Abstract
The microbiological characteristics associated with disease-suppressive peats are unclear. We used a bioassay for Pythium sylvaticum-induced damping-off of cress seedlings to identify conducive and suppressive peats. Microbial activity in unconditioned peats was negatively correlated with the counts of P. sylvaticum at the end of the bioassay. Denaturing gradient gel electrophoresis (DGGE) profiling and clone library analyses of small-subunit rRNA gene sequences from two suppressive and two conducive peats differed in the bacterial profiles generated and the diversity of sequence populations. There were also significant differences between bacterial sequence populations from suppressive and conducive peats. The frequencies of a number of microbial groups, including the Rhizobium-Agrobacterium group (specifically sequences similar to those for the genera Ochrobactrum and Zoogloea) and the Acidobacteria, increased specifically in the suppressive peats, although no single bacterial group was associated with disease suppression. Fungal DGGE profiles varied little over the course of the bioassay; however, two bands associated specifically with suppressive samples were detected. Sequences from these bands corresponded to Basidiomycete yeast genera. Although the DGGE profiles were similar, fungal sequence diversity also increased during the bioassay. Sequences highly similar to those of Cryptococcus increased in relative abundance during the bioassay, particularly in the suppressive samples. This study highlights the importance of using complementary approaches to molecular profiling of complex populations and provides the first report that basidiomycetous yeasts may be associated with the suppression of Pythium-induced diseases in peats.
Fungal plant pathogens are a persistent problem worldwide, often resulting in reduced yields and occasionally resulting in major crop damage. There have been increasing restrictions on the use of chemical fungicides, primarily on grounds of environmental effects and toxicity, and the development of disease-suppressive growing media has become a major goal of the horticultural industry (25). Some success in improving medium structure and disease suppression has been obtained with potting mixes based on composted bark and other materials (24, 44), but in Northern Europe most growing media are still peat-based.
Peats are generally considered conducive to soilborne pathogens (26), although some peats are suppressive to Pythium spp. (3, 34, 58, 61). Despite the development of a successful biocontrol product from an isolate of Streptomyces griseoviridis (33) from such a peat, the microbial and physical characteristics of peats that are associated with suppressiveness are not well defined. The level of organic decomposition of the peat, the development of an active microbial biomass, and the microbial composition of such a biomass (4, 11, 12, 25) have all been implicated as important factors in determining suppressiveness.
Predictive models based on microbial activity in media sampled immediately prior to use can predict disease suppression (28). Routine sampling in such a manner, however, is not economically practical in commercial horticulture. A test for suppressiveness applied to the raw components before a growing medium is blended would enable the production of more uniformly suppressive media. Alternatively, if the microbial groups associated with suppression are identified, then a suitable microbial inoculum with reliable disease suppression characters could be added to the peat during conditioning (31). The initial step in this process is to identify microbes particular to disease-suppressive peats to identify potential targets. A few studies based on bacterial culturing and fatty acid and total lipid profiling have been reported (4, 5, 16, 22, 57). Molecular techniques can also be used to characterize these systems and to identify the organisms present without the need to culture them, eliminating the introduction of culture-dependent bias into the comparisons. The few studies of peat with these methods usually have focused on bacterial populations or even specific groups, such as methanotrophs (15).
The objectives of this study were (i) to identify peats that were suppressive and conducive to Pythium sylvaticum-induced damping-off, (ii) to compare physical and chemical characteristics of disease-suppressive and -conducive peats before medium blending, and (iii) to characterize and compare the microbial populations present in selected suppressive and conducive peats before and after sowing of disease indicator plants and challenge with pathogen. We hypothesize that suppressive and conducive peats may contain different microbial populations and that specific microbial groups may be involved in disease suppression. By using molecular profiling techniques on DNA isolated directly from peats and then applying statistical testing directly to sequence data, we may identify differences in microbial populations between disease-suppressive and disease-conducive peats as well as novel suppressive microorganisms which would not be detectable if conventional plating techniques were used.
MATERIALS AND METHODS
Peat samples.
Thirty-nine peat samples from 16 different sites in Northern Europe were supplied by Bulrush Peat, Magherafelt, Ireland (Table 1). The upper particle size of the peat samples was standardized by sieving them through a mesh (10-mm pore size). Oversized particles were air dried before being resieved. Any remaining oversized fractions were milled (Culatti swing-hammer mill with a 2-mm round-hole screen; Glen Creston Ltd., Twickenham, United Kingdom) and sieved. For each sample, all sieved fractions were combined and stored at 5°C for further studies.
TABLE 1.
Sources, physical and microbiological properties, and Pythium sylvaticum damping-off in cress seedlings for 39 peat samples
| Sample (no. of samples)a | Origin | von Post value | Mean pH of raw peat | FDA hydrolysis activity (μg g−1 h−1)b
|
P. sylvaticum CFUb | Mean % damping-offb | |
|---|---|---|---|---|---|---|---|
| Raw peat | End of bioassay | ||||||
| Ai 1-6 (6) | Latvia | H3 | 4.5 | 1.9 ± 0.6 | 9.6 ± 1.4 | (4.9 ± 1.0) × 103 | 37 ± 6.6 |
| (Ai 4c) | 4.3 | 0.4 ± 0.1 | 10 ± 0.1 | (2.9 ± 1.2) × 103 | 59 ± 5.9 | ||
| Li 1-2 (2) | Latvia | H3 | 4.6 | 22 ± 7.6 | 9.2 ± 2.6 | (1.6 ± 0.8) × 103 | 22 ± 2.9 |
| Po 1-5 (5) | Denmark | H3 | 4.7 | 0.9 ± 0.3 | 5.9 ± 0.4 | (6.5 ± 1.2) × 103 | 51 ± 3.9 |
| (Po 1c) | 4.7 | 1.5 ± 0.6 | 6.6 ± 0.1 | (9.8 ± 1.8) × 103 | 66 ± 3.1 | ||
| TLS 1-2 (2) | Latvia | H2 | 4.5 | 40 ± 12 | 13 ± 0.3 | (0.9 ± 0.5) × 103 | 40 ± 12 |
| TLS 3-6 (4) | Latvia | H3 | 4.4 | 15 ± 4.7 | 11 ± 0.8 | (2.4 ± 0.4) × 103 | 22 ± 8.2 |
| TLS 7-9 (3) | Latvia | H4 | 4.3 | 5.8 ± 4.2 | 8.7 ± 1.1 | (3.1 ± 1.6) × 103 | 19 ± 4.5 |
| TLS 10 (1) | Latvia | H5 | 4.4 | 12 ± 1.7 | 7.3 ± 0.1 | (1.4 ± 0.3) × 103 | 22 ± 4.7 |
| Um 1-2 (2) | Denmark | H5 | 5.8 | 33 ± 16 | 3.1 ± 0.7 | (2.5 ± 1.0) × 103 | 11 ± 3.4 |
| Va 1-2 (2) | Latvia | H2 | 4.5 | 11 ± 8.1 | 9.5 ± 0.4 | (2.6 ± 1.0) × 103 | 25 ± 4.6 |
| Va 3-5 (3) | Latvia | H3 | 4.4 | 10 ± 7.4 | 7.4 ± 3.4 | (2.9 ± 1.1) × 103 | 14 ± 3.1 |
| (Va 4c) | 4.4 | 25 ± 0.8 | 12 ± 0.1 | (2.1 ± 1.4) × 103 | 2.7 ± 2.6 | ||
| Va 6 (1) | Latvia | H4 | 4.3 | 0.5 ± 0.1 | 7.4 ± 0.2 | (5.1 ± 3.9) × 103 | 41 ± 0.0 |
| Ve 1-4 (4) | Latvia | H2 | 4.2 | 10 ± 0.3 | 10 ± 3.5 | (1.3 ± 0.5) × 103 | 10 ± 5.9 |
| (Ve 2c) | 4.0 | 8.8 ± 0.2 | 11 ± 0.2 | (1.5 ± 1.3) × 103 | 6.3 ± 0.0 | ||
| Ef 1 (1) | Ireland | H2 | 4.8 | 26 ± 6.4 | 5.2 ± 1.3 | (5.2 ± 2.6) × 103 | 47 ± 11 |
| Ef 2 (1) | Ireland | H4 | 4.7 | 8.7 ± 1.9 | 1.6 ± 0.4 | (4.1 ± 2.8) × 103 | 18 ± 2.7 |
| We 1 (1) | Ireland | H2 | 4.6 | 45 ± 4.8 | 8.4 ± 1.1 | (5.2 ± 3.9) × 103 | 23 ± 1.8 |
| We 2 (1) | Ireland | H4 | 4.8 | 7.4 ± 1.1 | 0.1 ± 0.0 | (7.1 ± 5.4) × 103 | 44 ± 2.9 |
| Lev F2s (1) | Blend | NDd | ND | 4.9 ± 1.3 | (3.8 ± 2.5) × 103 | 47 ± 2.8 | |
Multiple samples from the same site were collected from stockpiles harvested at different times prior to sampling. Lev F2s is a commercial peat-based blended compost from Levingtons (Scotts).
Data are means ± standard errors based on two replicates each of multiple samples from each peat.
Repeated assays for specific peats used in investigations of microbial populations. Values are means ± standard errors based on two replicate samples in two independent bioassays.
ND, not determined.
Physical and chemical analysis of peats.
The pH, water content, bulk density, and mineral content of each peat were determined. For pH measurement, a 1:6 (vol/vol) mixture of peat and water was shaken at 200 rpm for 1 h on an orbital shaker; three pH measurements were taken over a 1-h period, and the average value was used. The water content was determined by weighing 80 ml of peat before and after drying to constant weight at 80°C. Bulk density and mineral content analyses for potassium, phosphorous, nitrate-N, and ammonium-N were performed by using industry standard methods (39). All peat samples were assessed for their degree of humification according to the von Post scale (59), which ranges from light, undecomposed peats, designated H1, to dark, fully degraded peats, designated H10. Measurements of the air-filled porosity (AFP) of each peat were taken immediately before the bioassay by using the protocol of Bragg and Chambers (6).
Microbiological analysis.
Samples of 10 g of peat were resuspended in 100 ml of sterile water and mixed at 200 rpm for 1 h on an orbital shaker. Tenfold serial dilutions were made with sterile water and plated in triplicate on appropriate media. Bacteria were enumerated on 1/10-strength tryptic soy agar (Oxoid Ltd., Basingstoke, United Kingdom), fluorescent Pseudomonas species were enumerated on P1 agar (29), and total fungi were enumerated on 1/4-strength potato dextrose agar (Oxoid) containing 10 mg liter−1 of chlorotetracycline (Sigma-Aldrich Company Ltd., Poole, United Kingdom), 2 ml liter−1 Triton X-100 (Sigma), and additional agar (Oxoid) (final concentration, 1.5% [wt/vol]). All plates were incubated for 7 days at 20°C, and plates with between 30 and 300 colonies per plate were counted.
Bioassay for disease suppression.
Suppressiveness was determined by measuring the occurrence of Pythium damping-off symptoms in cress (Lepidium sativum L.) seedlings in cocultured environment bioassays. Peat samples were first amended with Perlite (William Sinclair Horticulture Ltd., Lincoln, England) to standardize their AFP, and CaCO3 was added to a pH of 5.5 per UK Agricultural Advisory Service guidelines (1, 38). Subsequently, preparations of these mixes containing 1 liter amended peat, 1 g Bulrush Peat wetting agent, and 250 ml water were conditioned at room temperature (18 to 22°C) for 14 days prior to planting.
For pathogen inoculum preparation, 70 g of a sterile mixture of 800 g sand, 80 g medium-ground oatmeal, 250 ml Perlite, and 200 ml sterile water was placed in a sterile glass petri dish and inoculated with four 6-mm discs of an agar culture of Pythium sylvaticum (Campbell and Hendrix). After 7 days at 30°C, the inoculum was mixed thoroughly with sterile dry sand (10% of the wet weight), and sterile water was added (110 ml kg−1).
For each bioassay, 225 g of inoculum was loaded evenly into the bottom of a JFM-25 mushroom punnet (J. F. McKenna, Alderley, United Kingdom). Onto this was placed an array of cells (four by five; each cell was 24 by 24 by 35 mm) cut out from a P180 module tray (Plantpak, Maldon, United Kingdom) and filled with 250 ml of peat. Separate pathogen-inoculated and uninoculated arrays were prepared for each peat. Sterile sand containing 110 ml kg−1 water was used to replace the inoculum in uninoculated treatments, and a commercial standard peat (Levington F2; Scotts UK Ltd., Ipswich, United Kingdom) amended with only Perlite and water was assayed for comparison. Non-chemically treated cress (Lepidium sativum L.) seeds (Elsoms Seeds, Spalding, United Kingdom) were sown individually into the cells of the arrays and covered by sieving of the relevant peat mix over the tray. The arrays were watered lightly before being placed in contact with inoculum or sand. The assay mixtures were incubated for 14 days at 18°C with artificial lighting. Emergence and damping-off were recorded daily. Damping-off was defined as plant collapse through damage to the seedling hypocotyl at the soil surface. Two independent replicate bioassays were carried out sequentially. The emergence (%) and mean percentage of damping-off at the end of each assay were calculated, and colonization of the substrates by P. sylvaticum was determined by dilution plate counts (45). Suppressive peats were nominally defined as those with <20% damping-off, conducive peats had >35% damping-off, and intermediate peats had 20 to 35% damping-off, which is consistent with definitions used in similar studies (4, 14, 21, 27). Initial bioassays were performed with all 39 peats. More detailed studies were done with two conducive peats (Ai4 and Pol) and two suppressive peats (Va4 and Ve2) in two additional replicate bioassays (Table 1). Peats in these assays were sampled at four steps of the bioassay process, as follows: T0, unamended peat; T1, 2 weeks prior to sowing (peat amended with Perlite [Aquaculture Ltd., Sheffield, United Kingdom], CaCO3, and water prior to conditioning); T2, at sowing (peat conditioned for 2 weeks with wetting agent and water); and T3, at 2 weeks postsowing (at the end of the bioassay). Both P. sylvaticum-inoculated and uninoculated treatments were sampled at T3.
FDA analysis.
Microbial activity was assessed by measuring fluorescein diacetate (FDA) hydrolysis through adaptation of the procedure of Chen et al. (12), following the precautions of Inbar et al. (28). Briefly, 30-g peat samples were incubated in 100-ml conical flasks containing 100 ml of 60 mM sodium phosphate buffer (pH 7.6) and 20 μg ml−1 FDA (Merck Biosciences Ltd., Nottingham, United Kingdom) for 12 h at 90 rpm on an orbital shaker at room temperature. Samples without FDA were used as controls. The reaction was stopped by taking 10-ml subsamples and adding 10 ml acetone to each. The subsamples were passed through a glass-fiber syringe filter (1.6-μm nominal pore size; GD/X Whatman Inc., Clifton, New Jersey), and the A492 was measured. FDA hydrolysis was expressed in μg g (dry weight of peat)−1 h−1.
DNA isolation.
For DNA extraction, 0.5 g (wet weight) of each peat sample was homogenized in 1 ml of 120 mM K2HPO4 (pH 8.0) containing 10 mg liter−1 sodium dodecyl sulfate and 300 μl of sterile, acid-washed, 0.1-mm-diameter glass beads (BioSpect Products Inc., Bartlesville, Oklahoma). The homogenate was centrifuged (5,000 × g, 15 min, room temperature), and the supernatant was removed and mixed with 200 μl of 500 mM EDTA (pH 8.0) and 100 μl 5 M potassium acetate (pH 5.5). The mixture was cooled on ice for 15 min before centrifugation as described above. The supernatant was extracted with phenol-chloroform-isoamyl alcohol (25:24:1), and the phases were separated by centrifugation as described above. DNA was precipitated from the aqueous phase by adding a 0.5 volume of isopropanol, followed by incubation on ice for 30 min. DNA was pelleted by centrifugation (13,000 × g) for 30 min at room temperature. The pellet was washed with 70% (vol/vol) ethanol, air dried for 10 to 15 min, and resuspended in 100 μl of sterile water. The DNA was further purified with a Geneclean spin kit (Bio 101 Inc., Nottingham, United Kingdom) according to the manufacturer's instructions. The DNA from each sample was eluted in 50 μl of 10 mM Tris (pH 8.5) and stored at −70°C. Eluted DNA was diluted in water to identify appropriate concentrations for PCR.
PCR amplification.
PCR amplification of variable regions of the bacterial 16S or fungal 18S rRNA gene was performed with primers 341 (forward [5′-CCTACGGGAGGCAGCAG-3′]) (8) and 534 (reverse [5′-ATTACCGCGGCTGCTG-3′]) (8) for bacterial amplifications and EF4 (forward [5′-GGAAGGGRTGTATTTATTAG-3′]) (55) and Fung (reverse [5′-ATTCCCCGTTACCCGTTG-3′]) (36) in a novel combination for fungal amplifications. Both forward primers were modified to include a 40-base 3′ GC clamp (5′-CGCCCGCCGCGCGCGGCGGGCGGGGCGGGGGCACGGGGGG-3′) (40). Briefly, 10 to 50 ng of DNA template, 25 pmol of each primer, a 5 mM concentration of each deoxynucleoside triphosphate, 1.25 units Taq DNA polymerase (Roche Diagnostics GmbH, Penzberg, Germany), 1× reaction buffer, and 1.5 mM MgCl2 (ABgene, Epsom, United Kingdom) were used in a final reaction volume of 100 μl. PCR was performed for 1 cycle of 1 min at 95°C, 30 s at 55°C, and 45 s at 72°C; 38 cycles of 30 s at 95°C, 30 s at 55°C, and 45 s at 72°C; and 1 cycle of 30 s at 95°C, 30 s at 55°C, and 10 min at 72°C. Following amplification, products were cleaned using the Qiaquick PCR purification system (QIAGEN Ltd., Crawley, United Kingdom), eluted in 30 μl of elution buffer (QIAGEN), and visualized in 2% (wt/vol) agarose gels, and the amount of DNA was determined with a NanoDrop ND-1000 UV/Vis spectrophotometer (Labtech International, Ringmer, United Kingdom).
DGGE.
Denaturing gradient gel electrophoresis (DGGE) analysis was performed with a DCode mutation detection system (Bio-Rad, Hemel Hempstead, United Kingdom), using 160- by 160- by 1-mm gels of 8% (wt/vol) acrylamide (37:1 acrylamide:bisacrylamide) as described previously (8, 40). Linear gradients of between 20% and 70% denaturant for bacterial amplifications and between 30% and 60% denaturant for fungal amplifications were formed by using a gradient maker (BDH, Lutterworth, United Kingdom), with 100% denaturant defined as 7 M urea and 40% (vol/vol) formamide. Normally, 300 to 500 ng of PCR product was loaded into each lane of the gel. A 10-min initial migration of the samples from the wells was performed at 150 V, followed by electrophoresis at 60 V for 16.5 h in 0.5× TAE buffer (40 mM Tris, 20 mM acetic acid, 1 mM EDTA [pH 8.0]) at 60°C. Following electrophoresis, gels were stained for 30 min in 0.5× TAE containing 1.2 μg ml−1 of ethidium bromide and then washed with distilled water for 30 min prior to visualization under UV light (302 nm). Images were recorded with a BioDoc-It M-26 UV image capture system (UVP, Upland, California).
The relative positions of bands and their intensities (normalized to account for variations in DNA loading) were scored for three replicate DGGE patterns for each set of samples by using Phoretix 1D Advanced 4.01 image analysis software (Phoretix International Limited, Newcastle upon Tyne, United Kingdom).
Selected bands were excised from gels with a sterile scalpel blade and incubated in 50 μl of sterile water at 70°C for 1 h and then overnight at 4°C. A further round of PCR amplification was conducted as described previously, using 10 μl of the eluted DNA as the template and a non-GC-clamped version of the forward primer.
Construction of clone libraries.
Clone libraries representing bacterial and fungal populations in T0 and T3 samples from pathogen-inoculated bioassays and fungal populations in T3 samples from uninoculated bioassays were constructed as follows. Samples of the PCR products amplified for DGGE and of products reamplified from extracted DGGE bands were cloned into the vector pGEM-T Easy (Promega, Southampton, United Kingdom). The manufacturer's instructions were followed for ligation and the transformation of Escherichia coli DH10B cells (Invitrogen Life Technologies, Paisley, United Kingdom) by electroporation at 200 Ω, 2.5 kV, and 25 μF. An overnight incubation at 37°C on Luria-Bertani (LB) agar (Merck) containing 50 μg ml−1 ampicillin (Sigma) and 40 mg ml−1 X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside; Sigma) was used to select transformants. At least 50 transformants from each PCR used for DGGE and 20 transformants from each reamplified band were selected and cultured overnight at 37°C in 5 ml of LB broth containing 50 μg ml−1 ampicillin. Plasmid DNAs were extracted from the cultures by using the QIAGEN Miniprep spin system (QIAGEN). The presence of appropriate inserts was confirmed by digestion with EcoRI, followed by electrophoretic resolution of the fragments.
Sequence determination.
Standard PCR cycle sequencing was performed on both strands of plasmid DNA containing correctly sized inserts, as follows. Plasmid DNA (30 to 90 ng) was sequenced with 5 pmol T7 or T3 sequencing primer (Invitrogen) in a 10-μl reaction mixture containing 2 μl of ABI PRISM BigDye Terminator cycle sequence ready reaction mix (Perkin-Elmer Applied Biosystems, Warrington, United Kingdom) according to the manufacturer's instructions. Sequencing reactions were performed in a Hybaid PCR multiblock system (Hybaid Ltd., Middlesex, United Kingdom). Sequences were analyzed on an ABI PRISM 3700 DNA cycle sequencer (Perkin-Elmer) and were edited and assembled with the DNASTAR SeqMan II sequence analysis package (Lasergene Inc., Madison, Wisconsin). Sequences were characterized on the basis of >90% identity with 16S or 18S rRNA sequences from Ribosomal Database Project II, using the Sequence Match facility at http://rdp.cme.msu.edu/, or with sequences from the GenBank, EMBL, and DDBJ DNA databases, using the BLAST-n facility at http://www.ncbi.nlm.nih.gov/BLAST/BLAST.cgi.
Statistical analyses.
Correlation coefficients were calculated for pairwise comparisons of all physical, chemical, microbiological (counts), and disease data. An analysis of variance using Duncan's multiple-range tests with a significance limit of 0.05 was used for comparisons of cultured bacterial and fungal counts.
DGGE banding patterns were compared with Sammon's nonlinear mapping (51) and with dendrograms based on distance matrices (41), using the unweighted-pair group method using average linkages (56).
Differences between pairs of clone libraries were determined by calculating homologous and heterologous coverage curves and assessing the difference between the two curves, using the Cramér-von Mises statistic (ΔCXY) according to the method described by Singleton et al. (53). In brief, one library (X) can be compared to another library (Y) and vice versa, resulting in two test statistics (ΔCXY and ΔCYX). The significance of the statistics was determined with a randomization test in which sequences from the two samples were pooled and then randomly split (1,000 times) and the test statistics were recalculated and ranked from largest to smallest. The P value was estimated as the rank for the empirical value of the statistic divided by 1,000; two libraries were considered significantly different if the P value was <0.05. A significant P value for ΔCXY and a nonsignificant P value for ΔCYX indicated that the sequences in library Y were a subsample of the sequences in library X, and vice versa. Significant P values for both comparisons indicated that neither library was a subset of the other. Analyses were performed with the Perl program LIBSHUFF (version 1.2; Department of Microbiology, University of Georgia [http://www.arches.uga.edu/∼whitman/libshuff.html]).
Nucleotide sequence accession numbers.
All sequences have been deposited in the EMBL database under accession numbers DQ530671 to DQ530759 and DQ530761 to DQ531034.
RESULTS
There was no correlation between percentages of damping-off and initial levels of nitrate-N and ammonium-N (data not shown), pH, or the level of decomposition of the raw peat (von Post value) (Table 1). Furthermore, no significant differences were detected in seedling emergence (typically between 80 and 100%) between peat samples during the bioassays. Based on postemergence damping-off data, 15/39 peat samples were classified as conducive to disease, 10 were suppressive, and 14 were intermediate.
Mean bacterial counts from the peat samples (2.6 × 105 ± 1.1 × 105 CFU g dry weight−1) were consistently lower than mean fungal counts (3.7 × 106 ± 1.1 × 106 CFU g dry weight−1) and were correlated with the moisture contents of the peats (r = 0.64; P < 0.001; df, 77), which ranged from 20% to 71% (data not shown), whereas fungal counts were not.
Neither the bacterial and fungal counts nor the moisture content was correlated with the level of damping-off. Disease incidence was positively correlated with the level of P. sylvaticum at the end of the bioassay (r = 0.61; P < 0.05; df, 38), and lower pathogen levels at the end of bioassays correlated with higher recorded FDA hydrolysis levels in the raw peat samples (r = −0.66; P < 0.01; df, 38). No significant correlation was detected between disease levels and FDA hydrolysis, however, although a trend toward reduced disease levels with increasing FDA hydrolysis was noted.
Two conducive peats, Ai4 and Po1, and two suppressive peats, Va4 and Ve2, were selected for further investigation (Table 1). Bacterial counts in these four peats increased significantly (P < 0.05) at each sampling point, whereas fungal counts did not vary significantly with time. No significant differences in combined counts between the two suppressive peats and the two conducive peats were detected for either bacteria or fungi (data not shown).
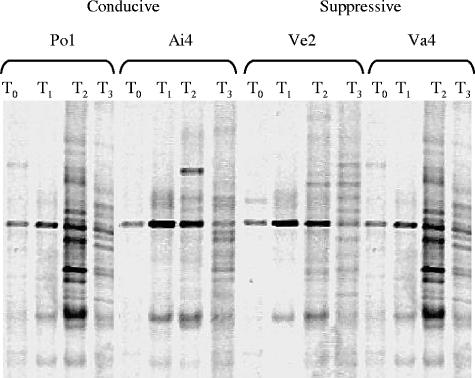
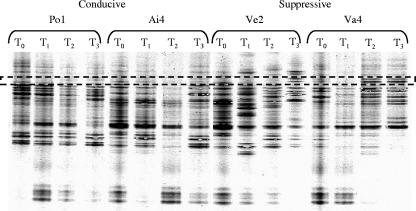
Despite an apparent increase in the number of bands observed in the bacterial DGGE patterns after peat conditioning, e.g., at T2 (Fig. 1), comparisons of both bacterial and fungal DGGE banding patterns showed no clear differences, either between peats at a given time point or within a peat sample over time (data not shown). Two bands observed in the fungal DGGE profiles were present in the T3 samples from both of the suppressive peats (and also in the T2 sample from Va4) but were either absent or very weakly represented in the conducive peats (Fig. 2). Strong bands migrating to similar positions were also present in the DGGE profiles of the Pythium sylvaticum-inoculated suppressive peat samples (data not shown). These bands were excised, cloned, and sequenced. Sequences from most of the clones (15/20 from the upper band and 20/20 from the lower band) corresponded to basidiomycetous yeasts. Sequences from the upper band had >97% sequence identity to 18S rRNA genes from the Ustilaginomycete genera Rhodotorula (10/20) and Tilletiopsis (5/20) (Exobasidiomycetidae). Those from the lower band corresponded to the Hymenomycete genera Cryptococcus (16/20) and Bullera (4/20) (Tremellomycetidae).
FIG. 1.
Negative image of PCR-DGGE banding patterns produced by amplification of the total DNAs of two conducive peats (Po1 and Ai4) and two suppressive peats (Ve2 and Va4), using 16S rRNA gene-specific primers 341 (forward) and 534 (reverse) at time points T0, T1, T2, and T3.
FIG. 2.
Negative image of PCR-DGGE banding patterns produced by amplification of the total DNAs of two conducive peats (Po1 and Ai4) and two suppressive peats (Ve2 and Va4) with 18S rRNA gene-specific primers EF4 (forward) and Fung (reverse) at time points T0, T1, T2, and T3. The hatched box indicates the positions of two bands that are commonly found in the suppressive peats (Va4 at T2 and T3 and Ve2 at T3) but are either rare or absent from the conducive samples. Only DGGE patterns for samples not inoculated with Pythium sylvaticum are shown. Patterns for inoculated samples were also produced and analyzed. Bands migrating to the same positions as those indicated were also present in the inoculated T3 samples of Va4 and Ve2 but were absent from the comparable conducive samples.
Collector's curves at 98% sequence similarity were used to confirm that clone libraries were good representations of the microbial populations, and comparisons of both fungal and bacterial sequence populations from both the T0 and T3 libraries were made with the Cramér-von Mises statistic (Table 2). For both the suppressive and conducive populations, the results show that both the bacterial and fungal sequences at T0 were likely a subset of the sequences at T3, indicating that the diversity of both 16S and 18S rRNA sequence populations in both types of peats increased over the course of the bioassay. Furthermore, at T0, bacterial sequences in the conducive sample were likely a subset of those in the suppressive sample, indicating that the populations in the unamended peats probably contained some sequences in common but that the populations in the suppressive peats were more diverse. By T3, however, neither set of sequences was a subset of the other, indicating that the populations had become distinct. Low levels of sequence diversity in the fungal populations meant that valid comparisons could not be made between suppressive and conducive populations at T0 or T3.
TABLE 2.
Comparisons of Cramér-von Mises statistics between clone library sequences from conducive and suppressive peats in unamended samples (T0) and samples after damping-off bioassay (T3)
| Sequence type | Time point | Test statistica | No. of sequences | P valueb |
|---|---|---|---|---|
| 16S rRNA (bacterial) | T0 | ΔCCS | 45 | 0.77 |
| ΔCSC | 45 | 0.036* | ||
| T3 | ΔCCS | 214 | 0.036* | |
| ΔCSC | 214 | 0.004** | ||
| T0 vs T3 (conducive) | 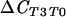 |
136 | 0.001*** | |
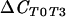 |
136 | 0.98 | ||
| T0 vs T3 (suppressive) | 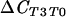 |
130 | 0.001*** | |
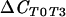 |
130 | 0.93 | ||
| 18S rRNA (fungal) | T0 vs T3 (all sequences) | 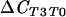 |
110 | 0.001*** |
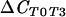 |
110 | 0.22 |
ΔCCS, test comparing suppressive population to conducive population; ΔCSC, reverse comparison of conducive population to suppressive population; 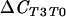 , test comparing population at T0 to that at T3;
, test comparing population at T0 to that at T3; 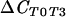 , reverse comparison of population at T3 to that at T0.
, reverse comparison of population at T3 to that at T0.
*, P < 0.05; **, P < 0.01; ***, P < 0.001.
Identification of sequences from the bacterial clone libraries (Table 3) showed that more bacterial groups were present at T3 than at T0, especially in the conducive samples, and that most of the groups identified at T0 were still present at T3. The relative proportions of some of the groups also changed between T0 and T3. The proportions of both gram-positive bacteria and γ-proteobacteria decreased from T0 to T3, with an accompanying increase in the proportion of β-proteobacteria. Representative sequences within some groups also changed between T0 and T3. Dominance in the gram-positive bacteria changed from sequences with high similarity to Acidimicrobium, Frankia, and Clavibacter (Actinobacteria) sequences to those with similarity to sequences from Bacillus and Paenibacillus species. For the γ-proteobacteria, sequences similar to those from Aeromonas and the enteric group were replaced by Pseudomonas-like sequences, although the relative abundances of these sequences did not alter greatly between the two sampling times. In contrast, the β-proteobacteria were dominated by sequences similar to Variovorax (Acidovorax group) sequences at both sampling times.
TABLE 3.
Representation of major bacterial groups identified from clone libraries of conducive and suppressive peat samples prior to conditioning (T0) and at the end of the damping-off bioassay (T3)
| Phylogenetic groupa | Representation (%) within each peat sample
|
|||
|---|---|---|---|---|
| Conducive
|
Suppressive
|
|||
| T0 | T3 | T0 | T3 | |
| Prosthecobacter group | —b | 4 | — | 1 |
| Acidobacterium | — | — | 15 | 1 |
| Bacteroidetes and Cytophaga | — | 31 | 4 | 34 |
| Cytophaga group I | — | 7 | 4 | 9 |
| Sphingobacterium group | — | 16 | — | 17 |
| Cyanobacteria | — | 1 | — | — |
| Gram-positive bacteria | 36 | 13 | 18 | 6 |
| Actinobacteria | 36 | 4 | 18 | — |
| Bacillus/Paenibacillus | — | 5 | — | 5 |
| α-Proteobacteria | — | 14 | 30 | 24 |
| Rhizobium-Agrobacterium group | — | 4 | 15 | 10 |
| Caulobacter group | — | 5 | 15 | 10 |
| Azospirillum group | — | 2 | — | — |
| β-Proteobacteria | — | 24 | 4 | 19 |
| Acidovorax group | — | 16 | 4 | 9 |
| γ-Proteobacteria | 46 | 8 | 25 | 13 |
| Aeromonas | 22 | — | 11 | — |
| Enterics and relatives | 11 | — | 7 | — |
| Pseudomonas and relatives | 9 | 6 | 4 | 6 |
| δ-Proteobacteria | — | 1 | — | 1 |
| Spirochaetes and relatives | — | — | 4 | — |
Bacterial identification was based on ≥90% sequence identity.
—, not detected.
The only bacterial group present exclusively in the suppressive samples at both T0 and T3 was the Acidobacterium group. Other sequences similar to those of the Rhizobium-Agrobacterium and Caulobacter groups (α-proteobacteria) were initially detected in the suppressive peats only but were detected in both samples at T3, although they were better represented in the suppressive samples. The sequences representing both of these groups also changed over time, notably for the Rhizobium-Agrobacterium group, where initial sequences with similarity to Methylobacterium species sequences were replaced by sequences with similarity to those of different species in the suppressive peats (Ochrobactrum and Zoogloea) from those in the conducive peats (Bradyrhizobium).
All of the fungal populations (Table 4) were dominated by Ascomycetes, particularly by sequences corresponding to the Sordariomycete genus Acremonium and by Aspergillus/Penicillium-like sequences, with sequences similar to those of the genera Phialophora and Sclerotium also being well represented. The second most abundant group was the Mesomycetozoa (Protists), which were dominated by sequences with similarity to those of Capsaspora owczarzaki (23) and Anurofeca richardsi (2). The other major group, the Basidiomycetes, was detected in similar proportions in both suppressive and conducive peats at T0. The proportion of these sequences, however, increased in the suppressive population during the bioassay, but not in the conducive population. The majority of sequences had similarity with those from the Ustilaginomycetes, particularly sequences representing the genus Exobasidium, and these were equally well represented in both samples at both time points. The relative proportion of the Hymenomycete subgroup, represented exclusively by sequences with homology to the genus Cryptococcus, was 1.8-fold higher in the suppressive peats than in the conducive ones at T0. This level increased between 1.5-fold (uninoculated) and 2.5-fold (Pythium-inoculated) in the suppressive peats, compared to between 1.3- and 2.0-fold, respectively, in the conducive peats. This corresponded to an overall 2.3-fold greater representation of these sequences in the Pythium-inoculated suppressive samples than in the corresponding conducive samples and a 2.1-fold greater representation in the absence of pathogen inoculation. The Cryptococcus-like sequences detected in the conducive samples were different from those detected in the suppressive samples.
TABLE 4.
Representation of the major eukaryotic groups identified from clone libraries of conducive and suppressive peat samples prior to conditioning (T0) and at the end of the damping-off bioassay from either uninoculated (T3u) or Pythium sylvaticum- inoculated (T3i) samples
| Phylogenetic groupa | Representation (%) within each peat sample
|
|||||
|---|---|---|---|---|---|---|
| Conducive
|
Suppressive
|
|||||
| T0 | T3u | T3i | T0 | T3u | T3i | |
| Ascomycota | 66 | 64 | 66 | 67 | 64 | 62 |
| Sordariomycetes | 41 | 35 | 38 | 40 | 39 | 31 |
| Acremonium | 33 | 32 | 33 | 32 | 32 | 26 |
| Other Ascomycota | 25 | 29 | 28 | 27 | 25 | 31 |
| Aspergillusb | 12 | 12 | 11 | 13 | 10 | 16 |
| Phialophora | 4 | 5 | 7 | 4 | 4 | 5 |
| Sclerotium | 8 | 7 | 4 | 7 | 5 | 5 |
| Basidiomycota | 13 | 14 | 11 | 14 | 18 | 20 |
| Hymenomycetes | 4 | 4 | 7 | 6 | 9 | 15 |
| Cryptococcus | 4 | 4 | 7 | 6 | 9 | 15 |
| Ustilaginomycetes | 9 | 10 | 4 | 8 | 9 | 5 |
| Exobasidium | 7 | 7 | 4 | 7 | 9 | 5 |
| Mesomycetozoa (protists) | 21 | 22 | 23 | 19 | 18 | 18 |
Identification was based on ≥90% sequence identity. Clone libraries from uninoculated T3 samples (T3u) were used to examine the effects of pathogen inoculation on phylogenetic groups potentially associated with suppression.
Sequences in this group also had high homology to Penicillium and related species.
DISCUSSION
The fact that neither the level of decomposition (von Post value) of the unamended peat, the pH, nor directly measured levels of nitrate and ammonium correlated with disease suppression indicates that the availability of these minerals in the unamended peat was not important in subsequent disease suppression. AFP can influence the severity of oomycete-induced root rots (44), and when AFP is <20%, it has a significant impact on the severity of other Pythium-induced diseases (T. R. Pettitt, unpublished data). The AFP levels in the unamended peats varied considerably but were kept at >20% in all peats during conditioning to minimize effects due to this variable.
The mean level of bacterial counts recovered by plating from these samples was lower than that previously reported (16, 17, 35); however, bacterial counts were positively correlated with the moisture contents of the peats, and the low levels may reflect the low moisture contents of some peats. The fact that neither bacterial nor fungal counts were correlated with the level of damping-off suggests that changes in the composition of microbial populations may be more important in disease suppression than gross changes in the number of microorganisms present. Microbial plate counts may represent only a small proportion of the soil microbe population, however, since many bacteria can exist in a viable but nonculturable state (30, 42) and others may not be cultured easily at all (43). Since estimates of microbial metabolic activity do not correlate with bacterial or fungal counts, a significant portion of the active microbial community is presumably not detected by plating. If the portion of the microbial population associated with disease suppression is not easily culturable, then even large changes in their numbers would not be detected by plate counts.
Commercial samples of peats very rarely contain detectable levels of pathogenic Pythium (the main source of the pathogen in commercial situations is contaminated water) (46). Since the initial samples were taken prior to contact with the pathogen inoculum, initial P. sylvaticum concentrations would have been negligible. Differences between levels of the pathogen in the suppressive and conducive samples at the end of the bioassay therefore represent a greater increase of Pythium levels in the conducive peats than in the suppressive peats rather than a reduction of pathogen levels in the suppressive samples. These differences in P. sylvaticum levels were positively correlated with the incidence of damping-off.
Although levels of FDA hydrolysis did not correlate with disease suppression, a trend toward lower pathogen levels at the end of bioassays with increasing FDA activity in the raw peat samples was observed, suggesting a general trend for increased disease control with increasing FDA activity in raw peats. This observation is consistent with a previously reported negative correlation between the severity of the Pythium-induced damping-off of cucumber seedlings and FDA hydrolysis in conditioned peat-based media (12). Measurements of microbial activity in unamended peat samples, using FDA hydrolysis, may therefore have some value as an indicator of potential disease-suppressive capacity.
The incidence of damping-off was used as the main criterion for the selection of four peats for more intensive study. The selected peats were among those rated the most suppressive and the most conducive to disease. We found considerable differences in their 16S DGGE banding patterns (Fig. 1). The most obvious changes occurred between T1 and T2, i.e., during conditioning just prior to sowing, with considerably more complex banding patterns observed in the latter samples than in the former samples. Further evidence for this increase in diversity comes from comparison of the sequence populations from the clone libraries using the Cramér-von Mises statistic. Based on this statistic, both the suppressive and conducive sequence populations at T0 were probably subsets of the sequences at T3, and therefore there were sequences present at T3 that were not detected at T0.
The tests also showed that bacterial sequences in the conducive sample probably were a subset of the suppressive sequences at T0. This result implies that sequences (and therefore bacteria) were present in the suppressive samples that were not present in the conducive peats at T0. The same relationship was not detected at T3, suggesting that the populations had differentiated enough to be considered distinct. This difference may be due to the previously noted general increase in bacterial diversity over time; however, the changes in frequency may mask changes in relative levels of various bacterial subpopulations, some of which may be associated with disease suppression. The identification of sequences from clone libraries was used to provide better insight into the presence of some of these subpopulations in the suppressive and conducive samples.
Sequence analysis showed that, consistent with both the DGGE profiles and the statistical comparison of the sequence data, considerable changes occurred in both the bacterial groups detected and the sequences that represented those groups between the populations at T0 and T3. A number of bacterial groups were potentially associated with disease suppression, based on their presence in the suppressive populations but not in the conducive ones (Table 3). The most promising candidate group was Acidobacterium, which was detected in the suppressive population but not in the conducive population at both T0 and T3. However, there are no recorded antimicrobial activities associated with this group, although it has been associated with escape from root rot disease in pine seedlings (20). Among the other groups potentially associated with suppression, there is no evidence that the organisms from the Caulobacter group exhibit antimicrobial behavior. Sequences from the Rhizobium-Agrobacterium group similar to those of Ochrobactrum and Zoogloea were detected in the final suppressive sample. One isolate of Ochrobactrum can significantly reduce Botrytis cinerea infection on tomato stems in vitro (13), while isolates of Zoogloea are associated with plant growth promotion (32), a potential disease evasion method (60). Methylobacterium species (Rhizobium-Agrobacterium group), which were present in the initial samples, are not known to have direct antimicrobial activity against fungal or oomycete pathogens, but mixed cultures have antagonistic effects on some of the more common bacterial plant pathogens, e.g., Xanthomonas, Pseudomonas, Erwinia, Clavibacter, and Agrobacterium species (50).
The high proportion of sequences with similarity to the mesomycetozoans detected in these samples is intriguing. The life cycles of these organisms are thought to require internalization by a host, with spore release outside the host organism (37). The levels of these sequences noted in the unamended peat samples may indicate a relatively large number of such spores in peats. The fact that the relative proportion of these organisms was maintained over time, however, suggests that they also could proliferate during the experiment. There is evidence that at least one species of Mesomycetozoa may have a life cycle without a mandatory host phase (37), and species with such a life cycle could account for our observations.
In contrast to the temporal increase in bacterial diversity, no such changes were observed in either fungal counts or the fungal DGGE profiles during the bioassay, suggesting that both fungal levels and the composition of the fungal population in the peats remained relatively constant throughout. Based on the comparison of sequences with the Cramér-von Mises statistic, however, fungal diversity increased significantly over the course of the bioassay. If different sequences have sufficiently similar electrophoretic mobilities so as to be unresolvable under a given set of conditions (52, 54), then the DGGE banding patterns will not be fully representative of the complexity of a sequence population and may explain this discrepancy.
Most of the increase in diversity was associated with increased numbers of basidiomycetes. The basidiomycetous yeasts may be of particular significance in disease suppression; three of the four Basidiomycete yeast genera identified from the excised DGGE bands (Cryptococcus, Rhodotorula, and Bullera) are also found on plate cultures from peat bogs or sphagnum wetland areas (47) and appear adaptable to the physiological and nutritional conditions found in peat. Sequences representing Cryptococcus were more abundant in the suppressive peats than in the comparable conducive samples at all times and increased to higher levels in the suppressive peats than in the conducive ones, particularly in the pathogen-inoculated suppressive samples. There are no reports of Cryptococcus species having biocontrol activity against Pythium, but activities against other fungal pathogens, e.g., Botrytis (7, 18) and Rhizoctonia (19), have been reported, as has suppression of postharvest infections of Penicillium (9, 10), Alternaria, and Rhizopus (48, 49).
The poorer representation of Rhodotorula-, Tilletiopsis-, and Bullera-like sequences detected from the cloned DGGE bands than the representation of Cryptococcus-like sequences may explain why, of the four genera, only Cryptococcus was found in the clone libraries. The Cryptococcus-like sequences in the conducive samples were different from those detected in the suppressive samples, which could explain why such sequences were detected in both the conducive and suppressive samples but the DGGE bands were associated only with the suppressive samples. Small sequence differences can result in products with different electrophoretic mobilities and denaturation characteristics and, hence, different band positions. The distinction between the Cryptococcus-like sequences in the suppressive and conducive samples also suggests that a specific subpopulation of these sequences may be associated with disease suppression.
In order to conclusively demonstrate the involvement of Cryptococcus or other basidiomycete yeasts in the suppression of Pythium-induced damping-off, such organisms need to be isolated, identified, and used to successfully suppress damping-off in new peat samples, i.e., satisfy Koch's postulates. Acquisition of such data presents a number of challenges. First, although many basidiomycetes can be cultured readily, there is no guarantee that the suppressive organisms are in this category. Assuming that culture is practical, the different Cryptococcus sequences from the suppressive and conducive peats suggest that at least species-level discrimination will be necessary to distinguish isolates. The short 18S rRNA sequence fragments used to generate the clone libraries will probably be too similar in related species to be useful as molecular probes, even under high-stringency conditions. Full-length 18S rRNA sequencing to identify isolates containing sequences identical to those detected from the suppressive peat samples is likely to be the only practical option. Finally, the candidate isolates must be tested in bioassays on samples of peat known to be conducive to damping-off. Since microbial populations are complex biological systems, they very likely vary in response to the introduction of another organism, which means that the widest possible range of peats would have to be used.
The ability to analyze microbial populations with several complementary techniques and at different levels of sensitivity increases the ability to probe changes in microbial populations. Although a number of microbial groups with the potential to suppress damping-off were detected, only the basidiomycetous yeasts were both well represented in the microbial population and associated with suppressive peats. This study is the first report of the potential involvement of these yeasts, in particular Cryptococcus, in the suppression of Pythium-induced disease. Although further work is required to determine the exact nature of the involvement, the larger numbers of basidiomycete yeast-like sequences in the suppressive samples, and in particular the pathogen-inoculated samples, combined with the high incidence of similar sequences in the clones produced from the excised DGGE bands and a history of antimicrobial activity against other fungal pathogens, all suggest that this group of organisms may suppress the growth of Pythium. The fact that differences between suppressive and conducive samples represented a smaller increase in P. sylvaticum levels in the suppressive peats, rather than an active reduction of pathogen levels from those detected in the conducive samples, also supports this suggestion. This work is a first step towards understanding the microbial populations found in suppressive peats, their interactions, and their potential for development as biocontrol inoculants, with the ultimate aim of providing reliable disease suppression.
Acknowledgments
We thank Defra for funding this work through projects HH1751SX and HH3207HX.
REFERENCES
- 1.Agriculture Development Advisory Service, UK. 1988. pH and conductivity measurements on soils, composts and liquids. Leaflet no. P3152. HMSO, London, United Kingdom.
- 2.Baker, G. C., T. J. C. Beebee, and M. A. Ragan. 1999. Prototheca richardsi, a pathogen of anuran larvae, is related to a clade of protistan parasites near the animal-fungal divergence. Microbiology 145:1777-1784. [DOI] [PubMed] [Google Scholar]
- 3.Boehm, M. J., and H. A. J. Hoitink. 1992. Sustenance of microbial activity in potting mixes and its impact on severity of Pythium root rot of poinsettia. Phytopathology 82:259-264. [Google Scholar]
- 4.Boehm, M. J., L. V. Madden, and H. A. J. Hoitink. 1993. Effect of organic matter decomposition level on bacterial species diversity and composition in relationship to Pythium damping-off severity. Appl. Environ. Microbiol. 59:4171-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borga, P., M. Nilsson, and A. Tunlid. 1994. Bacterial communities in peat in relation to botanical composition as revealed by phospholipid fatty acid analysis. Soil Biol. Biochem. 26:841-848. [Google Scholar]
- 6.Bragg, N. C., and B. J. Chambers. 1988. Interpretation and advisory applications of compost air-filled porosity (AFP) measurements. Acta Hortic. 221:35-44. [Google Scholar]
- 7.Calvo, J., V. Calvente, M. E. de Orellano, D. Benuzzi, and M. I. S. de Tosetti. 2003. Improvement in the biocontrol of postharvest diseases of apples with the use of yeast mixtures. Biocontrol 48:579-593. [Google Scholar]
- 8.Calvo-Bado, L. A., T. R. Pettitt, N. Parsons, G. M. Petch, J. A. W. Morgan, and J. M. Whipps. 2003. Spatial and temporal analysis of the microbial community in slow sand filters used for treating horticultural irrigation water. Appl. Environ. Microbiol. 69:2116-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castoria, R., F. de Curtis, G. Lima, and V. de Cicco. 1997. β-1,3-Glucanase activity of two saprophytic yeasts and possible mode of action as biocontrol agents against postharvest diseases. Postharvest Biol. Technol. 12:293-300. [Google Scholar]
- 10.Chand-Goyal, T., and R. A. Spotts. 1996. Postharvest biocontrol of blue mold of apple and brown rot of sweet cherry, by natural saprophytic yeasts alone or in combination with low doses of fungicides. Biol. Control 6:253-259. [Google Scholar]
- 11.Chen, W., H. A. J. Hoitink, and L. V. Madden. 1988. Microbial activity and biomass in container media for predicting suppressiveness to damping-off caused by Pythium ultimum. Phytopathology 78:1447-1450. [Google Scholar]
- 12.Chen, W., H. A. J. Hoitink, A. F. Schmitthenner, and O. H. Tuovinen. 1988. The role of microbial activity in suppression of damping-off caused by Pythium ultimum. Phytopathology 78:314-322. [Google Scholar]
- 13.Cook, D. W. M., P. G. Long, S. Ganesh, and L. H. Cheah. 1997. Attachment microbes antagonistic against Botrytis cinerea--biological control and scanning electron microscope studies in vivo. Ann. Appl. Biol. 121:503-518. [Google Scholar]
- 14.Cotxarrera, L., M. I. Trillas-Gay, C. Steinberg, and C. Alabouvette. 2002. Use of sludge compost and Trichoderma asperellum isolates to suppress Fusarium wilt of tomato. Soil Biol. Biochem. 34:467-476. [Google Scholar]
- 15.Dedysh, S. N., N. S. Panikov, and J. M. Tiedje. 1998. Acidophilic methanotrophic communities from sphagnum peat bogs. Appl. Environ. Microbiol. 64:922-929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dewey, F. M., Y. L. Wong, R. Seery, T. W. Hollins, and S. J. Gurr. 1999. Bacteria associated with Stagnospora (Septoria) nodorum increase pathogenicity of the fungus. New Phytol. 144:489-497. [DOI] [PubMed] [Google Scholar]
- 17.Dickinson, C. H., and M. J. Dooley. 1967. The microbiology of cut-away peat. I. Descriptive ecology. Plant Soil 27:172-186. [Google Scholar]
- 18.Elad, Y., J. Kohl, and N. J. Fokkema. 1994. Control of infection and sporulation of Botrytis cinerea on bean and tomato by saprophytic yeasts. Phytopathology 84:1193-1200. [Google Scholar]
- 19.El-Tarabily, K. A. 2004. Suppression of Rhizoctonia solani diseases of sugar beet by antagonistic and plant growth-promoting yeasts. J. Appl. Microbiol. 96:69-75. [DOI] [PubMed] [Google Scholar]
- 20.Filion, M., R. C. Hamelin, L. Bernier, and M. St.-Arnaud. 2004. Molecular profiling of rhizosphere microbial communities associated with healthy and diseased black spruce (Picea mariana) seedlings grown in a nursery. Appl. Environ. Microbiol. 70:3541-3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hadar, Y., and R. Mandelbaum. 1986. Suppression of Pythium aphanidermatum damping-off in container media containing composted liquorice roots. Crop Prot. 5:88-92. [Google Scholar]
- 22.Herlihy, M. 1973. Distribution of nitrifying and heterotrophic microorganisms in cutover peats. Soil Biol. Biochem. 5:621-628. [Google Scholar]
- 23.Hertel, L. A., C. J. Bayne, and E. S. Loker. 2002. The symbiont Capsaspora owczarzaki, nov. gen. nov. sp., isolated from three strains of the pulmonate snail Biomphalaria glabrata, is related to members of the Mesomycetozoea. Int. J. Parasitol. 32:1183-1191. [DOI] [PubMed] [Google Scholar]
- 24.Hoitink, H. A. J. 1980. Composted bark, a lightweight growth medium with fungicidal properties. Plant Dis. 64:142-147. [Google Scholar]
- 25.Hoitink, H. A. J., and M. J. Boehm. 1999. Biocontrol within the context of soil microbial communities: a substrate-dependent phenomenon. Annu. Rev. Phytopathol. 37:427-446. [DOI] [PubMed] [Google Scholar]
- 26.Hoitink, H. A. J., and G. A. Kuter. 1986. Effects of composts in growth media on soilborne pathogens, p. 289-306. In Y. Chen and Y. Avnimelech (ed.), The role of organic matter in modern agriculture. Martinus Nijhoff, Dordrecht, The Netherlands.
- 27.Huang, J. W., and E. G. Kuhlman. 1991. Formulation of a soil amendment to control damping-off of slash pine seedlings. Phytopathology 81:163-170. [Google Scholar]
- 28.Inbar, Y., M. J. Boehm, and H. A. J. Hoitink. 1991. Hydrolysis of fluorescein diacetate in sphagnum peat container media for predicting suppressiveness to damping-off caused by Pythium ultimum. Soil Biol. Biochem. 23:479-483. [Google Scholar]
- 29.Katoh, K., and I. Itoh. 1983. New selective media for Pseudomonas strains producing fluorescent pigment. Soil Sci. Plant Nutr. 29:525-532. [Google Scholar]
- 30.Kell, D. B., A. S. Kaprelyants, D. H. Weichart, C. R. Harwood, and M. R. Barer. 1998. Viability and activity in readily culturable bacteria: a review and discussion of the practical issues. Antonie Leeuwenhoek 73:169-187. [DOI] [PubMed] [Google Scholar]
- 31.Krause, M. S., L. V. Madden, and H. A. J. Hoitink. 2001. Effect of potting mix microbial carrying capacity on biological control of Rhizoctonia damping-off of radish and Rhizoctonia crown and root rot of poinsettia. Phytopathology 91:1116-1123. [DOI] [PubMed] [Google Scholar]
- 32.Ladha, J. K., F. J. de Bruijn, and K. A. Malik. 1997. Association of nitrogen-fixing, plant-growth-promoting rhizobacteria (PGPR) with kallar grass and rice. Plant Soil 194:37-44. [Google Scholar]
- 33.Lahdenpera, M. L., E. Simon, and J. Uoti. 1991. Mycostop—a novel biofungicide based on Streptomyces bacteria, p. 258-263. In A. B. R. Beemster, G. J. Bollen, M. Gerlagh, M. A. Ruissen, B. Schippers, and A. Tempel (ed.), Biotic interactions and soil-borne disease. Elsevier, Amsterdam, The Netherlands.
- 34.LeQuéré, D., S. Gagné, and M. Caron. 1990. Ability of sphagnum peat moss to suppress damping-off caused by Pythium ultimum. Can. J. Plant Pathol. 12:335-336. [Google Scholar]
- 35.Martin, N. J., J. Siwasin, and A. J. Holding. 1982. The bacterial population of blanket peat. J. Appl. Bacteriol. 53:35-48. [Google Scholar]
- 36.May, L. A., B. Smiley, and M. G. Schmidt. 2001. Comparative denaturing gradient gel electrophoresis analysis of fungal communities associated with whole plant corn silage. Can. J. Microbiol. 47:829-841. [DOI] [PubMed] [Google Scholar]
- 37.Mendoza, L., J. W. Taylor, and A. Libero. 2002. The class Mesomycetozoea: a heterogeneous group of microorganisms at the animal-fungal boundary. Annu. Rev. Microbiol. 56:315-344. [DOI] [PubMed] [Google Scholar]
- 38.Ministry of Agriculture Fisheries and Food, UK. 1985. Container-grown nursery stock: loamless compost nutrition. Leaflet no. P643. HMSO, London, United Kingdom.
- 39.Ministry of Agriculture Fisheries and Food, UK, and Agriculture Development Advisory Service, UK. 1986. The analysis of agricultural materials. Reference book 427, 3rd ed. HMSO, London, United Kingdom.
- 40.Muyzer, G., E. C. de Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified gene coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nei, M., and W. H. Li. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76:5269-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oliver, J. D. 2005. The viable but nonculturable state in bacteria. J. Microbiol. 43:93-100. [PubMed] [Google Scholar]
- 43.Olsen, R. A., and L. R. Bakken. 1987. Viability of soil bacteria: optimization of plate-counting technique and comparison between total counts and plate counts within different size groups. Microb. Ecol. 13:59-74. [DOI] [PubMed] [Google Scholar]
- 44.Ownley, B. H., and D. M. Benson. 1991. Relationship of matric potential and air-filled porosity of container media to development of Phytophthora root rot of rhododendron. Phytopathology 81:936-941. [Google Scholar]
- 45.Pettitt, T. R., A. J. Wakeham, M. F. Wainwright, and J. G. White. 2002. Comparison of serological, culture, and bait methods for detection of Pythium and Phytophthora zoospores in water. Plant Pathol. 51:720-727. [Google Scholar]
- 46.Pettitt, T. R., L. H. Hiltunen, S. R. Kenny, and J. G. White. May. 2001. Ornamentals: sources of Pythium inoculum, fungicide resistance and efficacy of surface sterilants. Final report of project PC97a. Horticulture Development Council, East Malling, United Kingdom.
- 47.Polyakova, A. V., I. L. Chernov, and N. S. Panikov. 2001. Yeast diversity in hydromorphic soils with reference to a grass-sphagnum wetland in western Siberia and a hummocky tundra region at Cape Barrow (Alaska). Mikrobiologiia 70:617-622. [PubMed] [Google Scholar]
- 48.Qin, G. Z., S. P. Tian, and Y. Xu. 2004. Biocontrol of postharvest diseases on sweet cherries by four antagonistic yeasts in different storage conditions. Postharvest Biol. Biotechnol. 31:51-58. [Google Scholar]
- 49.Qin, G. Z., S. P. Tian, Y. Xu, and Y. K. Wan. 2003. Enhancement of biocontrol efficacy of antagonist yeasts by salicylic acid in sweet cherry fruit. Physiol. Mol. Plant Pathol. 62:147-154. [Google Scholar]
- 50.Romanovskaya, V. A., S. M. Stolyar, Y. R. Malashenko, and T. N. Dodatko. 2001. The ways of plant colonization by Methylobacterium strains and properties of these bacteria. Mikrobiologiia 70:221-227. [PubMed] [Google Scholar]
- 51.Sammon, J. W. 1969. A nonlinear mapping for data structure analysis. IEEE Trans. Comput. C 18:401-409. [Google Scholar]
- 52.Schmalenberger, A., and C. C. Tebbe. 2003. Bacterial diversity in maize rhizospheres: conclusions on the use of genetic profiles based on PCR-amplified partial small subunit rRNA genes in ecological studies. Mol. Ecol. 12:251-262. [DOI] [PubMed] [Google Scholar]
- 53.Singleton, D. R., M. A. Furlong, S. L. Rathburn, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smalla, K., G. Wieland, A. Buchner, A. Zock, J. Parzy, S. Kaiser, N. Roskot, H. Heuer, and G. Berg. 2001. Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl. Environ. Microbiol. 67:4742-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Smit, E., P. Leeflang, B. Glandorf, J. D. van Elsas, and K. Wernars. 1999. Analysis of fungal diversity in the wheat rhizosphere by sequencing of cloned PCR-amplified genes encoding 18S rRNA and temperature gradient gel electrophoresis. Appl. Environ. Microbiol. 65:2614-2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sneath, P. H. A., and R. R. Sokal. 1973. Numerical taxonomy. W. H. Freeman and Company, San Francisco, Calif.
- 57.Sundh, I., M. Nilsson, and P. Borga. 1997. Variation in microbial community structure in two boreal peatlands as determined by analysis of phospholipid fatty acid profiles. Appl. Environ. Microbiol. 63:1476-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tahvonen, R. 1997. Microbes of peat and their use in plant disease control, p. 94-97. In Proceedings of Peat in Horticulture. International Peat Society, Jyväskylä, Finland.
- 59.von Post, L., and E. Granlund. 1926. Södra Sveriges Torvtillgångar. Sveriges Geologiska Undersökningar Public. Ser. C335 Arsbok 19:127. [Google Scholar]
- 60.Whipps, J. M. 2001. Microbial interactions and biocontrol in the rhizosphere. J. Exp. Bot. 52:487-511. [DOI] [PubMed] [Google Scholar]
- 61.Wolffhechel, H. 1988. The suppressiveness of sphagnum peat to Pythium spp. Acta Hortic. 221:217-222. [Google Scholar]