Abstract
The consumption of acetate in soils taken from a nutrient gradient in the northern Florida Everglades was studied by stable isotope probing. Bacterial and archaeal 16S rRNA gene clone libraries from eutrophic and oligotrophic soil microcosms strongly suggest that a significant amount of acetate is consumed by syntrophic acetate oxidation in nutrient-enriched soil.
We are interested in the routes through which acetate is mineralized in soils of the Florida Everglades, particularly Water Conservation Area 2-A (WCA-2A). Runoff from the adjacent agricultural area has resulted in a gradient of phosphorus in WCA-2A surface soils ranging from approximately 1,500 mg/kg in eutrophic regions to 400 mg kg−1 in the interior (3, 29, 38). Concomitant with increasing phosphorus concentrations in WCA-2A is an increase in primary productivity leading to higher numbers of microorganisms responsible for carbon cycling, including methanogens, fermenters, and sulfate-reducing prokaryotes (SRP) (2, 3-5, 7, 13, 35, 38).
A detailed understanding of the pathways leading to methanogenesis and of the mechanisms through which eutrophication impacts these pathways is crucial to developing global models of CH4 flux. In most wetland systems, acetate is considered to be the primary precursor for methane, accounting for approximately 70% of the methane produced globally (11). A few notable exceptions, including the Florida Everglades and northern sphagnum-rich bogs (17, 20, 25), exhibit significantly higher rates of hydrogenotrophic methanogenesis than acetoclastic methanogenesis. Specific pathways for methanogenesis are likely to depend on the specific system; however, syntrophic acetate oxidation has been shown to be more energetically competitive than acetoclastic methanogenesis in some subtropical sediments (27). A previous study suggested the presence of syntrophic acetate oxidizers in Everglades soils (8), so the current study was initiated to further investigate the microbial groups responsible for consumption of acetate as a function of nutrient enrichment in these soils.
Soil samples were collected in December 2004 from eutrophic stations F1 and F4 and oligotrophic station U3, as previously described (3, 8).
Stable isotope microcosms were established as previously described (8), using [13C]acetate (both carbon atoms labeled; Isotec, Miamisburg, OH) at a final concentration of 10 mM. A parallel set of microcosms contained unlabeled acetate. For more thorough labeling of acetate-utilizing assemblages, additional [13C]acetate or unlabeled acetate was added at 4 weeks to a final concentration of 10 mM to appropriate microcosms.
After 7 weeks of incubation, DNA was isolated from soil with an Ultra Clean Soil DNA mega kit (MoBio, Solana Beach, CA) (8). To evaluate possible mixing of [12C]DNA in 13C-labeled bands, 150 to 200 ng of unlabeled Escherichia coli DNA was mixed with experimental DNA prior to ultracentrifugation (8, 33). The purity of labeled DNA was confirmed by screening dense and light bands for added E. coli DNA as previously described (30). E. coli DNA was not detected in the [13C]DNA fractions but was detected in all [12C]DNA fractions (data not shown). Clone libraries were constructed from dense DNA bands and grouped into operational taxonomic units (OTUs) by restriction fragment length polymorphism classification, as described in previous reports (7, 8, 34). Two or three representatives of each OTU were sequenced and subjected to chimera evaluation (10); two putative chimeric sequences were eliminated from further analysis. Sequences were analyzed as described previously (7, 8).
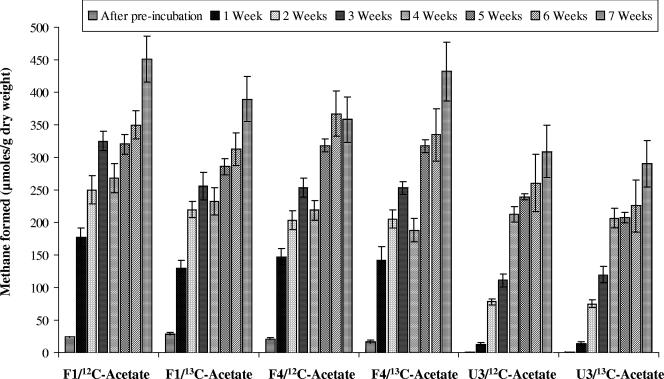
Weekly methanogenesis values obtained from depletion of [12C]acetate and [13C]acetate were determined by gas chromatography as previously reported (3) and are shown in Fig. 1. The average methanogenesis rates for stable isotope probing microcosms were similar for eutrophic soils F1 (46 μmol g−1 week−1) and F4 (46 μmol g−1 week−1), and lower rates were observed in U3 microcosms (25 μmol g−1 week−1) than in F1 or F4 microcosms (P ≤ 0.001) (Fig. 1). Predicted CH4 production from acetate and observed CH4 production from acetate also differed for all three soils. In theory, 1 mol of acetate utilization yields 1 mol of CH4 (31); assuming that all added acetate resulted in formation of CH4, the acetate-containing microcosms should have yielded 400 μmol of CH4. F1 [13C]acetate-containing microcosms formed 78% of the predicted CH4, F4 microcosms formed 89% of the predicted CH4, and U3 microcosms yielded 66% of the predicted CH4. The smaller amounts of CH4 formed in U3 microcosms are consistent with observations made with labeled fatty acids (8) and may indicate that the fate of acetate is more complex in U3 soils than in eutrophic soils.
FIG. 1.
Methanogenesis from [12C]acetate and [13C]acetate. F1, microcosm with soil from eutrophic site; F4, microcosm with soil from eutrophic site; U3, microcosm with soil from oligotrophic site. Analyses were conducted in duplicate, and the bars indicate mean values after conversion to a gram (dry weight)-of-soil basis; the error bars indicate ±1 standard deviation. After week 4, acetate was added to in all microcosms a second time.
For a total of 96 clones screened from each clone library, nine dominant OTUs were observed in F1 libraries, 11 dominant OTUs were observed in F4 libraries, and 12 dominant OTUs were observed in U3 libraries. The dominant sequences in these libraries represented deltaproteobacteria and low-G+C-content gram-positive bacteria (see Fig. S2 and S3 in the supplemental material), but they also included sequences representing alphaproteobacteria, betaproteobacteria, and gammaproteobacteria (Table 1). Distinct differences were observed between the acetate-utilizing microbial guilds in eutrophic and oligotrophic libraries.
TABLE 1.
Relative bacterial and archaeal phylotype abundance in F1, F4, and U3 soil microcosm libraries
| Closest phylogenetic relative from NCBI database | % in:
|
||
|---|---|---|---|
| F1 libraries | F4 libraries | U3 libraries | |
| Bacteriaa | |||
| Syntrophus sp. | 40 | 35 | 0 |
| Propionibacterium sp. | 15 | 0 | 0 |
| SRP (Deltaproteobacteria) | 15 | 15 | 12 |
| Streptococcus sp. | 8 | 0 | 0 |
| Idiomarina sp. | 5 | 0 | 0 |
| Caulobacter sp. | 4 | 5 | 6 |
| Thermoanaerobacter sp. | 4 | 6 | 9 |
| Thiobacillus sp. | 4 | 8 | 8 |
| Acidobacteria | 5 | 4 | 4 |
| Thermus sp. | 0 | 10 | 0 |
| Stenotrophomonas sp. | 0 | 5 | 0 |
| Chlorobi | 0 | 4 | 0 |
| Verrucomicrobia | 0 | 4 | 0 |
| Bacteroidetes sp. | 0 | 4 | 0 |
| Eubacterium sp. | 0 | 0 | 8 |
| Geobacter sp. | 0 | 0 | 20 |
| Clostridium sp. | 0 | 0 | 10 |
| Thermoleophilum sp. | 0 | 0 | 8 |
| Veillonella sp. | 0 | 0 | 6 |
| Chloroflexi | 0 | 0 | 5 |
| Actinobacterium | 0 | 0 | 4 |
| Archaeab | |||
| Methanosaeta sp. | 100 | 100 | 25 |
| Methanosarcina sp. | 0 | 0 | 69 |
| Novel NOB-1 like archaea | 0 | 0 | 3 |
| Crenarchaeota | 0 | 0 | 3 |
A total of 96 clones from each library were compared. Two or more representatives of each phylotype were sequenced, and 16S rRNA gene sequences were compared to their closest culturable phylogenetic relatives from the NCBI database.
A total of 40 clones from each library were compared.
In F1 and F4 clone libraries, 35 to 40% of the sequences clustered as a distinct clade within the Syntrophus group (Table 1; see Fig. S2 in the supplemental material). Syntrophus species are known to syntrophically oxidize a variety of compounds (1, 26, 36) but have not been shown to grow on acetate as a carbon source. Syntrophus spp. have been shown to grow syntrophically on benzoate in coculture with a hydrogen-using sulfate reducer (1, 14, 19) or methanogen (18, 21), and it may be that the Syntrophus-like sequences identified here represent strains that utilize acetate syntrophically. The second dominant group, unique to F1 clone libraries, clustered within the fermentative Propionibacterium (23). Other groups unique to F1 clone libraries included sequences clustering with Streptococcus spp. and Idiomarina spp. (Table 1). The groups unique to F4 clone libraries were relatively minor and clustered with Thermus spp., Verrucomicrobia, Stenotrophomonas spp., Bacteroidetes, and Chlorobi.
Dominant sequences in the U3 library clustered with Geobacter spp. and Clostridium spp. (Table 1), genera known to utilize acetate (6, 9). Clostridium spp. are primary fermenters, although some may participate in syntrophic associations. Clostridium bryantii oxidizes fatty acids or acetate in consortia with hydrogen-scavenging bacteria (32). The dominance of Geobacter spp. and Clostridium spp. in the U3 library indicated that acetate had a different fate in U3 microcosms than in eutrophic microcosms. The sequences common to all three sites included the sequences clustering with Acidobacteria, Thiobacillus spp., Caulobacter spp., and Thermoanaerobacter spp. (Table 1).
Sequences representing SRP formed significant proportions of all libraries (Table 1). The SRP sequences in F1 and F4 libraries clustered with Desulfovibrio spp., and F4 libraries contained sequences related to Desulfobacterium spp. (see Fig. S2 in the supplemental material). U3 microcosms contained sequences clustering with Desulfotomaculum spp. (see Fig. S3 in the supplemental material). These data strongly suggest that the dominant groups of Bacteria that utilize acetate are different in soils along the nutrient gradient. The significant representation of sequences clustering with SRP in all three libraries may have been due more to their potential role in syntrophic partnerships than to their role in sulfate reduction. It is unlikely that sufficient endogenous SO42− from soil was available as an electron acceptor after 10 days of preincubation. The sulfate concentrations at the time of sampling ranged from 0.13 to 0.46 mM in F1 and F4 soils and from 0.05 to 0.2 mM in U3 soils (A. Chauhan, H. F. Castro, K. Sand, K. R. Park, K. R. Reddy, and A. Ogram, Abstr. 105th Gen. Meet. Am. Soc. Microbiol., poster N-150, 2005); this sulfate was further diluted 1:1 (vol/vol) in BCYT-R medium during establishment of the microcosms. It is likely that SRP utilized acetate syntrophically with a methanogenic partner (28) or that Syntrophus (the dominant group in F1 and F4 libraries) syntrophically fermented acetate with consumption of CO2 by Desulfovibrio spp. (21). This differed from the situation in U3 libraries, in which dominance of Geobacter spp. and Clostridium spp. was observed, suggesting that there were syntrophic relationships, as previously reported for members of these genera (12, 32).
A total of 40 clones each of archaeal 16S rRNA gene sequences from F1, F4, and U3 microcosms were screened. Three OTUs were observed in the F1 library, three OTUs were observed in the F4 library, and five OTUs were observed in the U3 library. Two representatives of each OTU were sequenced and phylogenetically characterized (see Fig. S4 in the supplemental material), and the clone libraries from F1 and F4 microcosms included sequences clustering with acetotrophic Methanosaeta spp. (100%). U3 sequences were more diverse, with sequences clustering with Methanosarcina sp. and Methanosaeta spp. (Table 1; see Fig. S4 in the supplemental material). In addition, 3% of U3 sequences clustered with Methanocalculus sp., whose members require acetate for growth but are unable to utilize it as a methanogenic substrate (24). Methanosaeta spp. were the sole acetate-utilizing archaea in the F1 and F4 libraries; Methanosaeta spp. are competitive with Methanosarcina sp. in the presence of low acetate concentrations (7 to 70 μM). In contrast, the U3 library was dominated by Methanosarcina spp. (69%), and there was significantly lower representation of Methanosaeta spp. (25%). Methanosarcina spp. outcompete Methanosaeta spp. at high acetate concentrations ranging from 0.2 to 1.2 mM (22) and may utilize hydrogen as an electron donor (15, 37). The dominance of Methanosarcina spp. in the U3 library suggests that higher concentrations of acetate may accumulate in the U3 soil. In a previous study of soils of the northern Everglades, Drake et al. (13) suggested that acetate production by homoacetogens might be greater in soils similar to U3 soil than in eutrophic soils. In addition, Chauhan et al. (7) previously reported that the numbers of acetotrophic methanogens relative to the numbers of hydrogenotrophic methanogens were much higher in U3 soils than in F1 or F4 soils, which may also suggest that there is greater relative accumulation of acetate in U3 soils than in eutrophic soils.
Microcosms were incubated for 7 weeks, so that DNA from slowly growing syntrophs and methanogens might be labeled sufficiently to be separated from [12C]DNA. This may have been sufficient for labeling some non-acetate-utilizing groups. As with any stable isotope probing study, cross feeding or consumption of dead labeled biomass may have resulted in labeling of nonacetotrophs (16). Despite this limitation, dominant sequences identified here belong to genera that have the potential to utilize acetate in anoxic ecosystems, lending support to our contention that the dominant sequences were labeled as a result of direct consumption of acetate.
A second potential limitation in this study is the lack of incubation of replicated cores taken from each site. The within-site variability of methanogenic consortia at these sites is far less than the between-site variability (5). It is unlikely that additional replicates would have yielded different conclusions, although the precise proportions of groups shown in Table 1 may have varied somewhat.
Syntrophic oxidation of acetate appears to be a common theme among members of the Bacteria in all three soils, with Syntrophus-like sequences dominating libraries from eutrophic soils and Geobacter, Clostridium, and SRP sequences dominating the oligotrophic soil library. The dominance of Methanosarcina sequences in the U3 archaeal library suggests that there is greater accumulation of acetate in U3 soils than in F1 and F4 soils.
Nucleotide sequence accession numbers.
The partial 16S rRNA gene sequences obtained in this study have been deposited in the GenBank database under accession numbers DQ201572 to DQ201615 for bacterial sequences and under accession numbers DQ201616 to DQ201632 for archaeal sequences.
Acknowledgments
This study was supported by grant DEB-0078368 from the National Science Foundation.
We thank Sue Newman for assistance with sample collection.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Auburger, G., and J. Winter. 1995. Isolation and physiological characterization of Syntrophus buswellii strain GA from a syntrophic benzoate degrading, strictly anaerobic coculture. Appl. Microbiol. Biotechnol. 44:241-248. [Google Scholar]
- 2.Bachoon, D., and R. D. Jones. 1992. Potential rates of methanogenesis in sawgrass marshes with peat and marl soils in the Everglades. Soil Biol. Biochem. 24:21-27. [Google Scholar]
- 3.Castro, H., K. R. Reddy, and A. Ogram. 2002. Composition and function of sulfate-reducing prokaryotes in eutrophic and pristine areas of the Florida Everglades. Appl. Environ. Microbiol. 68:6129-6137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castro, H. F., A. Ogram, and K. R. Reddy. 2004. Phylogenetic characterization of methanogenic assemblages in eutrophic and oligotrophic areas of the Florida Everglades. Appl. Environ. Microbiol. 70:6559-6568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castro, H. F., S. Newman, K. R. Reddy, and A. Ogram. 2005. Distribution and stability of sulfate-reducing prokaryotic and hydrogenotrophic methanogenic assemblages in nutrient-impacted regions of the Florida Everglades. Appl. Environ. Microbiol. 71:2695-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cervantes, F. J., S. van der Velde, G. Lettinga, and J. A. Field. 2000. Competition between methanogenesis and quinone respiration for ecologically important substrates in anaerobic consortia. FEMS Microbiol. Ecol. 34:161-171. [DOI] [PubMed] [Google Scholar]
- 7.Chauhan, A., A. Ogram, and K. R. Reddy. 2004. Syntrophic-methanogenic associations along a nutrient gradient in the Florida Everglades. Appl. Environ. Microbiol. 70:3475-3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chauhan A., and A. Ogram. 2006. Fatty acid-oxidizing consortia along a nutrient gradient in the Florida Everglades. Appl. Environ. Microbiol. 72:2400-2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coates, J. D., D. J. Ellis, E. Roden, K. Gaw, E. L. Blunt-Harris, and D. R. Lovley. 1998. Recovery of humics-reducing bacteria from a diversity of sedimentary environments. Appl. Environ. Microbiol. 64:1504-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole, J. R., B. Chai, T. L. Marsh, R. J. Farris, Q. Wang, S. A. Kulam, S. Chandra, D. M. McGarrell, T. M. Schmidt, G. M. Garrity, and J. M. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conrad, R. 1999. Contribution of hydrogen to methane production and control of hydrogen concentrations in methanogenic soils and sediments. FEMS Microbiol. Ecol. 28:193-202. [Google Scholar]
- 12.Cord-Ruwisch, R., D. R. Lovley, and B. Schink. 1998. Growth of Geobacter sulfurreducens with acetate in syntrophic cooperation with hydrogen-oxidizing anaerobic partners. Appl. Environ. Microbiol. 64:2232-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drake, H. L., N. G. Aumen, C. Kuhner, C. Wagner, A. Grießhammer, and M. Schmittroth. 1996. Anaerobic microflora of Everglades sediments: effects of nutrients on population profiles and activities. Appl. Environ. Microbiol. 62:486-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elshahed, M. S., V. K. Bhupathiraju, N. Q. Wofford, M. A. Nanny, and M. J. McInerney. 2001. Metabolism of benzoate, cyclohex-1-ene carboxylate, and cyclohexane carboxylate by “Syntrophus aciditrophicus” strain SB in syntrophic association with H2-using microorganisms. Appl. Environ. Microbiol. 67:1728-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galagan, J. E., C. Nusbaum, A. Roy, M. G. Endrizzi, P. Macdonald, W. FitzHugh, S. Calvo, R. Engels, S. Smirnov, D. Atnoor, A. Brown, N. Allen, J. Naylor, N. Stange-Thomann, K. DeArellano, R. Johnson, L. Linton, P. McEwan, K. McKernan, J. Talamas, A. Tirrell, W. Ye, A. Zimmer, R. D. Barber, I. Cann, D. E. Graham, D. A. Grahame, A. M. Guss, R. Hedderich, C. Ingram-Smith, H. C. Kuettner, J. A. Krzycki, J. A. Leigh, W. Li, J. Liu, B. Mukhopadhyay, J. N. Reeve, K. Smith, T. A. Springer, L. A. Umayam, O. White, R. H. White, E. Conway de Macario, J. G. Ferry, K. F. Jarrell, H. Jing, A. J. Macario, I. Paulsen, M. Pritchett, K. R. Sowers, R. V. Swanson, S. H. Zinder, E. Lander, W. W. Metcalf, and B. Birren. 2002. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12:532-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallagher, E., L. McGuniness, C. Phelps, L. Y. Young, and L. Kerkhof. 2005. 13C-carrier DNA shortens the incubation time needed to detect benzoate-utilizing denitrifying bacteria by stable-isotope probing. Appl. Environ. Microbiol. 71:5192-5196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hines, M., K. Duddleston, and R. Kiene. 2001. Carbon flow to acetate and C1 compounds in northern wetlands. Geophys. Res. Let. 28:4251-4254. [Google Scholar]
- 18.Grabowski, A., D. Blanchet, and C. Jeanthon. 2005. Characterization of long-chain fatty-acid-degrading syntrophic associations from a biodegraded oil reservoir. Res. Microbiol. 156:814-821. [DOI] [PubMed] [Google Scholar]
- 19.Hopkins, B. T., M. J. McInerney, and V. Warikoo. 1995. Evidence for an anaerobic syntrophic benzoate degradation threshold and isolation of the syntrophic benzoate degrader. Appl. Environ. Microbiol. 61:526-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horn, M. A., C. Matthies, K. Kusel, A. Schramm, and H. L. Drake. 2003. Hydrogenotrophic methanogenesis by moderately acid-tolerant methanogens of a methane-emitting acidic peat. Appl. Environ. Microbiol. 69:74-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jackson, B. E., V. K. Bhupathiraju, R. S. Tanner, C. R. Woese, and M. J. McInerney. 1999. Syntrophus aciditrophicus sp. nov., a new anaerobic bacterium that degrades fatty acids and benzoate in syntrophic association with hydrogen-using microorganisms. Arch. Microbiol. 171:107-114. [DOI] [PubMed] [Google Scholar]
- 22.Jetten, M. S. M., A. J. M. Stams, and A. J. B. Zehnder. 1992. Methanogenesis from acetate: a comparison of the acetate metabolism in Methanothrix soehngenii and Methanosarcina spp. FEMS Microbiol. Ecol. 88:181-198. [Google Scholar]
- 23.Koussemon, M., Y. Combet-Blanc, and B. Ollivier. 2003. Glucose fermentation by Propionibacterium microaerophilum: effect of pH on metabolism and bioenergetic. Curr. Microbiol. 46:141-145. [DOI] [PubMed] [Google Scholar]
- 24.Lai M. C., S. C. Chen, C. M. Shu, M. S. Chiou, C. C. Wang, M. J. Chuang, T. Y. Hong, C. C. Liu, L. J. Lai, and J. J. Hua. 2002. Methanocalculus taiwanensis sp. nov., isolated from an estuarine environment. Int. J. Syst. Evol. Microbiol. 52:1799-1806. [DOI] [PubMed] [Google Scholar]
- 25.Landsdown, J. M., P. D. Quay, and S. L. King. 1992. CH4 production via CO2 reduction in a temperate bog: a source of 13C-depleted CH4. Geochim. Cosmochim. Acta 56:3493-3503. [Google Scholar]
- 26.Mountfort, D. O., and M. P. Bryant. 1982. Isolation and characterization of an anaerobic benzoate-degrading bacterium from sewage sludge. Arch. Microbiol. 133:249-256. [Google Scholar]
- 27.Nusslein, B., K.-J. Chin, W. Eckert, and R. Conrad. 2001. Evidence for anaerobic syntrophic acetate oxidation during methane production in the profundal sediment of subtropical Lake Kinneret (Israel). Environ. Microbiol. 3:460-470. [DOI] [PubMed] [Google Scholar]
- 28.Phelps, T. J., R. Conrad, and J. G. Zeikus. 1985. Sulfate-dependent interspecies H2 transfer between Methanosarcina barkeri and Desulfovibrio vulgaris during coculture metabolism of acetate or methanol. Appl. Environ. Microbiol. 50:589-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reddy, K. R., J. R. White, A. Wright, and T. Chua. 1999. Influence of phosphorus loading on microbial processes in the soil and water column of wetlands, p. 249-273. In K. R. Reddy, G. A. O'Connor, and C. L. Schelske (ed.), Phosphorus biogeochemistry in subtropical ecosystems. Lewis Publishers, New York, N.Y.
- 30.Sabat, G., P. Rose, W. J. Hickey, and J. M. Harkin. 2000. Selective and sensitive method for PCR amplification of Escherichia coli 16S rRNA genes in soil. Appl. Environ. Microbiol. 66:844-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schink, B. 1997. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol. Mol. Biol. Rev. 61:262-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schnurer, A., B. Schink, and B. H. Svensson. 1996. Clostridium ultunense sp. nov., a mesophilic bacterium oxidizing acetate in syntrophic association with a hydrogenotrophic methanogenic bacterium. Int. J. Syst. Bacteriol. 46:1145-1152. [DOI] [PubMed] [Google Scholar]
- 33.Singleton, D. R., S. N. Powell, R. Sangaiah, A. Gold, L. M. Ball, and M. D. Aitken. 2005. Stable-isotope probing of bacteria capable of degrading salicylate, naphthalene, or phenanthrene in a bioreactor treating contaminated soil. Appl. Environ. Microbiol. 71:1202-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uz, I., M. E. Rasche, T. Townsend, A. V. Ogram, and A. S. Lindner. 2003. Characterization of methanogenic and methanotrophic assemblages in landfill samples. Proc. R. Soc. Lond. B Biol. Lett. 270(Suppl.):S202-S205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vaithiyanathan, P., and C. J. Richardson. 1997. Nutrient profiles in the Everglades: examination along the eutrophication gradient. Sci. Total Environ. 205:81-95. [DOI] [PubMed] [Google Scholar]
- 36.Wallrabenstein, C., N. Gorny, N. Springer, W. Ludwig, and B. Schink. 1995. Pure culture of Syntrophus buswellii, definition of its phylogenetic status, and description of Syntrophus gentianae sp. nov. Syst. Appl. Microbiol. 18:62-66. [Google Scholar]
- 37.Whitman, W. B., T. L. Bowen, and D. R. Boone. 1992. Methanogenic bacteria, p. 719-767. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K. H. Schleifer (ed.), The prokaryotes. Springer-Verlag, New York, NY.
- 38.Wright, A. L., and K. R. Reddy. 2001. Heterotrophic microbial activities in northern Everglades wetland. Soil Sci. Soc. Am. J. 65:1856-1864. [Google Scholar]