Abstract
Combinatorial biosynthesis was applied to Streptomyces deoxysugar biosynthesis genes in order to reconstitute “unnatural natural gene clusters” for the biosynthesis of four d-deoxysugars (d-olivose, d-oliose, d-digitoxose, and d-boivinose). Expression of these gene clusters in Streptomyces albus 16F4 was used to prove the functionality of the designed clusters through the generation of glycosylated tetracenomycins. Three glycosylated tetracenomycins were generated and characterized, two of which (d-digitoxosyl-tetracenomycin C and d-boivinosyl-tetracenocmycin C) were novel compounds. The constructed gene clusters may be used to increase the capabilities of microorganisms to synthesize new deoxysugars and therefore to produce new glycosylated bioactive compounds.
Microorganisms, mainly actinomycetes, produce a huge variety of glycosylated compounds possessing biological activity, such as antibiotics or antitumors. In the last few years, remarkable efforts have been made to modify the sugar patterns of bioactive compounds by using combinatorial biosynthesis approaches (22, 38, 39). This requires the use of sugar donor flexible glycosyltransferases and the ability to provide microorganisms with the capability to synthesize new sugars. However, the extension of these studies is limited by the small variety of nucleotide diphosphate (NDP) sugars that can be produced by a single strain.
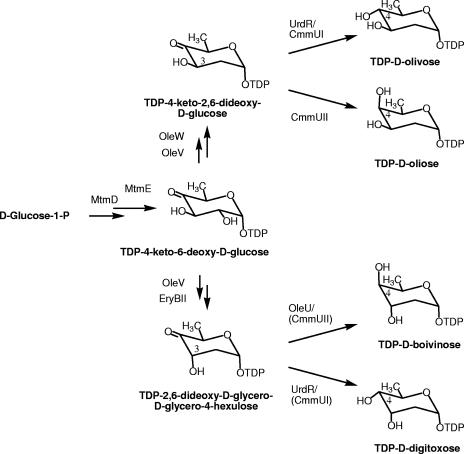
The saccharide moieties of bioactive glycosylated compounds are mainly deoxysugars. This is an important class of carbohydrates formed from common monosaccharides by replacement of one or more hydroxyl groups with hydrogen. Among them, 2,6-dideoxyhexoses (2,6-DOHs) are found ubiquitously in nature, but they are especially abundant in antibiotic or antitumor compounds, in which they play an important role in biological activity, participating in the interaction with the cell targets. Most 2,6-DOHs derive from glucose-1-phosphate, which is first activated to dTDP (TDP)-d-glucose and then undergoes a 4,6 dehydration to give rise to the common biosynthesis intermediate TDP-4-keto-6-deoxy-d-glucose (19, 40). Next, C-2 deoxygenation takes place, involving a dehydration step followed by a reduction reaction. Two different products can arise from this C-2 deoxygenation process, depending on the type of ketoreductase involved. When a TylC1-like ketoreductase is involved, TDP-2,6-dideoxy-d-glycero-d-glycero-4-hexulose is formed (7), whereas if a Gra-Orf26-like protein participates, the product will be TDP-4-keto-2,6-dideoxy-d-glucose (9). These two products differ only in the configuration at the C-3 hydroxyl group, which is axial in the former and equatorial in the latter. By further C-4 ketoreduction or C-3,5 epimerization and C-4 ketoreduction steps of these two biosynthesis intermediates, the different d- and l-2,6-DOHs are generated (14, 29, 32).
Four possible 2,6-d-DOHs exist, differing in the configuration of the hydroxyl groups at C-3 and C-4 (Fig. 1): d-olivose (also known as d-canarose or d-chromose C), d-oliose, d-digitoxose, and d-boivinose. Of these, d-olivose is the most commonly found in bioactive compounds. However, d-oliose, and especially d-digitoxose and d-boivinose, are quite unusual among bioactive compounds produced by microorganisms. Genes involved in the biosynthesis of d-olivose and d-oliose have been identified from different antibiotic producer microorganisms (9, 13, 15, 16, 20, 24, 40, 42, 43). Very recently, a gene cluster for the biosynthesis of d-digitoxose was also identified (5). However, no gene cluster involved in the biosynthesis of d-boivinose has been identified so far. Here, we report the use of combinatorial biosynthesis to reconstitute “unnatural natural gene clusters” for the biosynthesis of 2,6-d-DOH, which can be used to provide Streptomyces species with the capability to synthesize these four 2,6-d-DOHs. These DOH gene clusters were expressed in a bifunctional (Escherichia coli-Streptomyces) multicopy plasmid and were tested for the generation of glycosylated compounds using the sugar flexible glycosyltransferase ElmGT from the elloramycin cluster. Three glycosylated tetracenomycin derivatives were generated, two of which were new.
FIG. 1.
Proposed pathways for the biosynthesis of TDP-2,6-dideoxy-d-hexoses, showing the enzymes catalyzing the indicated biosynthetic steps. In parentheses are enzymes with similar functions that were unable to carry out the corresponding reaction here.
MATERIALS AND METHODS
Microorganisms, culture conditions, and vectors.
Streptomyces griseus subsp. griseus ATCC 13273 (a chromomycin producer) was used as the source of DNA. pFL942 and pFL943 (21) and pLN2 and pLNR (30) were used as sources of sugar genes. Streptomyces lividans 16F4 (6) was used as the host for gene expression. Growth was carried out on trypticase soy broth (Oxoid) or R5A medium (10) for product isolation. For sporulation, agar plates containing A medium (10) were used, and cultures were grown for 7 days at 30°C. Escherichia coli DH10B (Invitrogen) was used as a host for subcloning, and it was grown at 37°C in trypticase soy broth medium. pCRBlunt (Invitrogen) and pUC18 were used as vectors for subcloning experiments and DNA sequencing. pWHM3 (41) was used for expressing genes in Streptomyces. When antibiotic selection of transformants was needed, 50 μg/ml of thiostrepton, 25 μg/ml of apramycin, 50 μg/ml of kanamycin, or 100 μg/ml of ampicillin was used.
DNA manipulation, PCR amplification, and sequencing.
Plasmid DNA preparations, restriction endonuclease digestions, alkaline phosphatase treatments, ligations, and other DNA manipulations were performed according to standard procedures for Streptomyces (17) and for E. coli (33). Specific oligoprimers were used to amplify by PCR cmmUI (NMUI-U, 5′-AAAAACTAGTGTGTCTGAGAGGTGTCATG-3′; NMUI-R, 5′-AAAAGCTAGCTCAGGCGGAGGCCTCGG-3′) and cmmUII (NMUII-U, 5′-AAAAACTAGTGCATAGGAGCCACACCATG-3′; NMUII-R, 5′-AAAAGCTAGCTCAAGCCGGACTCGAAGG-3′). The PCR conditions were as follows: 100 ng of template DNA was mixed with 30 pmol of each primer and 1.25 units of Platinum-Pfx DNA Polymerase (Invitrogene) in a total reaction volume of 50 μl containing 1 mM MgSO4, 0.3 mM of each deoxynucleoside triphosphate, 1× Pfx buffer, and, in some cases, PCRx Enhancer Solution. The polymerization reactions were performed in a thermocycler (PT-100; MJ Research). The general conditions for PCR amplification were as follows: 2 min at 94°C; 30 cycles composed of 15 s at 94°C, 30 s at 55°C, and 1 min at 68°C; 5 min at 68°C; and 15 min at 4°C. The PCR products were purified with GFX PCR DNA and a Gel Band Purification Kit (Amersham Biosciences), subcloned into pCRBlunt, and sequenced. Sequencing was performed using the dideoxynucleotide chain terminator method (34) and the Thermo Sequenase Labeled Primer Cycle Sequencing Kit with 7-deaza-dGTP (Amersham Biosciences). Both DNA strands were sequenced with primers supplied in the kits or with internal oligoprimers (18-mer) using an ALF-express automatic DNA sequencer (Pharmacia). Computer-assisted database searching and sequence analyses were carried out using the University of Wisconsin Genetics Computer Group program package (8) and the BLAST program (4).
Plasmid constructs.
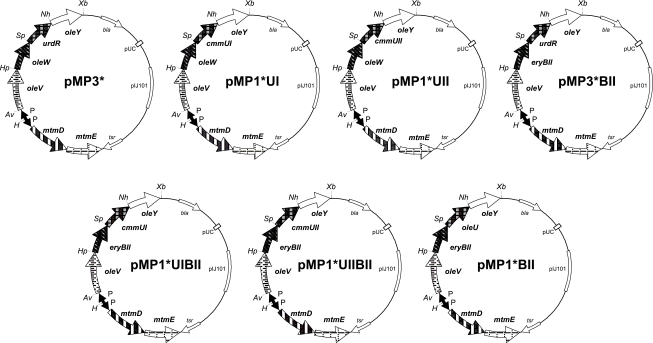
For the construction of pMP3*, first the SphI-XbaI polylinker fragment of pUC18 was used to replace the SphI-XbaI fragment in pLNR (30) containing the oleL, oleS, and oleE genes. Then, from the resultant construct, the 4.2-kb AvrII-XbaI fragment containing the oleV, oleW, urdR, and oleY genes was used to replace the AvrII-XbaI fragment in pFL942 (21), generating pMP3*. In this final construct, oleV, oleW, urdR, and oleY are under the control of ermE*p, and the mtmD and mtmE genes are divergently transcribed from ermEp.
Several other constructs were made using pMP3* as the starting plasmid, as shown in Table 1 and Fig. 2.
TABLE 1.
Plasmid constructs generated in this work
| Plasmid | Genes | Characteristics |
|---|---|---|
| pMP3* | mtmE, mtmD, oleV, oleW, urdR, oleY | Derived from pLNR (30) and pFL942 (21) |
| pMP1*UI | mtmE, mtmD, oleV, oleW, cmmUI, oleY | pMP3* was digested with SpeI and NheI, and the urdR gene was replaced by the cmmUI gene, flanked by the same restriction sites. |
| pMP1*UII | mtmE, mtmD, oleV, oleW, cmmUII, oleY | pMP3* was digested with SpeI and NheI, and the urdR gene was replaced by the cmmUII gene, flanked by the same restriction sites. |
| pMP3*BII | mtmE, mtmD, oleV, eryBII, urdR, oleY | pMP3* was digested with HpaI and SpeI, and the oleW gene was replaced by the eryBII gene, flanked by the same restriction sites. |
| pMP1*UIBII | mtmE, mtmD, oleV, eryBII, cmmUI, oleY | pMP1*UI was digested with HpaI and SpeI, and the oleW gene was replaced by the eryBII gene, flanked by the same restriction sites. |
| pMP1*UIIBII | mtmE, mtmD, oleV, eryBII, cmmUII, oleY | pMP3*BII was digested with SpeI and NheI, and the urdR gene was replaced by the cmmUII gene, flanked by the same restriction sites. |
| pMP1*BII | mtmE, mtmD, oleV, eryBII, oleU, oleY | pMP1*UIIBII was digested with SpeI and NheI, and the cmmUII gene was replaced by the oleU gene, flanked by the same restriction sites. |
| pMP1*BIIΔU | mtmE, mtmD, oleV, eryBII, oleY | pMP1*BII was digested with SpeI and NheI and religated to delete the oleU gene. |
FIG. 2.
Genetic organizations of plasmids for the biosynthesis of TDP-2,6-dideoxy-d-hexoses: TDP-d-olivose (pMP3* and pMP1*UI), TDP-d-oliose (pMP1*UII), TDP-d-digitoxose (pMP3*BII and pMP1*UIBII), and TDP-d-boivinose (pMP1*UIIBII and pMP1*BII). Abbreviations: P, erythromycin resistance promoter; bla, β-lactamase gene; tsr, thiostrepton resistance gene; Av, AvrII; H, HindIII; Hp, HpaI; Nh, NheI; Sp, SpeI; Xb, XbaI.
Production conditions and chromatographic techniques.
Spores of S. lividans 16F4 containing the different constructs were grown in R5A medium under conditions previously described (30). High-performance liquid chromatography (HPLC) analyses were performed as previously described (30). Pure compounds were used as standards for d-olivosyl-tetracenomycin C and 8-demethyl-tetracenomycin C comparisons. They were purified from cultures of S. lividans strains 16F4 and 16F4(pLNR) (6, 30).
For the isolation of new glycosylated tetracenomycins, S. lividans strains 16F4(pMP3*BII) and 16F4(pMP1*BII) were cultured for 7 days in 2-liter Erlenmeyer flasks with R5A medium, as previously described (27). In each case, 3.2 liters of culture was centrifuged, filtered, and extracted (10). The compounds were purified by preparative HPLC, using a μBondapak C18 radial compression cartridge (PrepPak Cartridge; 25 by 100 mm; Waters). An isocratic elution with a mixture of acetonitrile and 0.1% trifluoroacetic acid in water (35:65) was used in both cases. The peaks corresponding to the desired compounds were collected, diluted fourfold with water, applied to a reverse-phase extraction cartridge, washed with water to eliminate trifluoroacetic acid, and finally eluted with methanol. After lyophilization, 7.9 mg of d-digitoxosyl-tetracenomycin C (DDIG-TCMC) and 25.7 mg of d-boivinosyl-tetracenomycin C (DBOV-TCMC) were obtained. HPLC-mass spectrometry (MS) analysis was carried out as previously described (21).
NMR analysis.
The nuclear magnetic resonance (NMR) data (Tables 2 and 3) were recorded on a Varian Mercury 300 NMR spectrometer at a magnetic field strength of 7.05 T.
TABLE 2.
1H and 13C NMR data for DDIG-TCMC in pyridine-d5 at 300 MHz
| Carbon | 1H NMR δ (J in Hz) | 13C NMR (δ) | Multiplicity |
|---|---|---|---|
| 1 | 192.72 | C | |
| 2 | 6.01 br s | 101.37 | CH |
| 3 | 175.54 | C | |
| 3-OCH3 | 3.56 (s) | 57.06 | OCH3 |
| 4 | 5.77 br s | 71.74 | CH |
| 4a | 87.55 | C | |
| 5 | 195.71 | C | |
| 5a | 141.03 | C | |
| 6 | 8.18 (s) | 121.39 | CH |
| 6a | 130.34 | C | |
| 7 | 7.70 (s) | 112.01 | CH |
| 8 | 156.08 | C | |
| 9 | 130.08 | C | |
| 9-C = O | 168.59 | C | |
| 9-CH3CO | 4.01 | 53.15 | CH3 |
| 10 | 138.54 | C | |
| 10-CH3 | 2.89 (s) | 21.71 | CH3 |
| 10a | 121.96 | C | |
| 11 | 167.34 | C | |
| 11-OH | 14.03a br (s) | ||
| 11a | 110.67 | C | |
| 12 | 199.70 | C | |
| 12a | 86.01 | C | |
| 1′ | 6.27 (dd, J = 9, 3 Hz) | 97.37 | CH |
| 2′a | 2.24 (ddd, J = 13, 9, 3 Hz) | 39.08 | CH2 |
| 2′e | 2.60 (ddd, J = 13, 3, 3 Hz) | ||
| 3′ | 4.56 (ddd, J = 3, 3, 3 Hz) | 72.20 | CH |
| 4′ | 3.77 (dd, J = 9, 3 Hz) | 74.15 | CH |
| 5′ | 4.62 (dd, J = 9, 6 Hz) | 68.42 | CH |
| 6′ | 1.65 (d, J = 6) | 19.70 | CH3 |
Observable in d6-acetone.
TABLE 3.
1H and 13C NMR data for DBOV-TCMC in pyridine-d5 at 300 MHz
| Carbon | 1H NMR δ (J in Hz) | 13C NMR (δ) | Multiplicity |
|---|---|---|---|
| 1 | 192.76 | C | |
| 2 | 6.02 br s | 101.38 | CH |
| 3 | 175.57 | C | |
| 3-OCH3 | 3.56 (s) | 57.07 | OCH3 |
| 4 | 5.79 br s | 71.77 | CH |
| 4a | 87.59 | C | |
| 5 | 195.73 | C | |
| 5a | 141.07 | C | |
| 6 | 8.20 (s) | 121.39 | CH |
| 6a | 130.30 | C | |
| 7 | 7.74 (s) | 112.02 | CH |
| 8 | 156.12 | C | |
| 9 | 130.09 | C | |
| 9-C = O | 168.63 | C | |
| 9-CH3CO | 3.94 (s) | 53.10 | CH3 |
| 10 | 138.52 | C | |
| 10-CH3 | 2.91 (s) | 21.72 | CH3 |
| 10a | 121.94 | C | |
| 11 | 167.37 | C | |
| 11-OH | 14.03a br (s) | ||
| 11a | 110.65 | C | |
| 12 | 199.68 | C | |
| 12a | 86.02 | C | |
| 1′ | 6.28 (dd, J = 9, 3 Hz) | 98.08 | CH |
| 2′a | 2.80 (ddd, J = 13, 9, 3 Hz) | 35.29 | CH2 |
| 2′e | 2.38 (ddd, J = 13, 3, 3 Hz) | ||
| 3′ | 4.74 (ddd, J = 3, 3, 3 Hz) | 71.93 | CH |
| 4′ | 4.00 (dd, J = 3.0, 1.5) | 71.77 | CH |
| 5′ | 4.85 (dq, J = 6.0, 1.5) | 69.94 | CH |
| 6′ | 1.66 (d, J = 6.0) | 18.03 | CH3 |
Observable in d6-acetone.
RESULTS
In vivo reconstitution of gene clusters for the biosynthesis of TDP-d-olivose and TDP-d-oliose.
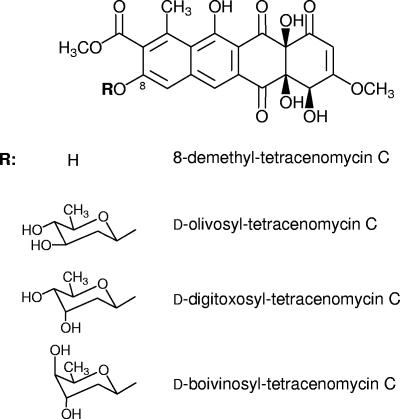
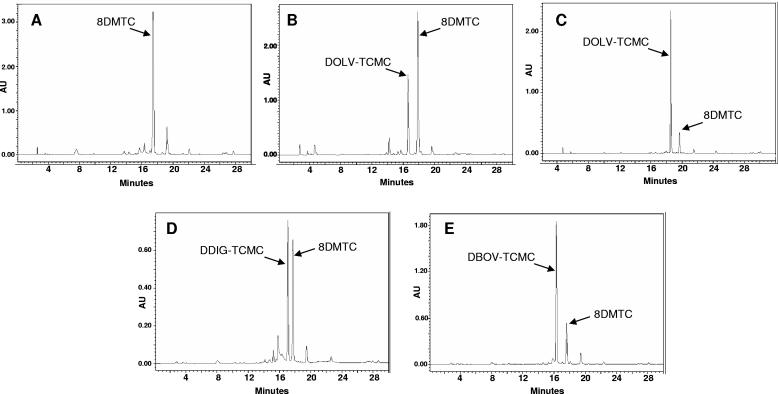
d-Olivose and d-oliose (Fig. 1) are both 2,6-d-DOHs with an equatorial C-3 hydroxyl group, but they differ in the configuration of the hydroxyl group at C-4: equatorial in the former and axial in the latter. d-Olivose is present in several antibiotic and antitumor compounds, such as mithramycin, chromomycin, urdamycin, and granaticin. d-Oliose is more unusual, but it is also found in mithramycin and, as a methylated or an acetylated derivative, in chromomycin A3. A gene cluster was constructed for the biosynthesis of TDP-d-olivose in plasmid pWHM3, generating pMP3* (Fig. 2). This construct contained all the gene functions required to synthesize TDP-d-olivose expressed under the control of two divergent erythromycin resistance promoters, one controlling the expression of two genes from the mithramycin gene cluster, mtmD (TDP-d-glucose synthase) and mtmE (TDP-d-hexose-4,6-dehydratase) (20), and the other controlling a set of four genes, three of them from the oleandomycin gene cluster, oleV (2,3-dehydratase), oleW (a 3-ketoreductase similar to Gra-orf26), and oleY (O-methyltransferase) (3), and a 4-ketoreductase gene (urdR) from the urdamycin gene cluster (15). To confirm that this gene cluster was able to direct the biosynthesis of TDP-d-olivose, pMP3* was introduced into Streptomyces lividans 16F4. This strain harbors cos16F4, a cosmid containing part of the elloramycin biosynthetic gene cluster from Streptomyces olivaceus Tü2353, and its expression in streptomycete hosts generates 8-demethyl-tetracenomycin C (8DMTC) (44) (Fig. 3). This cosmid also contains a gene for the sugar flexible glycosyltransferase ElmGT (6). Upon incubation of S. lividans 16F4(pMP3*) and analysis of cultures by HPLC-MS, in addition to the corresponding aglycone 8DMTC, a new peak (representing approximately 35% of all tetracenomycins) was detected (Fig. 4B). This peak was absent in a control experiment using S. lividans 16F4(pWHM3) (Fig. 4A). The new compound showed the same retention time as a pure sample of d-olivosyl-tetracenomycin C (DOLV-TCMC) and m/z values of 459 (corresponding to the 8DMTC aglycone fragment) and 589 (corresponding to DOLV-TCMC). The formation of this compound confirmed that pMP3* was directing the biosynthesis of TDP-d-olivose.
FIG. 3.
Chemical structures of glycosylated tetracenomycin derivatives.
FIG. 4.
HPLC analyses of cultures of S. lividans 16F4 harboring (A) pWHM3, (B) pMP3*, (C) pMP1*UI, (D) pMP3*BII, and (E) pMP1*BII. Peaks corresponding to the different compounds are indicated.
The biosynthesis of TDP-d-oliose differs from that of TDP-d-olivose only in the final ketoreduction step, which requires a 4-ketoreductase rendering a hydroxyl group at C-4 in axial configuration. To construct a gene cluster for the biosynthesis of TDP-d-oliose, it was necessary to replace the urdR gene (coding for a 4-ketoreductase) with another one able of generating the hydroxyl group in axial configuration. Chromomycin A3 contains both d-olivose and modified d-oliose moieties. In the chromomycin cluster, there are two putative 4-ketoreductase genes that could be involved in the biosynthesis of either d-olivose or d-oliose (24). These two genes, cmmUI and cmmUII, were independently used to replace urdR in pMP3*, generating pMP1*UI and pMP1*UII, respectively (Fig. 2). Upon introduction of pMP1*UI into S. lividans 16F4, most 8DMTC was converted into DOLV-TCMC (Fig. 4C). The formation of this compound confirmed that CmmUI is the 4-ketoreductase involved in d-olivose biosynthesis in the chromomycin pathway and pointed to CmmUII as the candidate for the 4-ketoreductase in d-oliose biosynthesis. However, when pMP1*UII was introduced into S. lividans 16F4, no oliosyl-tetracenomycin or glycosylated tetracenomycin was observed (data not shown). In order to prove the functionality of pMP1*UII, i.e., its capability to direct the biosynthesis of d-oliose, we expressed the plasmid in Streptomyces argillaceus M7U1, a mithramycin nonproducer mutant in which d-oliose biosynthesis is affected (13). Complementation of this mutant was achieved, indicating that d-oliose biosynthesis was recovered and therefore that genes in pMP1*UII were functional (data not shown).
In vivo reconstitution of gene clusters for the biosynthesis of TDP-d-digitoxose and TDP-d-boivinose and generation of novel glycosylated derivatives of tetracenomycin C.
d-Digitoxose and d-boivinose are both 2,6-d-DOHs with an axial C-3 hydroxyl group, but they differ in the configuration of the hydroxyl group at C-4: equatorial in the former and axial in the latter (Fig. 1). d-Digitoxose is well known as a constituent of plant cardiac and other steroidal glycosides (1, 2). d-Boivinose is an unusual DOH. As far as we know, it has been described only as a component of two flavone C-glycosides from Zea mays with glycation-inhibitory activities (37). As a constituent of antibiotic or antitumour compounds, d-digitoxose is present in the apoptosis inducer ammocidin produced by Saccharothrix sp. (25) and in the antibacterial saccharomicin produced by Saccharotrix espanaensis (18, 35). Very recently, a gene cluster for the biosynthesis of TDP-d-digitoxose was identified (5). However, no gene cluster for the biosynthesis of TDP-d-boivinose has been identified in antibiotic-producing microorganisms. We anticipated that a pathway for the biosynthesis of 2,6-d-DOH with a hydroxyl group at C-3 in axial configuration would require the same enzymatic steps needed to synthesize d-olivose and d-oliose but involving a TylC1-like ketoreductase to render the sugar intermediate TDP-2,6-dideoxy-d-glycero-d-glycero-4-hexulose (Fig. 1). Consequently, we decided to test this hypothesis.
d-Olivose differs from d-digitoxose only in the stereochemistry at C-3 (Fig. 1). To construct a gene cluster for the biosynthesis of TDP-d-digitoxose, we used as starting plasmids pMP3* and pMP1*UI, which direct the biosynthesis of d-olivose. We replaced in these plasmids the oleW 3-ketoreductase gene with the TylC1-homologous 3-ketoreductase gene eryBII (12, 36), generating pMP3*BII and pMP1*UIBII, respectively (Fig. 2). After expressing pMP1*UIBII in S. lividans 16F4, only a peak corresponding to 8DMTC was observed (data not shown). However, when pMP3*BII was used, approximately 55% of the aglycone 8DMTC was converted into a novel compound eluting at 17.0 min (Fig. 4D). The compound in this peak showed an m/z value in positive mode of 589 (with a fragmentation ion of m/z 459, corresponding to 8DMTC). This compound was purified, and NMR analysis (see below) confirmed that it corresponded to the novel compound DDIG-TCMC (Fig. 3). The formation of this compound confirmed that pMP3*BII was directing the biosynthesis of TDP-d-digitoxose. These results also indicated that, as has been reported (15, 28), the UrdR 4-ketoreductase shows a certain degree of substrate flexibility and can be used to generate 2,3,6-trideoxyhexoses and 2,6-dideoxyhexoses with different stereochemistries at C-3. On the other hand, the 4-ketoreductase CmmUI shows low substrate flexibility, since it is not able to act on a substrate differing from its natural one in the stereochemistry at C-3.
To construct a gene cluster for the biosynthesis of TDP-d-boivinose, it was necessary to incorporate, in addition to eryBII, a gene coding for a 4-ketoreductase that would render a hydroxyl group in axial configuration. We therefore replaced in pMP3*BII the 4-ketoreductase urdR gene (which renders hydroxyl groups in equatorial configuration) with the cmmUII gene. The resultant plasmid pMP1*UIIBII was expressed in S. lividans 16F4, but no glycosylated tetracenomycin was detected (data not shown). This suggested that, similarly to CmmUI, the CmmUII ketoreductase was unable to act on the substrate TDP-2,6-dideoxy-d-glycero-d-glycero-4-hexulose, which has stereochemistry at C-3 opposite to that of its normal substrate. Alternatively, the possibility exists that ElmGT is unable to transfer d-boivinose. To clarify this situation, we decided to use a different 4-ketoreductase, OleU, which is involved in the biosynthesis of l-oleandrose in the oleandomycin pathway (3). A first indication that OleU could be suitable for the biosynthesis of d-boivinose arose from the analysis of cultures of S. lividans 16F4 expressing pFL1012 (21). This plasmid derives from pFL943, which has been shown to direct the biosynthesis of TDP-l-olivose and TDP-l-rhamnose. pFL1012 contains the same gene functions as pFL943, but it lacks the oleL 3,5-epimerase gene, which prevents it from directing the biosynthesis of l-DOH (21). Notably, cultures of S. lividans 16F4(pFL1012) produced a glycosylated tetracenomycin with the same mass as DOLV-TCMC or DDIG-TCMC but showing a different retention time. This result prompted us to think that this compound could correspond to DBOV-TCMC. Consequently, we used the oleU 4-ketoreductase to replace cmmUII in pMP1*UIIBII. Upon transforming S. lividans 16F4 with the resultant construct, pMP1*BII (Fig. 2), a major peak (representing 76% of all tetracenomycins) was detected (Fig. 4E), eluting at 16.2 min and showing an m/z value of 589 (with a fragmentation ion of m/z 459, corresponding to 8DMTC). This mass was in accordance with the presence of a 2,6-DOH attached to 8DMTC. The compound from this peak was purified, and NMR analysis (see below) revealed that it corresponded to the novel compound DBOV-TCMC (Fig. 3). The formation of this compound confirmed that pMP1*BII was directing the biosynthesis of TDP-d-boivinose. To eliminate the possibility that a 4-ketoreductase gene from the S. lividans 16F4 host strain was participating in the biosynthesis of d-boivinose, a control experiment was run using pMP1*BIIΔU, a derivative of pMP1*BII in which oleU was deleted. Using this construct, no DBOV-TCMC was detected, confirming that OleU was the 4-ketoreductase involved in the formation of this boivinosyl derivative. This result also indicated that OleU shows a certain degree of flexibility, since, although it normally reduces a 2,6-l-DOH intermediate, it can also act on a 2,6-d-DOH substrate.
Structure elucidation of d-digitoxosyl-tetracenomycin C and d-boivinosyl-tetracenomycin C.
The two novel tetracenomycins were characterized by liquid chromatography-MS (see below) and NMR spectroscopy in comparison with various previously described 8-position glycosylated tetracenomycins and elloramycins (11, 31, 44).
The positive atmospheric pressure chemical ionization (APCI) mass spectrum of DDIG-TCMC showed two major mass fragments, at m/z 589 (M+) and at m/z 459 (M+-sugar). While the former confirms the deduced molecular formula of C28H28O14, the latter is consistent with the fragment obtained from the aglycon after cleavage of a 2,6-DOH unit. This pattern of M+ and M-sugar fragmentations is typical of the glycosylated tetracenomycins and elloramycins. The 1H NMR data (Table 2) revealed a β-glycosidically bound d-sugar (deduced from the large coupling of 1′-H of 9 Hz), as well as a sugar in 4C1 conformation typical of d-sugars (from the overall H-H coupling pattern observed in the sugar unit, particularly the two axial protons in the 1" and 5′ positions). The observed stereochemistry at the 3′ position (a multiple resulting from three small 3JH-H couplings with neighboring protons reveals an axially attached OH group) and in the 4′ position (dd, J = 9, 3 Hz, revealing an equatorially attached OH group) is consistent with digitoxose stereochemistry. This suggests the structure of 8-demethyl-8-β-d-digitoxosyltetracenomycin C for DDIG-TCMC, which was fully confirmed by the 13C NMR data (Table 2).
The positive APCI mass spectrum of DBOV-TCMC gave the same molecular formula (C28H28O14) and major fragmentation (M+ at m/z 589; M-sugar at m/z 459) found for DDIG-TCMC, also consistent with a tetracenomycin derivative with a 2,6-dideoxysugar attached in the 8 position (for the APCI mass spectrum and the major fragmentation in the APCI mass spectrum, see the supplemental material). The 1H NMR spectrum (Table 3) revealed the difference from DDIG-TCMC. While we also observed a β-glycosidically bound d-sugar in 4C1 conformation, the signals of 3′-H (δ 4.74, ddd, J = 3.0, 3.0, 3.0 Hz) and 4′-H (δ 4.00, dq, J = 6.0, 1.5 Hz) of the sugar moiety clearly show that the OH groups in these two positions are attached axially in both cases, which gives rise to a boivinose stereochemistry. Therefore, the structure of 8-demethyl-8-β-d-boivinosyl-tetracenomycin C could be deduced for DBOV-TCMC. This was fully confirmed by the 13C NMR data (Table 3).
DISCUSSION
Many bioactive compounds produced by actinomycetes are glycosylated, and in many cases, the saccharide moieties are essential for bioactivity. Therefore, efforts to modify the glycosylation pattern of a certain compound could have practical implications, since they could lead to the generation of a more active compound or one with improved pharmacological characteristics. This type of modification can be approached by combinatorial biosynthesis, providing that flexible glycosyltransferases exist, together with the possibility of using different nucleotide DOHs. In this context, the isolation of natural gene clusters and the reconstitution of “unnatural gene clusters” for the biosynthesis of DOHs have allowed their use for the generation of new glycosylated compounds (23, 26, 38). In this study, we have reconstituted “unnatural natural gene clusters” for the biosynthesis of four different activated 2,6-d-DOHs, namely, TDP-d-olivose, TDP-d-oliose, TDP-d-digitoxose, and TDP-d-boivinose. To achieve this, we combined, in a rational way, the required genes involved in sugar biosynthesis from different antibiotic and antitumor biosynthetic gene clusters: the antitumor agents mithramycin, chromomycin, and urdamycin and the antibiotics oleandomycin and erythromycin. Since the biosynthesis of 2,6-d-DOH proceeds through three common initial steps, all the gene clusters constructed contain three genes, which code for the enzymatic activities involved in those steps: the glucose synthase gene mtmD and the 4,6-dehydratase gene mtmE from the mithramycin cluster (20) and the 3,4-dehydratase gene oleV from the oleandomycin cluster (3), whose enzymatic activities had already been proven in vitro (20) or in vivo (30). In addition, gene clusters for the biosynthesis of TDP-d-olivose and TDP-d-oliose include the 3-ketoreductase gene oleW from the oleandomycin gene cluster, which had been shown to be involved in l-olivose biosynthesis reducing the keto group at C-3 with the formation of a hydroxyl group in equatorial configuration (3, 30). On the other hand, gene clusters for the biosynthesis of TDP-d-digitoxose and TDP-d-boivinose contain the eryBII C-3 ketoreductase from the erythromycin gene cluster, involved in l-mycarose biosynthesis (12, 36). Previous experiments have suggested that EryBII was able to introduce hydroxyl groups at C-3 in axial configuration (21). The fact that pMP3*BII and pMP1*BII were able to direct the biosynthesis of TDP-d-digitoxose and TDP-d-boivinose, respectively, confirms that function for EryBII and also indicates that this process takes place on a nucleotide sugar intermediate of d configuration.
All 2,6-d-DOH gene clusters contain a 4-ketoreductase gene. Thus, those for the biosynthesis of TDP-d-olivose and TDP-d-digitoxose (with hydroxyl groups at C-4 in equatorial configuration) include the 4-ketoreductase gene urdR from the urdamycin cluster, which has been shown to code for a 4-ketoreductase in d-olivose biosynthesis (15, 30). A gene cluster (present in pMP1*UI) for the biosynthesis of TDP-d-olivose was also constructed using a different 4-ketoreductase gene, cmmUI, a gene from the chromomycin gene cluster, which was proposed to code for a 4-ketoreductase involved in sugar biosynthesis (24). The fact that pMP1*UI directed the biosynthesis of d-olivose demonstrates that CmmUI is the 4-ketoreductase for d-olivose biosynthesis in the chromomycin pathway. On the other hand, in the gene cluster for the biosynthesis of TDP-d-boivinose, we included the oleU gene from the oleandomycin gene cluster, which has been shown to be involved in the biosynthesis of l-olivose (3). The fact that OleU can also be used for the biosynthesis of d-boivinose indicates that OleU shows some substrate flexibility, since although its natural substrate is a nucleotide sugar intermediate in l configuration, the results obtained demonstrate that OleU is also able to act on d-DOH biosynthesis intermediates. Finally, the gene cluster for the biosynthesis of TDP-d-oliose (present in pMP1*UII) includes the 4-ketoreductase cmmUII from the chromomycin gene cluster.
Validation of the functionality of the described gene clusters was achieved by expressing them in Streptomyces albus 16F4 and the subsequent formation of the corresponding glycosylated tetracenomycin. However, when plasmid pMP1*UII was used, no glycosylated tetracenomycin was produced. Two possible hypotheses exist to explain this result: (i) CmmUII is not the 4-ketoreductase for TDP-d-oliose and therefore pMP1*UII is not able to direct the biosynthesis of this deoxysugar, and consequently no glycosylated tetracenomycin is formed, or (ii) even though the biosynthesis of TDP-d-oliose is directed by pMP1*UII, the possibility exists that this DOH was not recognized by the ElmGT glycosyltransferase. We favor the second explanation, since pMP1*UII directs the biosynthesis of d-oliose, as it was able to complement the non-mithramycin-producing mutant S. argilllaceus M7U1, in which d-oliose biosynthesis is affected. Moreover, previous experiments also suggested that the glycosyltransferase ElmGT was not able to recognize TDP-d-oliose as a substrate. Thus, it has been shown that by expressing cosmid 16F4 in the mithramycin producer S. argillaceus, no d-oliosyl-tetracenomycin C was obtained, despite the fact that this microorganism synthesizes TDP-d-oliose (44).
In this work, two novel glycosylated tetracenomycins were generated, DDIG-TCMC and DBOV-TCMC. The formation of these two new compounds extends the NDP-sugar donor substrate profile usable by the ElmGT glycosyltransferase by two. ElmGT has been previously shown to be able to transfer nine different sugars (23).
Gene clusters for the biosynthesis of TDP-d-boivinose in antibiotic producers had not been described or isolated previously. The reconstituted gene clusters described here for the biosynthesis of this DOH, and also those for TDP-d-digitoxose, TDP-d-olivose, and TDP-d-oliose, will be very useful for the generation of new glycosylated derivatives of bioactive compounds by providing host strains with the capability of synthesizing these d-DOHs, which then could be potentially transferred by existing glycosyltransferases of the host to an aglycone. In addition, the information obtained from the reconstituted gene clusters for the biosynthesis of TDP-d-boivinose could help to predict which gene functions should be present in a natural gene cluster for this deoxysugar.
Supplementary Material
Acknowledgments
This work was supported by grants from the Spanish Ministry of Education and Science (BMC2002-03599 and BIO2005-04115) to C.M. and from the U.S. National Institutes of Health (CA 91901 and CA 102102) to J.R. M.P. was the recipient of a predoctoral fellowship from the Spanish Ministry of Science and Technology. We thank Obra Social Cajastur for financial support of F.L.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Abe, F., Y. Mori, H. Okabe, and T. Yamauchi. 1994. Steroidal constituents from the roots and stems of Aclepias fruticosa. Chem. Pharm. Bull. 42:1777-1783. [DOI] [PubMed] [Google Scholar]
- 2.Abe, F., and T. Yamauchi. 2000. 5,11-Epoxymegastigmanes from the leaves of Asclepias fruticosa. Chem. Pharm. Bull. 48:1908-1911. [DOI] [PubMed] [Google Scholar]
- 3.Aguirrezabalaga, I., C. Olano, N. Allende, L. Rodríguez, A. F. Braña, C. Méndez, and J. A. Salas. 2000. Identification and expression of genes involved in L-oleandrose biosynthesis and its intermediate L-olivose in the oleandomycin producer Streptomyces antibioticus. Antimicrob. Agents Chemother. 44:1266-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Bihlmaier, C., E. Welle, C. Hofmann, K. Welzel, A. Vente, E. Breitling, M. Muller, S. Glaser, and A. Bechthold. 2006. Biosynthetic gene cluster for the polyenoyltetramic acid alpha-lipomycin. Antimicrob. Agents Chemother. 50:2113-2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanco, G., E. Pérez-Patallo, A. F. Braña, J. Rohr, C. Méndez, and J. A. Salas. 2001. Identification of a sugar flexible glycosyltransferase from Streptomyces olivaceus, the producer of the antitumor polyketide elloramycin. Chem. Biol. 8:253-263. [DOI] [PubMed] [Google Scholar]
- 7.Chen, H., G. Agnihotri, Z. Guo, N. L. S. Que, X. H. Chen, and H.-w. Liu. 1999. Biosynthesis of mycarose: isolation and characterization of enzymes involved in the C-2 deoxygenation. J. Am. Chem. Soc. 121:8124-8125. [Google Scholar]
- 8.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acid Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Draeger, G., S. H. Park, and H. G. Floss. 1999. Mechanism of the 2-deoxygenation step in the biosynthesis of the deoxyhexose moieties of the antibiotics granaticin and oleandomycin. J. Am. Chem. Soc. 121:2611-2612. [Google Scholar]
- 10.Fernández, E., U. Weissbach, C. S. Reillo, A. F. Braña, C. Méndez, J. Rohr, and J. A. Salas. 1998. Identification of two genes from Streptomyces argillaceus encoding glycosyltransferases involved in transfer of a disaccharide during biosynthesis of the antitumor drug mithramycin. J. Bacteriol. 180:4929-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer, C., L. Rodríguez, E. P. Patallo, F. Lipata, A. F. Braña, C. Méndez, J. A. Salas, and J. Rohr. 2002. Digitoxosyltetracenomycin C and glucosyltetracenomycin C, two novel elloramycin analogues obtained by exploring the sugar donor substrate specificity of glycosyltransferase ElmGT J. Nat. Prod. 65:1685-1689. [DOI] [PubMed] [Google Scholar]
- 12.Gaisser, S., G. A. Bohm, J. Cortes, and P. F. Leadlay. 1997. Analysis of seven genes from the eryAI-eryK region of the erythromycin biosynthetic gene cluster in Saccharopolyspora erythraea. Mol. Gen. Genet. 256:239-251. [DOI] [PubMed] [Google Scholar]
- 13.González, A., L. L. Remsing, F. Lombó, M. J. Fernández, L. Prado, A. F. Braña, E. Kunzel, J. Rohr, C. Méndez, and J. A. Salas. 2001. The mtmVUC genes of the mithramycin gene cluster in Streptomyces argillaceus are involved in the biosynthesis of the sugar moieties. Mol. Gen. Genet. 264:827-835. [DOI] [PubMed] [Google Scholar]
- 14.He, X., and H. W. Liu. 2002. Mechanisms of enzymatic C-bond O-bond cleavages in deoxyhexose biosynthesis. Curr. Opin. Chem. Biol. 6:590-597. [DOI] [PubMed] [Google Scholar]
- 15.Hoffmeister, D., K. Ichinose, S. Domann, B. Faust, A. Trefzer, G. Draeger, A. Kirschning, C. Fischer, E. Künzel, D. Bearden, J. Rohr, and A. Bechthold. 2000. The NDP-sugar co-substrate concentration and the enzyme expression level influence the substrate specificity of glycosyltransferases: cloning and characterization of deoxysugar biosynthetic genes of the urdamycin biosynthetic gene cluster. Chem. Biol. 7:821-831. [DOI] [PubMed] [Google Scholar]
- 16.Ichinose, K., D. J. Bedford, D. Tornus, A. Bechthold, M. J. Bibb, W. P. Revill, H. G. Floss, and D. A. Hopwood. 1998. The granaticin biosynthetic gene cluster of Streptomyces violaceoruber Tü22: sequence analysis and expression in a heterologous host. Chem. Biol. 5:647-659. [DOI] [PubMed] [Google Scholar]
- 17.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. (Norwich, United Kingdom: The John Innes Foundation).
- 18.Kong, F., N. Zhao, M. M. Siegel, K. Janota, J. S. Ashcroft, F. E. Koehn, D. B. Borders, and G. T. Carter. 1998. Saccharomicins, novel heptadecaglycoside antibiotics effective against multi-resistant bacteria. J. Am. Chem. Soc. 120:13301-13311. [Google Scholar]
- 19.Liu, H. W., and J. S. Thorson. 1994. Pathways and mechanisms in the biogenesis of novel deoxysugars by bacteria. Annu. Rev. Microbiol. 48:223-256. [DOI] [PubMed] [Google Scholar]
- 20.Lombó, F., K. Siems, A. F. Braña, C. Méndez, K. Bindseil, and J. A. Salas. 1997. Cloning and insertional inactivation of Streptomyces argillaceus genes involved in the earliest steps of biosynthesis of the sugar moieties of the antitumor polyketide mithramycin. J. Bacteriol. 179:3354-3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lombó, F., M. Gibson, L. Greenwell, A. F. Braña, J. Rohr, J. A. Salas, and C. Méndez. 2004. Engineering biosynthetic pathway for deoxysugars: branched-chain sugar pathways and novel derivatives from the antitumor tetracenomycin. Chem. Biol. 11:1709-1718. [DOI] [PubMed] [Google Scholar]
- 22.Méndez, C., and J. A. Salas. 2001. Altering the glycosylation pattern of bioactive compounds. Trends in Biotechnol. 19:449-456. [DOI] [PubMed] [Google Scholar]
- 23.Mendez, C., and J. A. Salas. 2005. Engineering glycosylation in bioactive compounds by combinatorial biosynthesis. Ernst Schering Res. Found. Workshop. 51:127-146. [DOI] [PubMed] [Google Scholar]
- 24.Menéndez, N., M. Nur-e-Alam, A. F. Braña, J. Rohr, J. A. Salas, and C. Méndez. 2004. Biosynthesis of the antitumor chromomycin A3 in Streptomyces griseus: analysis of the gene cluster and rational design of novel chromomycin analogues. Chem. Biol. 11:21-32. [DOI] [PubMed] [Google Scholar]
- 25.Murakami, R., T. Tomikawa, K. Shin-ya, J. Shinozaki, T. Kajiura, H. Seto, and Y. Hayakawa. 2001. Ammocidin, a new apoptosis inducer in ras-dependent cells from Saccharothrix sp. II. Physico-chemical properties and structure elucidation. J. Antibiot. 54:714-717. [DOI] [PubMed] [Google Scholar]
- 26.Olano, C., N. Lomovskaya, L. Fonstein, J. T. Roll, and C. R. Hutchinson. 1999. A two-plasmid system for the glycosylation of polyketide antibiotics: bioconversion of epsilon-rhodomycinone to rhodomycin D. Chem. Biol. 6:845-855. [DOI] [PubMed] [Google Scholar]
- 27.Patallo, E. P., G. Blanco, C. Fischer, A. F. Braña, J. Rohr, C. Méndez, and J. A. Salas. 2001. Deoxysugar methylation during biosynthesis of the antitumor polyketide elloramycin by Streptomyces olivaceus: characterization of three methyltransferase genes. J. Biol. Chem. 276:18765-18774. [DOI] [PubMed] [Google Scholar]
- 28.Pérez, M., F. Lombó, L. Zhu, M. Gibson, A. F. Braña, J. Rohr, J. A. Salas, and C. Méndez. 2005. Combining sugar biosynthesis gene cassettes to generate two glycosylated antitumor tetracenomycins. Chem. Comm. 12:1604-1606. [DOI] [PubMed] [Google Scholar]
- 29.Piepersberg, W. 1994. Pathway engineering in secondary metabolite-producing actinomycetes. Crit. Rev. Biotech. 14:251-285. [DOI] [PubMed] [Google Scholar]
- 30.Rodríguez, L., I. Aguirrezabalaga, N. Allende, A. F. Braña, C. Méndez, and J. A. Salas. 2002. Engineering deoxysugar biosynthetic pathways from antibiotic-producing microorganisms: a tool to produce novel glycosylated bioactive compounds. Chem. Biol. 9:721-729. [DOI] [PubMed] [Google Scholar]
- 31.Rodríguez, L., C. Oelkers, I. Aguirrezabalaga, A. F. Braña, J. Rohr, C. Méndez, and J. A. Salas. 2000. Generation of hybrid elloramycin analogs by combinatorial biosynthesis using genes from anthracycline-type and macrolide biosynthetic pathways. J. Mol. Microbiol. Biotechnol. 2:271-276. [PubMed] [Google Scholar]
- 32.Salas, J. A., and C. Méndez. 2005. Biosynthesis pathways for deoxysugars in antibiotic-producing actinomycetes: isolation, characterization and generation of novel glycosylated derivatives. J. Molec. Microbiol. Biotechnol. 9:77-85. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. (Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press).
- 34.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain terminating inhibitors. Proc. Natl. Acad. Sci. USA(USA). 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh, M. P., P. J. Petersen, W. J. Weiss, F. Kong, and M. Greenstein. 2000. Saccharomicins, novel heptadecaglycoside antibiotics produced by Saccharothrix espanaensis: antibacterial and mechanistic activities Antimicrob. Agents Chemother. 44:2154-2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Summers, R. G., S. Donadio, M. J. Staver, E. Wendt-Pienkowski, C. R. Hutchinson, and L. Katz. 1997. Sequencing and mutagenesis of genes from the erythromycin biosynthetic gene cluster of Saccharopolyspora erythraea that are involved in L-mycarose and D-desosamine production. Microbiol. 143:3251-3262. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki, R., Y. Okada, and T. Okuyama. 2003. Two flavone C-glycosides from the style of Zea mays with glycation inhibitory activity. J. Nat. Prod. 66:564-565. [DOI] [PubMed] [Google Scholar]
- 38.Tang, L., and R. McDaniel. 2001. Construction of desosamine containing polyketide libraries using a glycosyltransferase with broad substrate specificity. Chem. Biol. 8:547-555. [DOI] [PubMed] [Google Scholar]
- 39.Trefzer, A., J. A. Salas, and A. Bechthold. 1999. Genes and enzymes of deoxysugar biosyntheses. Nat. Prod. Rep. 16:283-299. [DOI] [PubMed] [Google Scholar]
- 40.Trefzer, A., S. Pelzer, J. Schimana, S. Stockert, C. Bihlmaier, H. P. Fiedler, K. Welzel, A. Vente, and A. Bechthold. 2002. Biosynthetic gene cluster of simocyclinone, a natural multihybrid antibiotic Antimicrob. Agents Chemother. 46:1174-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vara, J., M. Lewandowska-Skarbek, Y. G. Wang, S. Donadio, and C. R. Hutchinson. 1989. Cloning of genes governing the deoxysugar portion of the erythromycin biosynthesis pathway in Saccharopolyspora erythraea (Streptomyces erythreus). J. Bacteriol. 171:5872-5881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weitnauer, G., A. Mühlenweg, A. Trefzer, D. Hoffmeister, R. D. Süssmuth, G. Jung, K. Welzel, A. Vente, U. Girreser, and A. Bechthold. 2001. Biosynthesis of the orthosomycin antibiotic avilamycin A: deductions from the molecular analysis of the avi biosynthetic gene cluster of Streptomyces viridochromogenes Tu57 and production of new antibiotics. Chem. Biol. 8:569-581. [DOI] [PubMed] [Google Scholar]
- 43.Westrich, L., S. Domann, B. Faust, D. Bedford, D. A. Hopwood, and A. Bechthold. 1999. Cloning and characterization of a gene cluster from Streptomyces cyanogenus S136 probably involved in landomycin biosynthesis. FEMS Microbiol. Lett. 170:381-387. [DOI] [PubMed] [Google Scholar]
- 44.Wohlert, S.-E., G. Blanco, F. Lombó, E. Fernández, A. F. Braña, S. Reich, G. Udvarnoki, C. Méndez, H. Decker, J. A. Salas, and J. Rohr. 1998. Novel hybrid tetracenomycins through combinatorial biosynthesis using a glycosyltransferase encoded by the elm-genes in cosmid 16F4 which shows a broad sugar substrate specificity. J. Am. Chem. Soc. 41:10596-10601. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.