The expression of several genes participating in protein maintenance and repair, i.e., msrB, clpB, and cpxA, was affected by the presence of AI-2.
One-third of the affected genes are involved in intermediary metabolism. Genes related to the biosynthesis or turnover of several amino acids, i.e., methionine (metA, metB, metE, and map), arginine (argB and argD), and glycine (gcvP, which may be involved in the homeostasis of the one-carbon pool needed for methionine biosynthesis), were regulated to increase the pool of these amino acids. By contrast, polyamine biosynthesis genes (speD and speE) were repressed in presence of AI-2. The increased expression of operon plu4566/plu4568, which presumably codes for the synthesis or turnover of a secondary metabolite involving both methionine and arginine, may also be connected to this observation.
Finally, several genes that may be associated with virulence were altered in the luxS-deficient mutant: a bacteriocin operon (plu0884/plu0888), insecticidal toxin genes (tcdA1 and tccC1), and bacteriophage protein operons (e.g., plu1665/plu1666, plu3026/plu3037, and plu3428/plu3429), as well as genes encoding a hemolysin secretion protein and a toxin ABC transporter.
Comparative analysis of expression profiling and two-dimensional protein patterns of wild-type and luxS-deficient strains during mid-exponential growth.
In some bacteria, including P. luminescens, AI-2 starts being produced during exponential growth (7, 12, 65). A second study was therefore performed at the middle of the exponential growth phase, combining the transcriptome and proteome experiments. The proteome approach was chosen as a complementary experiment since during this growth phase we would be able to visualize newly synthesized proteins, in contrast to the case for stationary phase, in which variations are limited due to protein stability.
This second transcriptome experiment was performed on a new set of arrays. During this growth phase, there is less AI-2 binding protein of its ABC transporter in the luxS-deficient strain (data not shown), so we added 19 μM of AI-2 to help identify authentic AI-2-regulated genes. After statistical analysis, we identified 74 genes with an expression level in the luxS-deficient strain different from that in the wild-type strain. Among these, the expression levels of 23 genes were at least partially restored to their wild-type levels after the addition of AI-2 (see Table S1 in the supplemental material). Among the 74 genes found, only six had been previously identified in the study at the end of the exponential growth phase. These were a nitrogen availability regulator (ptsN), luxS, three genes involved in the construction of the cell envelope (mrfA, plu3120, and plu0356), and an unknown gene (plu0361). All displayed a lower differential expression during this growth phase (see Table S1 in the supplemental material).
Most of the AI-2-sensitive genes were repressed in the presence of AI-2. These included (i) several transcriptional regulators, most not functionally characterized; (ii) several genes involved in the cell envelope metabolism; (iii) two genes related to iron transport and storage (fecC and bfr); (iv) two stress-related genes (hns and cspE); and (v) several phage-related and toxin-coding genes. By contrast, mrfA and a gene involved in resistance to oxidative stress (idgB) were induced (Table 1; see Table S1 in the supplemental material).
The proteome modifications generated by the disruption of luxS were explored using two-dimensional electrophoresis. The overall profile of total soluble proteins (see reference maps in reference 72) extracted from the luxS-deficient and wild-type strains displayed significant differences. By using the PhotoList database (http://genolist.pasteur.fr/PhotoList/) (15) and MALDI-TOF mass spectrometry, 41 spots, corresponding to soluble proteins in the range of pH 4 to 7 that varied by a factor of at least two, were identified without ambiguity (Table 2). Of these, 29 were up-regulated and the others were down-regulated, with one, LuxS, being absent as expected. Among these proteins, only four had been predicted to vary according to the transcriptome study. We performed RT-PCR for three unexpected genes (sodA, gor, and tpx) and found no differences in transcript levels between the wild-type and luxS-deficient strains (data not shown). This confirmed the results of the transcriptome experiment and suggested that the expression of these genes is posttranscriptionally regulated. The levels of several proteins involved in protein biosynthesis or processing were modified. These included four elongation factors, several chaperones and tRNA synthetases, and some proteins involved in metabolic pathways (CysK, SucCD, LeuA, and ArgG) (Table 2). Interestingly, many of the proteins known to play an essential role in resistance to oxidative stress had their levels altered in the luxS-deficient strain, including alcohol dehydrogenase (60), AhpC (56), iron transporters (73), Gor (20), SodA (71), and Tpx (28) (Table 2).
TABLE 2.
Proteins with altered level of synthesis in the luxS mutant PL2102
| plu no. | Function | Gene | Theoretical isoelectric point | Theoretical molecular mass (kDa) | Sequence coverage (%) | Induction ratioa (wild type/luxS mutant) |
|---|---|---|---|---|---|---|
| plu0075 | Superoxide dismutase | sodA | 6.52 | 23.5 | 39 | >3,000 |
| plu0158 | Putative l-arginine:lysine amidinotransferase | 5.43 | 42.2 | 38 | >800 | |
| plu0373 | Hemolysin-coregulated protein | 5.91 | 19.0 | 22 | 0.45 | |
| plu0375/plu3556 | Glutathione oxidoreductase/putative aminomethyltransferase | gor/- | 6.24/6.17 | 49.5/36.3 | 35/16 | 3.80 |
| plu0431 | Elongation factor G | fusA | 4.78 | 77.7 | 25 | 0.37 |
| plu0495 | Phosphoribosylaminoimidazolecarboxamide formyltransferase and IMP cyclohydrolase | purH | 6.16 | 57.7 | 48 | 3.75 |
| plu0565 | Threonine synthase | thrC | 4.91 | 47.8 | 32 | 0.50 |
| plu0692 | Prolyl-tRNA synthetase | proS | 5.15 | 63.7 | 42 | 2.25 |
| plu0831 | Beta-lactamase class C | ampC | 6.43 | 42.5 | 21 | 2.04 |
| plu0872 | 3-Methyl-2-oxobutanoate hydroxymethyltransferase | panB | 5.35 | 28.8 | 21 | 4.80 |
| plu0912 | CTP synthetase | pyrG | 6.14 | 60.2 | 27 | 11.60 |
| plu1174 | Iron compound ABC transporter substrate binding protein | 8.63 | 34.2 | 31 | >100 | |
| plu1253b | AI-2 production protein LuxS | luxS | 5.77 | 19.2 | 35 | Absent in luxS |
| plu1395/plu0375 | Cysteine synthase A/glutathione oxidoreductase | cysK/gor | 6.18/6.24 | 34.1/49.5 | 32/19 | >500 |
| plu1432 | Succinyl-CoA synthetase beta chain | sucC | 5.07 | 41.3 | 33 | >400 |
| plu1433 | Succinyl-CoA synthetase alpha chain | sucD | 6.25 | 29.9 | 26 | 3.37 |
| plu1750/plu2853 | Aspartate aminotransferase/solute binding periplasmic protein of iron-siderophore ABC transporter | aspC/- | 6.19/6.54 | 43.6/40.7 | 16/21 | 2.50 |
| plu1840 | Unknown | 4.67 | 40.0 | 25 | 2.04 | |
| plu2059 | Unknown | 4.30 | 54.8 | 20 | 4.21 | |
| plu2579 | Thiol peroxidase | tpx | 4.36 | 17.7 | 57 | 2.00 |
| plu2595 | Pyridoxamine kinase | pdxY | 6.23 | 31.7 | 61 | 2.08 |
| plu2665/plu3598 | Phenylalanyl-tRNA synthetase alpha chain/aminomethyltransferase | pheS/gcvT | 5.73/5.60 | 37.2/40.1 | 53/25 | 3.40/0.61 |
| plu2861 | Elongation factor P-like protein | 4.97 | 21.3 | 21 | 0.49 | |
| plu3147 | AI-2-processing aldolase | lsrF | 6.14 | 31.8 | 40 | 3.70 |
| plu3291 | Serine hydroxymethyltransferase | glyA | 6.32 | 45.3 | 33 | 2.90 |
| plu3621 | Dihydrolipoamide dehydrogenase | lpdA | 6.07 | 50.5 | 29 | 5.55 |
| plu3622 | Dihydrolipoamide acetyltransferase component of pyruvate dehydrogenase complex (E2) | aceF | 4.85 | 56.8 | 21 | 3.73 |
| plu3673 | 2-Isopropylmalate synthase | leuA | 5.52 | 57.5 | 24 | >1,000 |
| plu3739b | Aldehyde dehydrogenase B | aldB | 5.28 | 54.2 | 46 | 0.47 |
| plu3795 | Unknown | 4.72 | 16.5 | 62 | 2.08 | |
| plu3870b | Trigger factor (chaperone) | tig | 4.46 | 48.6 | 29 | 0.33 |
| plu3907 | Alkyl hydroperoxide reductase small subunit (antioxidant) | ahpC | 6.40 | 22.3 | 64 | 2.60 |
| plu4064 | Putative modulator of DNA gyrase | tldD | 5.10 | 51.4 | 42 | 0.50 |
| plu4098 | Leucine-specific binding protein precursor | livK | 6.79 | 39.6 | 35 | 0.30 |
| plu4130 | Elongation factor P | efp | 4.58 | 20.7 | 27 | 5.00 |
| plu4134 | 60-kDa chaperonin | groEL | 4.60 | 57.5 | 23 | 0.50 |
| plu4332 | Alcohol dehydrogenase class III | adhC | 6.23 | 39.3 | 28 | <300 |
| plu4547b | Malate dehydrogenase | mdh | 5.50 | 32.6 | 46 | >500 |
| plu4730 | Elongation factor TufA | tufA | 4.90 | 43.2 | 37 | 3.20 |
| plu4742 | Arginosuccinate synthase | argG | 5.36 | 44.79 | 36 | 2.02 |
| plu4871 | 87% similarity with alpha helix protein YicC of E. coli | 4.78 | 33.6 | 32 | 0.50 |
Induction ratios were calculated from the mean of three values after normalization. In the normalization method used, the raw quantity of each spot in a member gel is divided by the total intensity value of all the pixels in the image.
Gene found in the equivalent transcriptome experiment.
Autoregulation of AI-2 synthesis, coregulation with the polyamines synthesis pathway, and bioluminescence.
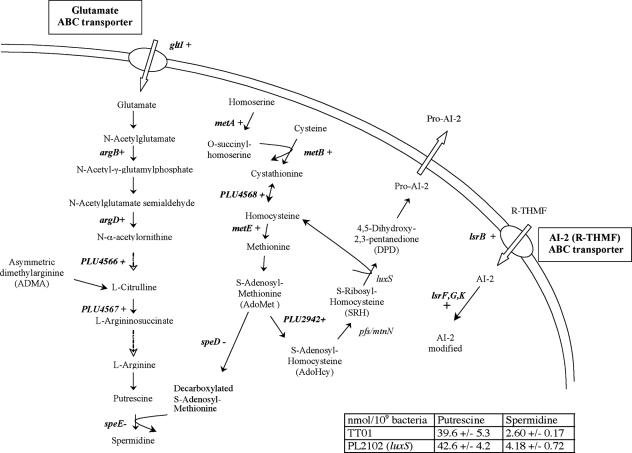
We found that several processes important for survival and possibly virulence depend on the presence of AI-2. Thus, we focused our study on those that consistently altered the regulation of groups of genes involved in a common process (Table 1). The first genes characterized were those that are targets of AI-2 towards the end of the exponential growth phase (Table 1). In addition to the AI-2 ABC transporter gene and the genes involved in its internal processing (lsrBFG and lsrK), many genes linked to AdoMet metabolism, a precursor of AI-2, were also affected: the methionine biosynthesis pathway (metA, metB, and metE), which leads to AdoMet, and an AdoMet-dependent methyltransferase gene (plu2942) were activated. By contrast, the genes of the spermidine synthesis pathway (speDE), which consumes AdoMet, were repressed (Fig. 1 and Table 3). The expression of all these genes was partially or completely restored by adding AI-2 (Tables 1 and 3). However, AdoHcy and homocysteine, the substrate and the secondary product of in vitro AI-2 synthesis, were also present with AI-2 in the medium supplement. To exclude a contribution of these molecules to the observed effect, we performed quantitative RT-PCR experiments on the genes displayed in Fig. 1 with a luxS-deficient strain culture supplemented with AdoHcy and homocysteine (20 μM and 4.25 μM, respectively), which confirmed that their restoration level essentially came from AI-2 alone (Table 3). This suggested that AI-2 induced its own synthesis, transport, and processing by activating AdoMet synthesis while inhibiting the use of AdoMet for spermidine synthesis. Furthermore, by activating glutamate transport and the genes involved in arginine and putrescine biosynthesis (Fig. 1), it could increase the putrescine/spermidine ratio in the polyamine cellular content, suggesting differential metabolism of these two polyamines in this bacterium. To challenge this hypothesis, the intracellular putrescine and spermidine contents were measured. We observed no significant difference for the putrescine content (Fig. 1). By contrast, the intracellular spermidine content increased by more than 60% in the luxS-deficient strain (Fig. 1), confirming the inhibition of spermidine synthesis by a LuxS-dependent process.
FIG. 1.
Polyamine and AI-2 metabolic pathways in Photorhabdus luminescens. +, genes whose expression is induced by AI-2; −, genes whose expression is repressed by AI-2. The dashed arrows represent multiple steps. The table describes the intracellular polyamine contents of the two strains.
TABLE 3.
Quantitative RT-PCR analysis of the genes regulated by AI-2 and involved in AI-2 and polyamine synthesis
| Gene | Expression ratio
|
||
|---|---|---|---|
| TT01/PL2102 | TT01/(PL2102 + AI-2) | TT01/(PL2102 + AdoHcy + homocysteine) | |
| argB | 1.93 | 1.09 | 1.91 |
| argD | 1.96 | 1.13 | 1.95 |
| gltI | 2.70 | 1.80 | 2.53 |
| lsrB | 3.75 | 1.61 | 3.27 |
| lsrK | 2.77 | 1.26 | 2.41 |
| metA | 1.78 | 0.72 | 1.55 |
| metB | 1.84 | 1.35 | 1.85 |
| metE | 22.09 | 15.14 | 25.27 |
| plu2942 | 2.32 | 0.97 | 5.21 |
| plu4566 | 2.40 | 1.87 | 4.95 |
| speD | 0.46 | 1.16 | 0.25 |
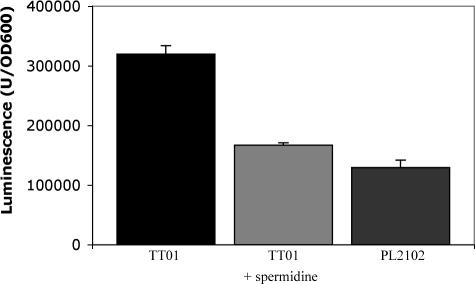
This observation suggested that polyamines were involved in an AI-2-related process. Spermidine is particularly reactive towards some aldehydes, the substrates of the luciferase responsible for bioluminescence (46, 79). These aldehydes are of limited availability in P. luminescens (31, 55). Thus, variations in the spermidine level could influence the bioluminescence intensity under conditions in which we observed no AI-2-dependent regulation of the lux operon, which encodes luciferase, in our expression profiling studies (confirmed with RT-PCR). Therefore, we measured light emission from both the wild-type and luxS-deficient strains. In both strains, luminescence began to be induced during exponential growth and reached a maximum at the end of this phase, when the cell density was between 5 and 8 OD600 units. However, the luminescence in the luxS-deficient strain was 2.5 times lower than that in the wild-type strain (Fig. 2), supporting the association of AI-2 synthesis and light production in P. luminescens. Therefore, we investigated the effect on bioluminescence of adding putrescine or spermidine to the medium. Putrescine had no significant effect on luminescence intensity (data not shown), whereas the addition of 1 mM spermidine to culture medium of the wild-type strain reduced the luminescence intensity to almost that observed in the luxS-deficient strain with no added spermidine (Fig. 2). Therefore, it can be concluded that the level of spermidine participates in the control of the bioluminescence intensity by AI-2.
FIG. 2.
Luminescence of P. luminescens strains. Strains were grown in Schneider medium. Spermidine was used at 1 mM. The values correspond to the maximal specific luminescence. Error bars indicate standard deviations.
The luxS-deficient strain is less resistant to oxidative stress.
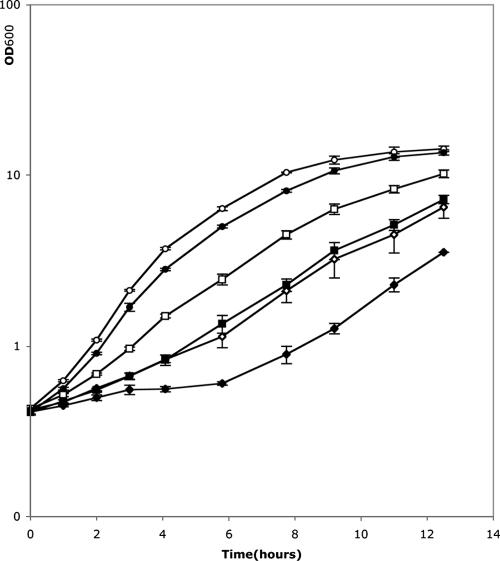
There have been several hypotheses for the function of bioluminescence. The favorite suggests that photon production is tightly coupled to a process that consumes oxygen, thus protecting the cell against the highly damaging oxygen derivatives (66). Interestingly, the reported expression profiling studies showed that genes associated with the oxidative stress response were affected in the luxS-deficient strain. Indeed, the proteome analysis revealed that five proteins involved in oxidative stress resistance were repressed in a luxS-deficient strain (AhpC [56], iron transporters [73], Gor [20], SodA [71], and Tpx [28]), whereas AdhC was induced. In transcriptome analyses, seven genes involved in the same process were also repressed (iscA [49], iscU [69], msrB [25], rseC [34], uvrC [43], idgB [51], and adhE [18]), whereas bfr (8) and speD and speE (70) were activated. As most of the resistance genes were repressed in the absence of luxS, this suggested that the mutant would have a lower resistance to oxidative stress. Therefore, we investigated the sensitivity of both the wild-type and the luxS-deficient (PL2102) strains to oxidative stress to determine the role of AI-2 in oxidative stress resistance. We studied the effect of hydrogen peroxide (H2O2) on bacterial growth in Schneider medium. The growth of both strains was slower when hydrogen peroxide was added to the medium. As expected, the luxS-deficient strain was sensitive to two times lower H2O2 concentration than the wild type (Fig. 3), consistent with how the genes involved in this process are regulated.
FIG. 3.
Growth of P. luminescens strain TT01 (open symbols) and luxS-deficient strain PL2102 (solid symbols) in Schneider medium. Concentrations of hydrogen peroxide were 0 mM (circles), 0.5 mM (squares), and 1 mM (rhombuses). Error bars indicate standard deviations.
AI-2 regulates motility and biofilm processes.
The analysis of differential gene expression in transcriptome experiments, particularly at the end of the exponential growth phase, suggested that AI-2 may regulate the synthesis of the main cell appendages. We found that several genes involved in synthesis of flagella (flgN, fliI, hdfR, and flhC), type IV and type V pili (e.g., the pilPSUV operon), and three fimbriae (plu0417, plu0782/plu0783, and plu1734) were all induced in the luxS-deficient strain, whereas only one gene involved in the synthesis of fimbriae (mrfA) was repressed (Table 1). These three types of appendage are involved in motility and biofilm synthesis, two processes that are regulated by AI-2 in many bacteria (2, 19, 62). Furthermore, the Cpx signaling pathway, which is known to increase adhesion for several bacteria, was repressed in the luxS-deficient strain; indeed, the level of the sensor CpxA decreased, whereas the level of the negative regulator CpxP increased (47). Together, these data suggested that the absence of luxS may increase motility and modify biofilm formation. Therefore, we studied the swimming ability and biofilm formation in both wild-type (TT01) and luxS-deficient (PL2102) strains.
Swimming motility assays were carried out at 30°C on semisolid medium supplemented with 0.2% NaCl. After 20 h, we observed no halo for the wild-type strain, only an 8-mm-diameter colony, whereas we observed a clear 12-mm-diameter halo, characteristic of a motility process, for the luxS-deficient strain. Biofilm formation was assessed after 4 days at 30°C. The biofilms formed on borosilicate glass tubes stained with crystal violet that are shown in Fig. 4 are representative of the results obtained. We observed that there was less biofilm for the luxS-deficient strain (1.23 versus 2.05 OD570 units) (Fig. 4).
FIG. 4.
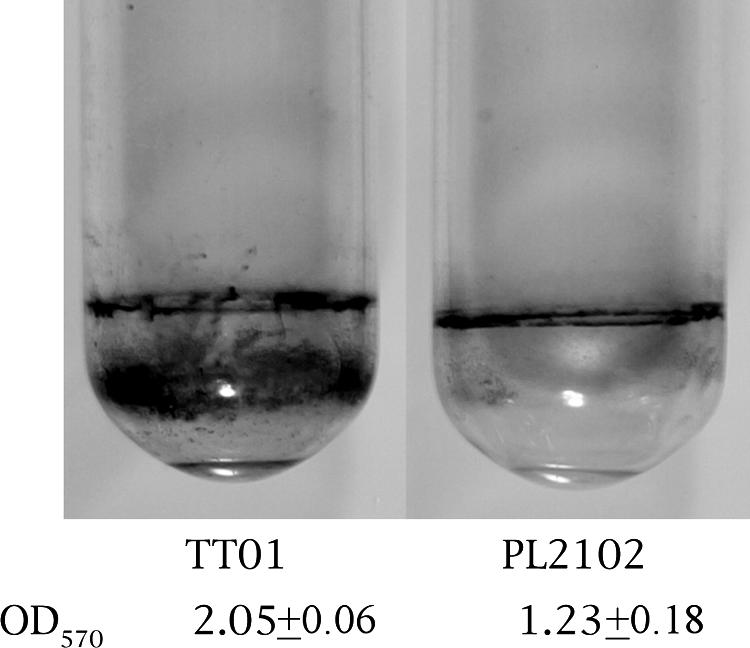
Biofilms made by P. luminescens strains TT01 (wild type) and PL2102 (luxS deficient). Strains were grown in Schneider medium for 4 days at 30°C and stained with crystal violet. The crystal violet staining was expressed as the OD570.
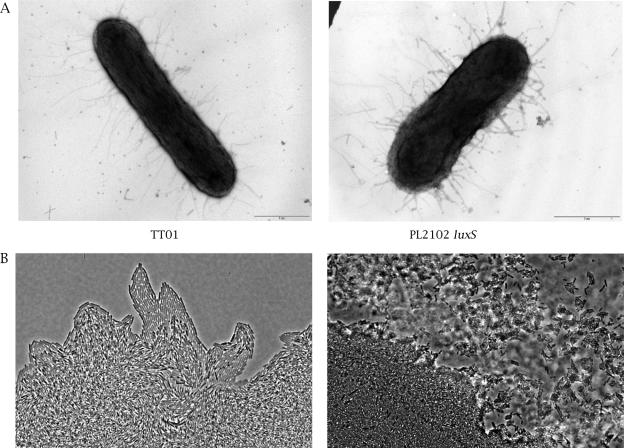
Both results involved the AI-2-regulation of envelope-associated protein synthesis. Therefore, bacteria were stained with uranyl acetate and visualized with an electron microscope to better characterize the structures involved. Remarkably, while we observed flagella in a hyperflagellated mutant (see Materials and Methods), we observed no flagella either in the wild-type or the luxS-deficient strain. This implied that some epistatic process prevented the observed down-regulation of the four genes involved in flagellum synthesis, to restore the flagellum content, in Schneider medium. By contrast, we observed numerous fimbriae/pili (Fig. 5A). This suggested that the observed motility of the luxS-deficient strain probably does not depend on flagella but possibly depends on the presence of pili. In Pseudomonas aeruginosa, type IV and type V pili are involved in a specific motile behavior called twitching motility (50, 59). Therefore, we used a slide culture assay and light microscopy to study twitching motility (Fig. 5B). A significant difference was observed between the two strains. At the leading edge of the twitching zone, the luxS-deficient strain PL2102 showed large rafts moving away from the colony edge and a lattice-like network behind the rafts, characteristic of type IV and type V pilus-dependent twitching motility (50, 59). By contrast, none of these structures was observed with the wild-type strain. Thus, the increased motility observed for the luxS-deficient strain was due to a type IV/type V pilus-dependent twitching motility.
FIG. 5.
Microscopy images. A. Morphology of P. luminescens by electron microscopy. After growth in Schneider medium with 10 μM Na borate, cells were stained with uranyl acetate. B. Light microscopy with phase contrast of expansion zones of twitching motility by slide culture assay, obtained at the interstitial surface between glass cover and medium after 16 h of incubation at 30°C. Magnification, ×40.
Virulence is attenuated in a luxS-deficient strain.
Finally, we found that several genes and pathways involved in virulence in related bacteria were repressed in the luxS-deficient strain: biofilm formation, the cytotoxic protein CcdB, insecticidal toxin (tcdA1), phage-related proteins, and resistance to oxidative stress, the first line of defense of insects. Therefore, we expected the mutant strain (PL2102) to be less virulent. To study this process, both P. luminescens strains were tested for their capacity to kill insects. Wild-type (TT01) and luxS-deficient (PL2102) cells were injected directly into the hemocoel of Spodoptera littoralis larvae and the insect mortality monitored. When 10,000 bacteria were injected into the insects, the time required to kill 50% of the insects (LT50) was delayed for more than 7 hours with the luxS strain (LT50 varying from 35.5 to 46 h, depending on the experiment) compared to the wild-type strain (calculated LT50 of 28 h), but no significant difference was observed for the mortality rate. This showed that the absence of AI-2 in P. luminescens reduced virulence.
DISCUSSION
The type 2 quorum-sensing molecule AI-2 is responsible for a major signaling mechanism in response to population density, i.e., quorum sensing. It has been proposed as an universal signaling molecule involved in interspecies communication (9). AI-2 is synthesized from AdoMet after this has been converted to AdoHcy by methyl transferases. This pathway involves 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase (Pfs/MtnN) and the autoinducer 2 synthesis protein (LuxS) (Fig. 1). Expression profiling studies of a luxS-deficient P. luminescens strain unable to produce AI-2 allowed us to determine more precisely the involvement of AI-2 in this bacterium.
At the end of the exponential growth phase, the transcription of 221 genes was significantly altered in the absence of luxS. The effect of luxS was already visible at an earlier growth stage, with 74 genes being affected during the mid-exponential growth phase. The addition of exogenous AI-2 restored the expression level of 108 of the 221 genes at the end of the exponential growth phase and of 23 of the 74 genes during the mid-exponential growth phase, confirming the involvement of AI-2 in regulation (see Table S1 in the supplemental material). The affected genes were mostly involved in cell envelope-related processes and in intermediary metabolism, although some were involved in information pathways (e.g., transcriptional regulators) and in prophage and toxin synthesis. We found that radA, gyrA, and rsmB were not affected by the addition of AI-2. The induction of these genes in the absence of luxS may reflect their ability to adapt to the accumulation of AdoHcy, which inhibits AdoMet-dependent DNA methyltransferases (52). We compared the proteomes of the luxS-deficient and wild-type strains under condition in which differential protein synthesis could be monitored to correlate transcription with the resulting protein level. Two-dimensional gel electrophoresis revealed that accumulation of 45 proteins was affected during the mid-exponential growth phase in the absence of luxS. These were essentially involved in protein biosynthesis or processing and in resistance to oxidative stress (Table 2). Unexpectedly, the cpm operon, previously shown to be regulated by AI-2 in P. luminescens (12), was not found to be differentially expressed. This was due to these genes being weakly expressed, precluding their differential identification from the background noise.
From the expression profiling studies, we found that the AI-2 control molecule is located high in the hierarchy of regulation, as many transcriptional regulators are regulated. The presence of AI-2 has been detected early in the exponential growth phase (12) and may regulate different targets depending of the state of growth. For example, the differential expression of the six genes found in both assay conditions (mid-exponential and early stationary growth phases) increased at the end of exponential growth, when luxS is more strongly expressed, which is consistent with a dose-dependent AI-2-mediated regulation.
Two signaling pathways mediated by AI-2 via two different AI-2 membrane binding proteins have been recently characterized. In Vibrio harveyi, the receptor LuxP binds (2S,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran-borate (S-THMF-borate), a derivative of a precursor of AI-2 (4,5-dihydroxy-2,3-pentanedione [DPD]), leading to a phosphorylation cascade via LuxP, LuxO, LuxU, and LuxQ (1, 9). In S. enterica serovar Typhimurium, the Lsr ABC transporter, which binds an unborated DPD derivative, (2R,4S)-2-methyl-2,3,3,4-tetrahydroxytetrahydrofuran (R-THMF), a precursor of AI-2, allows the internalization of AI-2 in the cell and its processing via enzymes encoded by lsrFG and lsrK (with a 5 mM borate concentration partially inhibiting the AI-2 response) (41, 67, 68). These pathways appear to be mutually exclusive. As in S. enterica serovar Typhimurium (67, 68), we found in P. luminescens that AI-2 activated four genes (plu3141 and plu3146 to -3148) homologous to and having a conserved synteny with lsrB, which encodes the AI-2 ABC transport binding protein, and with lsrFG and lsrK. This suggests that AI-2 is internalized in the cell by an Lsr ABC transporter, as is found in S. enterica serovar Typhimurium. However, we were not able to decide whether AI-2 is used as a borated or unborated molecule, as 100 μM borate had no effect on the wild-type strain's growth and bioluminescence, a process regulated by AI-2 in P. luminescens (see below), while addition of 1 mM borate led to a reduced bioluminescence, but in parallel with an important decrease of growth (data not shown).
In S. enterica serovar Typhimurium, AI-2 does not seem to regulate its own synthesis, because although metE expression is sensitive to the presence of luxS, its level was not restored upon the addition of AI-2 (68). By contrast, in P. luminescens, we found that AI-2 increased its own synthesis and transport by inducing the availability of its precursor AdoMet, with the methionine biosynthesis pathway (metA, metB, and metE) being activated and the spermidine synthesis pathway (speD and speE) being repressed, as well as inducing its ABC transporter and processing enzymes (lsr operon) (Fig. 1); all these genes were affected by the addition of exogenous AI-2. On an other hand, AI-2 activates glutamate transport (gltI) and arginine biosynthesis (e.g., argB and argD), suggesting that arginine and methionine biosyntheses are coregulated. This may be connected to the activation of the operon plu4568/plu4565, which presumably encodes proteins involved in the synthesis or turnover of a secondary metabolite involving both methionine and arginine.
In Vibrio, AI-2 regulates bioluminescence via the transcription of the luxCDABE operon, which encodes the luciferase that is necessary for light emission (26, 35). In the present work, we showed that the bioluminescence of P. luminescens was activated by AI-2 but not at the transcriptional level of the luxCDABE operon. Long-chain aliphatic aldehydes (hexanal or longer) are the limiting substrates of luciferase in P. luminescens (31, 55). Spermidine, the level of which is increased in the absence of AI-2, has a greater reactivity than other polyamines towards aldehydes and decreases the level of hexanal in vitro (46, 79), making it an excellent candidate regulator. Indeed, we observed that the addition of spermidine decreased the luminescence of the wild-type strain to almost that observed in luxS-deficient strains with no added spermidine. We suggest that AI-2 promotes bioluminescence through an original pathway by reducing the spermidine level limiting the aldehyde availability to luciferase.
It was recently shown that luciferase deficiency in Vibrio harveyi severely impairs growth in the presence of oxidants, suggesting that luminescence plays a physiological role in protecting against oxidative stress (66). However, AI-2 has not been shown to be involved in oxidative stress resistance in this organism. By contrast, luxS-deficient strains of Porphyromonas gingivalis and Streptococcus mutans exhibited increased survival rates in the presence of H2O2 (75, 78). In our expression profiling studies, many proteins involved in the resistance to oxidative stress were affected by AI-2 (e.g., Gor, SodA, IscA, and IscU) (Table 2). A further indication of an AI-2 protective effect can be suggested by the induction of the proteins LpdA and AhpC, which are involved in antioxidant defense against peroxynitrite, a strongly reactive compound generated by the reaction of NO and superoxide (6, 38). Moreover, we found that AI-2 repressed pathways that lead to the production of H2O2 by repressing fumarate reductase and activating the two subunits of succinyl coenzyme A (CoA) synthetase (40) and by increasing the aldehyde availability to luciferase, leading to the consumption of reduced flavin mononucleotide for light and not H2O2 production (31). With an in vivo oxidative stress assay, we showed that wild-type P. luminescens resists the presence of H2O2 better than its luxS-deficient counterpart. AI-2 may further control ROS protection by regulating metabolic pathways that cause an increase in the intracellular pools of glutamate, cysteine, and glycine, the three precursors of glutathione. This may also be related to the induction of luxS and the biosynthetic pathway of the sulfur-containing amino acids (cysteine and methionine) that has been observed under oxidative stress in Bacillus subtilis (43). In conclusion, unlike for P. gingivalis and S. mutans, AI-2 activates the resistance to oxidative stress in P. luminescens.
AI-2 has been shown to regulate flagella, motility, and biofilm production in Streptococcus gordonii, Campylobacter jejuni, and Escherichia coli O157:H7 (2, 19, 62). We also found that AI-2 regulates both motility and the formation of biofilm in P. luminescens. Indeed, a luxS-deficient strain exhibited type IV and type V pili, leading to a twitching motility. To our knowledge, this is the first time that AI-2 has been shown to repress motility, although in a flagellum-independent manner. By contrast, AI-2 activates biofilm formation, possibly via the induction of the Cpx signaling pathway and MrfA fimbriae involved in bacterial adhesion.
This latter observation could be related to alteration of virulence, as it has been recently suggested that MrfA plays an important role in the pathogenic process of Photorhabdus temperata (39). In P. luminescens, the luxS-deficient strain exhibited an attenuated and delayed pathogenicity, consistent with the reduced virulence observed in luxS-deficient strains of Neisseria meningitidis, Streptococcus pneumoniae, and Vibrio vulnificus (32, 64, 77). The reduced capacity of the luxS-deficient P. luminescens to make a biofilm could be involved in the virulence decrease (61). Several other genes regulated by AI-2 could also be involved in the reduced virulence process, as many identified gene products are located in the envelope and numerous transporters, pili, and fimbriae are repressed by AI-2 (see Table S1 in the supplemental material). In P. luminescens, a reduced level of outer membrane proteins is associated with an increase in lipopolysaccharide release into the hemolymph, a process related to hemocyte damage (17). AI-2 also activates AceF and LpdA, two genes necessary for the full virulence of Haemophilus influenzae (27). The decrease in resistance to oxidative stress at mid-exponential phase growth could also contribute to the delayed virulence of the luxS-deficient strain, as ROS have been suggested to mediate an early step that induces the innate immune response upon infection with gram-negative bacteria (21, 45). Finally, many phage genes and genes that code for bacteriocins were up-regulated by AI-2: the former encode or transmit virulence factors in many Enterobacteriaceae, whereas the latter act specifically against bacteria and could be involved in the out-competing of other microorganisms after the insect host death. Finally, only one gene of the four major loci of toxins, Tc, was affected, possibly explaining why the virulence of the luxS-deficient strain was only attenuated.
Supplementary Material
Acknowledgments
We are grateful to Sandrine Auger for performing Ellman's test, to Christelle Jaspart for technical assistance, to Sylvie Pagès for performing insect pathology assays, to Mathieu Brochet for helpful advice on quantitative RT-PCR experiments, and to Emmett Johnson and Philippe Bertin for critical reading of the manuscript.
Financial support came from the Institut Pasteur, the Pasteur-Genopole-Ile-de-France, the Centre National de la Recherche Scientifique (URA 2171), and the French ASG program involving Bayer CropScience, the Institut Pasteur, and INRA and supported by the Ministry of Industry.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bassler, B. L., M. Wright, and M. R. Silverman. 1994. Multiple signalling systems controlling expression of luminescence in Vibrio harveyi: sequence and function of genes encoding a second sensory pathway. Mol. Microbiol. 13:273-286. [DOI] [PubMed] [Google Scholar]
- 2.Blehert, D. S., R. J. Palmer, Jr., J. B. Xavier, J. S. Almeida, and P. E. Kolenbrander. 2003. Autoinducer 2 production by Streptococcus gordonii DL1 and the biofilm phenotype of a luxS mutant are influenced by nutritional conditions. J. Bacteriol. 185:4851-4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boemare, N., A. Givaudan, M. Brehelin, and C. Laumond. 1997. Symbiosis and pathogenicity of nematode-bacterium complexes. Symbiosis 22:21-45. [Google Scholar]
- 4.Bowen, D., T. A. Rocheleau, M. Blackburn, O. Andreev, E. Golubeva, R. Bhartia, and R. H. Ffrench-Constant. 1998. Insecticidal toxins from the bacterium Photorhabdus luminescens. Science 280:2129-2132. [DOI] [PubMed] [Google Scholar]
- 5.Brugirard-Ricaud, K., E. Duchaud, A. Givaudan, P. Girard, F. Kunst, N. Boemare, M. Brehelin, and R. Zumbihl. 2005. Site-specific antiphagocytic function of the Photorhabdus luminescens type III secretion system during insect colonization. Cell. Microbiol. 7:363-371. [DOI] [PubMed] [Google Scholar]
- 6.Bryk, R., C. D. Lima, H. Erdjument-Bromage, P. Tempst, and C. Nathan. 2002. Metabolic enzymes of mycobacteria linked to antioxidant defense by a thioredoxin-like protein. Science 295:1073-1077. [DOI] [PubMed] [Google Scholar]
- 7.Burgess, N. A., D. F. Kirke, P. Williams, K. Winzer, K. R. Hardie, N. L. Meyers, J. Aduse-Opoku, M. A. Curtis, and M. Camara. 2002. LuxS-dependent quorum sensing in Porphyromonas gingivalis modulates protease and haemagglutinin activities but is not essential for virulence. Microbiology 148:763-772. [DOI] [PubMed] [Google Scholar]
- 8.Chen, C. Y., and S. A. Morse. 1999. Neisseria gonorrhoeae bacterioferritin: structural heterogeneity, involvement in iron storage and protection against oxidative stress. Microbiology 145:2967-2975. [DOI] [PubMed] [Google Scholar]
- 9.Chen, X., S. Schauder, N. Potier, A. V. Dorsselaer, I. Pelcser, B. L. Bassler, and F. M. Hughson. 2002. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 415:545-549. [DOI] [PubMed] [Google Scholar]
- 10.Daborn, P. J., N. Waterfield, M. A. Blight, and R. H. Ffrench-Constant. 2001. Measuring virulence factor expression by the pathogenic bacterium Photorhabdus luminescens in culture and during insect infection. J. Bacteriol. 183:5834-5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daborn, P. J., N. Waterfield, C. P. Silva, C. P. Au, S. Sharma, and R. H. Ffrench-Constant. 2002. A single Photorhabdus gene, makes caterpillars floppy (mcf), allows Escherichia coli to persist within and kill insects. Proc. Natl. Acad. Sci. USA 99:10742-10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derzelle, S., E. Duchaud, F. Kunst, A. Danchin, and P. Bertin. 2002. Identification, characterization, and regulation of a cluster of genes involved in carbapenem biosynthesis in Photorhabdus luminescens. Appl. Environ. Microbiol. 68:3780-3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derzelle, S., S. Ngo, E. Turlin, E. Duchaud, A. Namane, F. Kunst, A. Danchin, P. Bertin, and J. F. Charles. 2004. AstR-AstS, a new two-component signal transduction system, controls swarming, adaptation to stationary phase and phenotypic variation in Photorhabdus luminescens. Microbiology 150:897-910. [DOI] [PubMed] [Google Scholar]
- 14.Derzelle, S., E. Turlin, E. Duchaud, S. Pages, F. Kunst, A. Givaudan, and A. Danchin. 2004. The PhoP-PhoQ two-component regulatory system of Photorhabdus luminescens is essential for virulence in insects. J. Bacteriol. 186:1270-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duchaud, E., et al. 2003. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 21:1307-1313. [DOI] [PubMed] [Google Scholar]
- 16.Dudoit, S., J. P. Shaffer, and J. C. Boldrick. 2003. Multiple hypothesis testing in microarray experiments. Stat. Sci. 18:71-103. [Google Scholar]
- 17.Dunphy, G. 1995. Physicochemical properties and surface components of Photorhabdus luminescens influencing bacterial interaction with non-self response systems of nonimmune Galleria mellonella larvae. J. Invertebr. Pathol. 65:25-34. [Google Scholar]
- 18.Echave, P., J. Tamarit, E. Cabiscol, and J. Ros. 2003. Novel antioxidant role of alcohol dehydrogenase E from Escherichia coli. J. Biol. Chem. 278:30193-30198. [DOI] [PubMed] [Google Scholar]
- 19.Elvers, K. T., and S. F. Park. 2002. Quorum sensing in Campylobacter jejuni: detection of a luxS encoded signalling molecule. Microbiology 148:1475-1481. [DOI] [PubMed] [Google Scholar]
- 20.Feige, U., R. Morimoto, I. Yahara, and B. Polla. 1996. Transcriptional regulators for oxidative stress inducible genes in prokaryotes, p. 240-243. In D. B. Catloging (ed.), Stress-inducible cellular responses. Deutsche Bibliothek, Frankfurt, Germany.
- 21.Foley, E., and P. H. O'Farrell. 2003. Nitric oxide contributes to induction of innate immune responses to gram-negative bacteria in Drosophila. Genes Dev. 17:115-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forst, S., and D. Clarke. 2002. Bacteria-nematode symbiosis, p. 57-77. In R. Gaugler (ed.), Entomopathogenic nematology. CAB International, Wallingford, United Kingdom.
- 23.Frangeul, L., P. Glaser, C. Rusniok, C. Buchrieser, E. Duchaud, P. Dehoux, and F. Kunst. 2004. CAAT-box, contigs-assembly and annotation tool-box for genome sequencing projects. Bioinformatics 20:790-797. [DOI] [PubMed] [Google Scholar]
- 24.Gaboriau, F., R. Havouis, J. P. Moulinoux, and J. G. Delcros. 2003. Atmospheric pressure chemical ionization-mass spectrometry method to improve the determination of dansylated polyamines. Anal. Biochem. 318:212-220. [DOI] [PubMed] [Google Scholar]
- 25.Grimaud, R., B. Ezraty, J. Mitchell, D. Lafitte, C. Briand, P. Derrick, and F. Barras. 2001. Repair of oxidized proteins. Identification of a new methionine sulfoxide reductase. J. Biol. Chem. 276:48915-48920. [DOI] [PubMed] [Google Scholar]
- 26.Henke, J. M., and B. L. Bassler. 2004. Three parallel quorum-sensing systems regulate gene expression in Vibrio harveyi. J. Bacteriol. 186:6902-6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herbert, M., A. Kraiss, A. K. Hilpert, S. Schlor, and J. Reidl. 2003. Aerobic growth deficient Haemophilus influenzae mutants are non-virulent: implications on metabolism. Int. J. Med. Microbiol. 293:145-152. [DOI] [PubMed] [Google Scholar]
- 28.Herren, C., E. Rocha, and C. Smith. 2003. Genetic analysis of an important oxidative stress locus in the anaerobe Bacteroides fragilis. Gene 316:167-175. [DOI] [PubMed] [Google Scholar]
- 29.Hommais, F., E. Krin, C. Laurent-Winter, O. Soutourina, A. Malpertuy, J. P. Le Caer, A. Danchin, and P. Bertin. 2001. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 40:20-36. [DOI] [PubMed] [Google Scholar]
- 30.Hommais, F., C. Laurent-Winter, V. Labas, E. Krin, C. Tendeng, O. Soutourina, A. Danchin, and P. Bertin. 2002. Effect of mild acid pH on the functioning of bacterial membranes in Vibrio cholerae. Proteomics 2:571-579. [DOI] [PubMed] [Google Scholar]
- 31.Inouye, S. 1994. NAD(P)H-flavin oxidoreductase from the bioluminescent bacterium, Vibrio fischeri ATCC 7744, is a flavoprotein. FEBS Lett. 347:163-168. [DOI] [PubMed] [Google Scholar]
- 32.Kim, S., S. Lee, Y. Kim, C. Kim, P. Ryu, H. Choy, S. Chung, and J. Rhee. 2003. Regulation of Vibrio vulnificus virulence by the LuxS quorum-sensing system. Mol. Microbiol. 48:1647-1664. [DOI] [PubMed] [Google Scholar]
- 33.Kim, T., and Y. Kim. 2005. Overview of innate immunity in Drosophila. J. Biochem. Mol. Biol. 38:121-127. [DOI] [PubMed] [Google Scholar]
- 34.Koo, M., J. Lee, S. Rah, W. Yeo, J. Lee, K. Lee, Y. Koh, S. Kang, and J. Roe. 2003. A reducing system of the superoxide sensor SoxR in Escherichia coli. EMBO J. 22:2614-2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuo, A., S. Callahan, and P. Dunlap. 1996. Modulation of luminescence operon expression by N-octanoyl-l-homoserine lactone in ainS mutants of Vibrio fischeri. J. Bacteriol. 178:971-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu, D., S. Burton, T. Glancy, Z. S. Li, R. Hampton, T. Meade, and D. J. Merlo. 2003. Insect resistance conferred by 283-kDa Photorhabdus luminescens protein TcdA in Arabidopsis thaliana. Nat. Biotechnol. 21:1222-1228. [DOI] [PubMed] [Google Scholar]
- 37.Livak, K., and T. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Δ Δ C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 38.Master, S. S., B. Springer, P. Sander, E. C. Boettger, V. Deretic, and G. S. Timmins. 2002. Oxidative stress response genes in Mycobacterium tuberculosis: role of ahpC in resistance to peroxynitrite and stage-specific survival in macrophages. Microbiology 148:3139-3144. [DOI] [PubMed] [Google Scholar]
- 39.Meslet-Cladiere, L. M., A. Pimenta, E. Duchaud, I. B. Holland, and M. A. Blight. 2004. In vivo expression of the mannose-resistant fimbriae of Photorhabdus temperata K122 during insect infection. J. Bacteriol. 186:611-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Messner, K. R., and J. A. Imlay. 2002. Mechanism of superoxide and hydrogen peroxide formation by fumarate reductase, succinate dehydrogenase, and aspartate oxidase. J. Biol. Chem. 277:42563-42571. [DOI] [PubMed] [Google Scholar]
- 41.Miller, S. T., K. B. Xavier, S. R. Campagna, M. E. Taga, M. F. Semmelhack, B. L. Bassler, and F. M. Hughson. 2004. Salmonella typhimurium recognizes a chemically distinct form of the bacterial quorum-sensing signal AI-2. Mol. Cell. 15:677-687. [DOI] [PubMed] [Google Scholar]
- 42.Milohanic, E., P. Glaser, J. Y. Coppee, L. Frangeul, Y. Vega, J. Vazquez-Boland, F. Kunst, P. Cossart, and C. Buchrieser. 2003. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 47:1613-1625. [DOI] [PubMed] [Google Scholar]
- 43.Mostertz, J., C. Scharf, M. Hecker, and G. Homuth. 2004. Transcriptome and proteome analysis of Bacillus subtilis gene expression in response to superoxide and peroxide stress. Microbiology 150:497-512. [DOI] [PubMed] [Google Scholar]
- 44.Munier, H., A. M. Gilles, P. Glaser, E. Krin, A. Danchin, R. Sarfati, and O. Barzu. 1991. Isolation and characterization of catalytic and calmodulin-binding domains of Bordetella pertussis adenylate cyclase. Eur. J. Biochem. 14:469-474. [DOI] [PubMed] [Google Scholar]
- 45.Nappi, A. J., E. Vass, F. Frey, and Y. Carton. 1995. Superoxide anion generation in Drosophila during melanotic encapsulation of parasites. Eur. J. Cell Biol. 68:450-456. [PubMed] [Google Scholar]
- 46.Niitsu, M., T. Ohya, X. S. Xu, and K. Samejima. 1995. Identification of N4-(2-propenal)spermidine as a major reaction product of malondialdehyde and spermidine. Biol. Pharm. Bull. 18:1162-1164. [DOI] [PubMed] [Google Scholar]
- 47.Otto, K., and T. Silhavy. 2002. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc. Natl. Acad. Sci. USA 99:2287-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petersohn, A., M. Brigulla, S. Haas, J. D. Hoheisel, U. Volker, and M. Hecker. 2001. Global analysis of the general stress response of Bacillus subtilis. J. Bacteriol. 183:5617-5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pomposiello, P. J., and B. Demple. 2002. Global adjustment of microbial physiology during free radical stress. Adv. Microb. Physiol. 46:319-341. [DOI] [PubMed] [Google Scholar]
- 50.Rashid, M. H., and A. Kornberg. 2000. Inorganic polyphosphate is needed for swimming, swarming, and twitching motilities of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 97:4885-4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Reverchon, S., C. Rouanet, D. Expert, and W. Nasser. 2002. Characterization of indigoidine biosynthetic genes in Erwinia chrysanthemi and role of this blue pigment in pathogenicity. J. Bacteriol. 184:654-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rollins, C. M., and F. W. Dahlquist. 1980. Methylation of chemotaxis-specific proteins in Escherichia coli cells permeable to S-adenosylmethionine. Biochemistry 19:4627-4632. [DOI] [PubMed] [Google Scholar]
- 53.Saux, M. F.-L., V. Viallard, B. Brunel, P. Normand, and N. E. Boemare. 1999. Polyphasic classification of the genus Photorhabdus and proposal of new taxa: P. luminescens subsp. luminescens subsp. nov., P. luminescens subsp. akhurstii subsp. nov., P. luminescens subsp. laumondii subsp. nov., P. temperata sp. nov., P. temperata subsp. temperata subsp. nov. and P. asymbiotica sp. nov. Int. J. Syst. Bacteriol. 49:1645-1656. [DOI] [PubMed] [Google Scholar]
- 54.Schauder, S., K. Shokat, M. G. Surette, and B. L. Bassler. 2001. The LuxS family of bacterial autoinducers: biosynthesis of a novel quorum-sensing signal molecule. Mol. Microbiol. 41:463-476. [DOI] [PubMed] [Google Scholar]
- 55.Schmidt, T. M., K. Kopecky, and K. H. Nealson. 1989. Bioluminescence of the insect pathogen Xenorhabdus luminescens. Appl. Environ. Microbiol. 55:2607-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seaver, L., and J. Imlay. 2001. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J. Bacteriol. 183:7173-7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Seiler, N. 1970. Use of the dansyl reaction in biochemical analysis. Methods Biochem. Anal. 18:259-337. [DOI] [PubMed] [Google Scholar]
- 58.Sekowska, A., V. Denervaud, H. Ashida, K. Michoud, D. Haas, A. Yokota, and A. Danchin. 2004. Bacterial variations on the methionine salvage pathway. BMC Microbiol. 4:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Semmler, A., C. Whitchurch, A. Leech, and J. Mattick. 2000. Identification of a novel gene, fimV, involved in twitching motility in Pseudomonas aeruginosa. Microbiology 146:1321-1332. [DOI] [PubMed] [Google Scholar]
- 60.Shenton, D., and C. Grant. 2003. Protein S-thiolation targets glycolysis and protein synthesis in response to oxidative stress in the yeast Saccharomyces cerevisiae. Biochem. J. 2003:519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Solano, C., B. Sesma, M. Alvarez, T. J. Humphrey, C. J. Thorns, and C. Gamazo. 1998. Discrimination of strains of Salmonella enteritidis with differing levels of virulence by an in vitro glass adherence test. J. Clin. Microbiol. 36:674-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sperandio, V., A. G. Torres, J. A. Giron, and J. B. Kaper. 2001. Quorum sensing is a global regulatory mechanism in enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 183:5187-5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Storey, J. D. 2002. A direct approach to false discovery rates. J. R. Stat. Soc. Ser. B 64:479-498. [Google Scholar]
- 64.Stroeher, U., A. Paton, A. Ogunniyi, and J. Paton. 2003. Mutation of luxS of Streptococcus pneumoniae affects virulence in a mouse model. Infect. Immun. 71:3206-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Surette, M. G., and B. L. Bassler. 1999. Regulation of autoinducer production in Salmonella typhimurium. Mol. Microbiol. 31:585-595. [DOI] [PubMed] [Google Scholar]
- 66.Szpilewska, H., A. Czyz, and G. Wegrzyn. 2003. Experimental evidence for the physiological role of bacterial luciferase in the protection of cells against oxidative stress. Curr. Microbiol. 47:379-382. [DOI] [PubMed] [Google Scholar]
- 67.Taga, M. E., S. T. Miller, and B. L. Bassler. 2003. Lsr-mediated transport and processing of AI-2 in Salmonella typhimurium. Mol. Microbiol. 50:1411-1427. [DOI] [PubMed] [Google Scholar]
- 68.Taga, M. E., J. L. Semmelhack, and B. L. Bassler. 2001. The LuxS-dependent autoinducer AI-2 controls the expression of an ABC transporter that functions in AI-2 uptake in Salmonella typhimurium. Mol. Microbiol. 42:777-793. [DOI] [PubMed] [Google Scholar]
- 69.Thibessard, A., F. Borges, A. Fernandez, B. Gintz, B. Decaris, and N. Leblond-Bourget. 2004. Identification of Streptococcus thermophilus CNRZ368 genes involved in defense against superoxide stress. Appl. Environ. Microbiol. 70:2220-2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tkachenko, A., L. Nesterova, and M. Pshenichnov. 2001. The role of the natural polyamine putrescine in defense against oxidative stress in Escherichia coli. Arch. Microbiol. 176:155-157. [DOI] [PubMed] [Google Scholar]
- 71.Touati, D. 1992. Regulation and protective role of microbial superoxide dismutases, p. 231-261. In J. G. Scandalios (ed.), Molecular biology of free radical scavenging systems. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 72.Turlin, E., G. Pascal, J.-C. Rousselle, P. Lenormand, S. Ngo, A. Danchin, and S. Derzelle. 2006. Proteome analysis of the phenotypic variation process in Photorhabdus luminescens. Proteomics 6:2705-2725. [DOI] [PubMed] [Google Scholar]
- 73.Venisse, J., M. Barny, J. Paulin, and M. N. Brisset. 2003. Involvement of three pathogenicity factors of Erwinia amylovora in the oxidative stress associated with compatible interaction in pear. FEBS Lett. 27:198-202. [DOI] [PubMed] [Google Scholar]
- 74.Waterfield, N., S. Kamita, B. Hammock, and R. Ffrench-Constant. 2005. The Photorhabdus Pir toxins are similar to a developmentally regulated insect protein but show no juvenile hormone esterase activity. FEMS Microbiol. Lett. 245:47-52. [DOI] [PubMed] [Google Scholar]
- 75.Wen, Z., and R. Burne. 2004. LuxS-mediated signaling in Streptococcus mutans is involved in regulation of acid and oxidative stress tolerance and biofilm formation. J. Bacteriol. 186:2682-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Williams, J., M. Thomas, and D. J. Clarke. 2005. The gene stlA encodes a phenylalanine ammonia-lyase that is involved in the production of a stilbene antibiotic in Photorhabdus luminescens TT01. Microbiology 151:2543-2550. [DOI] [PubMed] [Google Scholar]
- 77.Winzer, K., Y. Sun, A. Green, M. Delory, D. Blackley, K. Hardie, T. Baldwin, and C. Tang. 2002. Role of Neisseria meningitidis luxS in cell-to-cell signaling and bacteremic infection. Infect. Immun. 70:2245-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yuan, L., J. Hillman, and A. Progulske-Fox. 2005. Microarray analysis of quorum-sensing-regulated genes in Porphyromonas gingivalis. Infect. Immun. 73:4146-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhou, S., and E. A. Decker. 1999. Ability of amino acids, dipeptides, polyamines, and sulfhydryls to quench hexanal, a saturated aldehydic lipid oxidation product. J. Agric. Food Chem. 47:1932-1936. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.