Abstract
Pseudomonas putida KT2440 metabolizes a wide range of carbon and nitrogen sources, including many amino acids. In this study, a σ54-dependent two-component system that controls the uptake and metabolism of acidic amino acids was identified. The system (designated aau, for acidic amino acid utilization) involves a sensor histidine kinase, AauS, encoded by PP1067, and a response regulator, AauR, encoded by PP1066. aauR and aauS deletion mutants were unable to efficiently utilize aspartate (Asp), glutamate (Glu), and glutamine (Gln) as sole sources of carbon and nitrogen. Growth of the mutants was partially restored when the above-mentioned amino acids were supplemented with glucose or succinate as an additional carbon source. Uptake of Gln, Asp, and asparagine (Asn) by the aauR mutant was moderately reduced, while Glu uptake was severely impaired. In the absence of glucose, the aauR mutant even secreted Glu into the medium. Furthermore, disruption of aauR affected the activities of several key enzymes of Glu and Asp metabolism, leading to the intracellular accumulation of Glu and greatly reduced survival times under conditions of nitrogen starvation. By a proteomics approach, four major proteins were identified that are downregulated during growth of the aauR mutant on Glu. Two of these were identified as periplasmic glutaminase/asparaginase and the solute-binding protein of a Glu/Asp transporter. Transcriptional analysis of lacZ fusions containing the putative promoter regions of these genes confirmed that their expression is indeed affected by the aau system. Three further periplasmic solute-binding proteins were strongly expressed during growth of the aauR deletion mutant on Glu but downregulated during cultivation on glucose/NH4+. These systems may be involved in amino acid efflux.
Microorganisms often encounter a rapidly changing environment to which they exhibit a variety of adaptive responses. These responsive changes are mediated by a number of different regulatory mechanisms. One of these involves signal transduction proteins belonging to the family of two-component regulatory systems. A typical bacterial two-component system consists of a sensor protein kinase residing in the inner membrane and a cognate response regulator present in the cytoplasm (19). This specific arrangement enables the cells to sense environmental stimuli, which then trigger autophosphorylation of the sensor kinase at a conserved histidine residue. The cognate response regulator then catalyzes the transfer of histidine-bound phosphate to one of its own aspartate residues. Response regulators are made up of a receiver domain that contains the phosphorylation site and an output domain, usually a DNA binding module, that affects the transcription of other genes (23).
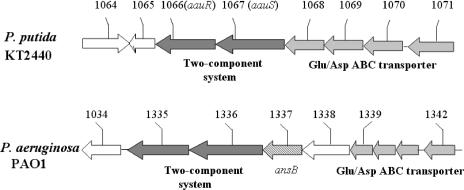
The present knowledge of bacterial amino acid metabolism and its regulation is based mainly on findings with enterobacteria, whereas much less is known about the modes of amino acid utilization in pseudomonads. Pseudomonas putida KT2440, an efficient root colonizer, is widely applied in biotransformation, bioremediation, and agriculture. It metabolizes a wide range of carbon and nitrogen sources, including many amino acids. During growth of P. putida KT2440 on glutamate as a sole source of carbon and nitrogen, a number of proteins are upregulated, including a periplasmic glutaminase/asparaginase (PGA) and two components of an ATP-binding cassette (ABC)-type transporter, i.e., its ATP-binding subunit (encoded by PP1068) and the cognate periplasmic solute-binding protein (encoded by PP1071) (17). A BLAST search in the Comprehensive Microbial Resource database (http://cmr.tigr.org/tigr-scripts/CMR/CmrHomePage.cgi) indicated that the ABC transporter encoded by PP1071 to PP1068 is most probably involved in the uptake of glutamate and/or aspartate. A pair of genes (PP1067 and PP1066) located immediately adjacent to PP1071 to PP1068 encode a yet-uncharacterized σ54-dependent two-component system (Fig. 1) consisting of a sensor histidine kinase (PP1067) and the cognate response regulator (PP1066). In the P. aeruginosa PAO1 genome, the closest orthologues of PP1071 to PP1068 and PP1067-PP1066 (i.e., PA1342 to PA1339 and PA1336-PA1335, respectively) cluster with genes that code for enzymes of glutamate metabolism, i.e., PGA (PA1337) and γ-glutamyl transferase (PA1338) (Fig. 1), suggesting that the two-component system encoded by PA1336-PA1335 (or PP1066-PP1067) might be involved in the regulation of glutamate metabolism as well. The two-component system was therefore designated aau (for acidic amino acid utilization). Here, PP1067 will be referred to as aauS, since it encodes the sensor kinase, while PP1066 is called aauR. The aauR open reading frame has a translation initiation codon that overlaps with the aauS termination codon, suggesting that expression of these two genes is probably translationally coupled.
FIG. 1.
Genetic organizations of the aau two-component systems in the genomes of P. putida KT2440 and P. aeruginosa PAO1 (see the text).
The aim of the present study was to screen P. putida KT2440 for targets of the aau regulatory system. Wild-type cells and aau deletion mutants were compared with respect to growth and survival on various carbon and nitrogen sources. In addition, rates of amino acid uptake and the activities of key enzymes of glutamate and aspartate metabolism were studied. Finally, a proteomics approach was used to identify proteins differentially expressed in wild-type P. putida KT2440 and its deletion mutant, KTaauR. The results confirmed that the aau system is a major regulator of acidic amino acid uptake and utilization.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. P. putida KT2440 was grown at 30°C either on Luria-Bertani (LB) agar plates, in LB broth (Gibco BRL), or on M9 minimal medium (14) supplemented with different carbon and nitrogen sources. Escherichia coli strains were grown at 37°C on LB agar plates or in LB broth. Media for plasmid and mutant selection were supplemented with kanamycin (30 μg/ml), chloramphenicol (50 μg/ml), streptomycin (300 μg/ml), or ampicillin (200 μg/ml). To examine the effects of nutrients on growth and PGA activity, cells were first grown overnight in M9 medium supplemented with 19 mM NH4Cl and 22 mM glucose (M9+), collected by centrifugation, washed with M9 salt solution, and then transferred to M9 medium supplemented with (i) NH4+-glucose as described above, (ii) amino acids (5 mM or 10 mM) as the sole source of carbon and nitrogen, or (iii) amino acids supplemented with 22 mM glucose, 10 mM succinate, or 19 mM NH4+. Bacterial growth was monitored by absorption at 595 nm (A595). Growth curves (A595 versus time) were analyzed by fitting a logistic model to the absorption-versus-time profile (26).
TABLE 1.
Strains and plasmids used
| Strain or plasmid | Description, origin, or sequence | Source or reference |
|---|---|---|
| P. putida strains | ||
| KT2440 | mt-2 hsdR1 (r− m+) | 1 |
| KTaauR | aauR derivative of KT2440 | This work |
| KTaauS | aauS derivative of KT2440 | This work |
| KTansB::lacZ | KT2440 containing the PP2453 promoter region fused to a promoterless lacZ gene | This work |
| KTABC::lacZ | KT2440 containing the PP1071 promoter fused to a promoterless lacZ gene | This work |
| KTaauRansB::lacZ | KTaauR containing the PP2453 promoter fused to a promoterless lacZ gene | This work |
| KTaauRABC::lacZ | KTaauR containing the PP1071 promoter fused to a promoterless lacZ gene | This work |
| E. coli strains | ||
| HB101 | supE44 hsdS20 (rB− mB−) recA13 ara-14 proA21 acY1 galK2 rpsL20 | Amersham Biosciences |
| S-17 | 15 | |
| Plasmids | ||
| pK18 | Cloning vector, kanamycin resistance | 12 |
| PDN19lacΩ | Promoterless lacZ oriV oriT Tetr Strr Ω fragment |
For studies of survival in nitrogen-free medium, cells were grown overnight in M9+, washed with M9 salt solution, and incubated in M9 minimal medium containing 5 mM Glu or Asp for up to 6 weeks. Samples (0.1 ml) were removed at regular intervals and spread on LB plates after appropriate dilution. Colonies were counted after 24 h. Protein concentrations were determined with a 2-D Quant Kit (Amersham Biosciences) and bicinchoninic acid assay (Pierce) with bovine serum albumin as the standard protein.
Construction of plasmids and strains.
To knock out the aauR gene, a 640-bp internal fragment of the gene (335 to 975 bp relative to the translation start site) was amplified by PCR using P. putida KT2440 chromosomal DNA as a template and the following gene-specific primers designed to add a restriction site at the ends: aauRFor (5′-CGCGGATCCGCCTGGTCGAACGTGGTACG-3′; the BamHI site is underlined, and the base shown in boldface was introduced to generate a frameshift) and aauRRev (5′-CCCAAGCTTGATGTCTTCACGGCGCTCAC-3′; the HindIII site is underlined). The PCR product generated in this way was cloned into the compatible BamHI and HindIII sites of plasmid pK18 (12) to yield pK18aauR. This construct was then inserted into the P. putida KT2440 chromosome by homologous recombination, followed by selection of kanamycin-resistant clones. Such clones must carry the plasmid integrated into the genome, since pK18 derivatives cannot otherwise replicate in P. putida. They contain fragments of the target gene separated by the plasmid sequence: (i) a short truncated fragment and (ii) a fragment containing the frameshift mutation at the 5′ end. The frameshift mutation was necessary, as in the second case, the transconjugants might contain a wild-type copy of the genes after the recombination. aauS was inactivated by a similar strategy using the primers aauSFor (5′-CGCGGATCCCGAATACCCTTGAAGGCCTGA-3′; the BamHI site is underlined, and the additional base shown in boldface was introduced to generate a frameshift) and aauSRev (5′-CCCAAGCTTTCAGTTTTTCCACACCATCG-3′; the HindIII site is underlined). Here, a 506-bp internal fragment of the gene (312 to 817 bp relative to the translation start site) was used for PCR amplification.
Complementation of the aauR phenotype.
In order to ascertain that the observed aauR phenotype was due to inactivation of that gene, the KTaauR deletion mutation was complemented using a pMMB67EH vector. The coding sequence of aauR was amplified from P. putida KT2440 DNA using the primers cAAURFor (5′-CGCGGATCCATGAACCAAGCGCCTCTTACCG-3′; the BamHI site is underlined) and cAAURRev (5′-CCCAAGCTTTCAGGCGAGGCCGTATTTTTTCAC-3′; the HindIII site is underlined). Due to the lack of useful restriction sites, it was necessary to introduce BamHI and HindIII sites (underlined) into the forward and reverse primers, respectively. The amplified, digested product was cloned into pMMB67EH immediately downstream of an inducible tac promoter to generate plasmid pMMaauR. To verify the plasmid construct, nucleotide sequence information was obtained from plasmid pMMaauR using primers cAAURFor and cAAURRev. The mutant strain KTaauR was transformed with the plasmid construct pMMaauR to generate strain KTaauR+. Positive clones were selected on carbenicillin-containing plates.
Reporter gene constructs.
Putative promoter regions of ansB (PP2453) and PP1071 (450 to 500 bp upstream of the translation start site) were amplified from the KT2440 genome by PCR using Pfu polymerase (Stratagene) and the primers ansBFor (5′-CGCGTCGAATTCGGCGAGGCTAAGCGAGGAAATG-3′; the EcoRI site is underlined) and ansBRev (5′-ACGCATGGATCCGTGCGCTTGGGCGAAGGTTTTCA-3′; the BamHI site is underlined) in the case of ansB. The PP1071 promoter was amplified using ABCFor (5′-CGCGTCGAATTCCGCATCGAGCCTTTCCGGTTGGGC-3′; the EcoRI site is underlined) and ABCRev (5′-ACGCATGGATCCTGCAGCCGCGATGGCTGCGCCCA-3′; the BamHI site is underlined). The amplified products were cloned upstream of the promoterless lacZ into pDN19lacΩ to yield KTansB::lacZ and KTABC::lacZ, respectively. The fidelity of the constructs was verified by sequencing using primers ansBFor and ABCFor, respectively. The plasmids were transformed into P. putida KT2440, the deletion mutant (KTaauR), and the complemented strain (KTaauR+). The cells were grown to late log phase (A600 = 0.6 to 1.0) in M9 medium containing 5 mM Glu and streptomycin. At this point, cells were harvested and assayed for β-galactosidase activity as described by Miller (10). Each experiment was performed in triplicate.
Preparation of protein extracts for two-dimensional (2-D) gel electrophoresis.
Bacteria from 100 ml of culture were harvested 6 h after transfer to fresh medium and collected by centrifugation. The cell pellets were washed twice with M9 salt solution and resuspended in ice-cold homogenization buffer (50 mM Tris, 2.5 mM EDTA, 250 mM sucrose, 5 mM MgCl2, pH 7.2) containing a protease inhibitor cocktail (Roche Diagnostics, Lewes, United Kingdom). The cells were disrupted by sonication (Ultrasonics Inc.) four times for 1 min each time, with 1 min on ice between sonications. After removal of cell debris, the protein samples were precipitated by the addition of 5 volumes of ice-cold acetone (analytical grade) and incubated overnight at −20°C. The fine precipitate was pelleted by centrifugation at 4°C (14,000 rpm; 20 min).
Analysis of cellular proteins by 2-D gel electrophoresis.
For isoelectric focusing, the protein precipitate was solubilized in a rehydration solution (8 M urea, 2 M thiourea, 4% [wt/vol] CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 50 mM dithiothreitol, 0.5% pharmalytes, pH 3 to 10). About 400 μg protein was loaded on Immobiline DryStrips (IPG strips; 18 cm; Amersham Biosciences) with linear gradients of pH 3 to 11. The strips were passively rehydrated overnight in a reswelling tray under low-viscosity paraffin oil and then subjected to isoelectric focusing with the following voltage/time profile: linear increase from 0 to 500 V for 1,000 V · h, 500 V for 2,000 V · h, linear increase from 500 to 3,500 V for 10,000 V · h, and a final phase of 3,500 V for 35,000 V · h to a total of 56,000 V · h. The strips were equilibrated twice for 15 min in an equilibration solution (50 mM Tris-HCl, pH 6.0, 6 M urea, 30% [vol/vol] glycerol, 4% [wt/vol] sodium dodecyl sulfate [SDS]) containing 3.5 mg · ml−1 dithiothreitol for the first step and 45 mg · ml−1 iodoacetamide in the second step. Second-dimension SDS-polyacrylamide gel electrophoresis was carried out with 12.5% SDS-polyacrylamide gels at 20 W per gel in a Multicell Protean II XL system (Bio-Rad). Routinely, proteins were visualized by silver staining. For identification of the protein spots, the gels were stained with PhastGel Coomassie R250 according to the manufacturer's instructions (Amersham Biosciences). Scanned images were analyzed with Phoretix software (Nonlinear Dynamics, United Kingdom). Three gels for each condition were analyzed, and only those spots showing similar patterns (i.e., up- or downregulation) were labeled.
Protein identification by peptide mass fingerprinting.
Selected protein spots were excised from preparative 2-D gels, and the slices were destained and digested with trypsin (Promega). Peptide samples were extracted, purified using ZipTip (μC18) pipette tips according to the manufacturer's instructions (Millipore), and eluted with 0.1% trifluoroacetic acid prepared in 50% (vol/vol) acetonitrile. Mass spectrometric analysis of the tryptic digests was accomplished by a hybrid quadrupole time-of-flight instrument (QSTAR; Applied Biosystems, CA) equipped with a zero matrix-assisted laser desorption ionization source. Fragment ion data generated by information-dependent acquisition via QSTAR were searched against the protein sequence database using the Mascot database search.
Enzyme assays.
PGA activity was measured by a published procedure using l-aspartic β-hydroxymate as the substrate (2). One unit of PGA activity is the amount of enzyme that hydrolyzes 1 μmol of substrate per min under these conditions. Aspartase activity was measured by a UV spectrophotometer as described elsewhere (24). One unit of aspartase activity is the amount of enzyme that converts 1 μM l-aspartate to fumarate per min under these conditions. Glutamate dehydrogenase (GDH) activity was determined as described before by monitoring NADPH consumption in a continuous enzymatic assay (16). Glutamine synthetase was assayed by its γ-glutamyltransferase activity according to the method of Meyer and Stadtman (9). Glutamate synthase (GOGAT) activity was also determined by monitoring NADPH consumption (7).
Determination of intracellular levels of amino acids.
Amino acid levels in the media or within the cells were analyzed by high-performance liquid chromatography (HPLC) after precolumn derivatization with o-phthalaldehyde according to the protocol of Suresh et al. (20). The cells were collected by centrifugation (14,000 rpm for 10 min). The resulting supernatants were used to assay residual amino acid levels, whereas the cell pellets were used to determine intracellular amino acid concentrations. The pellets were washed, suspended in 200 μl 80% ethanol, and incubated at 100°C for 10 min. After centrifugation at 14,000 rpm, 100-μl samples were derivatized by adding freshly prepared o-pthalaldehyde reagent (25 μl) and 50 μl sodium borate buffer (0.5 M, pH 10.5). After incubation at 25°C for 10 min, the volume was adjusted to 1 ml with water. Standard amino acids were treated in the same way; 50-μl volumes of the derivatized samples were analyzed by reversed-phase HPLC using a LichroSphere Select B column (Merck, Darmstadt, Germany) fitted to a Merck-Hitachi HPLC system. Fluorescence was detected by a Shimadzu RF-551 fluorescence detector with excitation and emission wavelengths set to 330 nm and 450 nm, respectively.
RESULTS
aau-deficient mutants of P. putida KT2440 (KTaauR and KTaauS) were obtained by an approach described previously (18). Briefly, frameshift mutations were introduced into aauR (PP1066) or aauS (PP1067), and the mutant genes were reintroduced into the P. putida KT2440 genome via homologous recombination (see Materials and Methods). The resulting transconjugants and wild-type cells were characterized with respect to their growth behaviors and a number of other physiological and biochemical properties.
Growth properties of aau deletion mutants on different carbon and nitrogen sources.
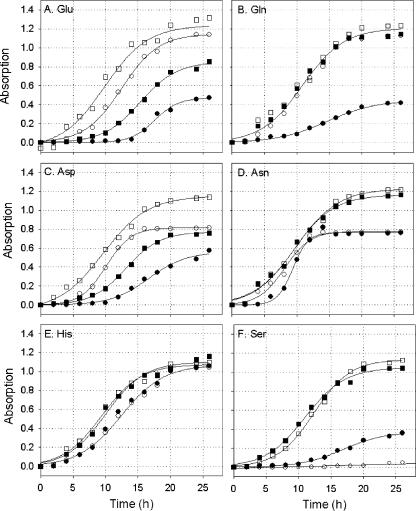
Acidic amino acids are excellent growth substrates for P. putida KT2440 (16). This ability, however, depends on a functional aau system (see Table S1 in the supplemental material) (Fig. 2). Wild-type cells and KTaauR grew equally well on M9 medium supplemented with glucose and NH4+ (data not shown). Growth of the mutant on Glu, Gln, and Asp, on the other hand, was impaired when these amino acids were provided as sole sources of carbon and nitrogen (Fig. 2A to C). Under these conditions, the aau-deficient cells did not proliferate at all for 10 to 15 h after transfer. In addition, the final cell densities attained by the mutants after 24 h were lower than those of wild-type cells. Glucose as an additional carbon source partially or fully restored growth of the KTaauR mutant on Glu, Gln, and Asp. By contrast, NH4+ supplied as an additional nitrogen source did not significantly improve the growth behavior of the mutant (data not shown). Asn or Asn-glucose supported normal growth of KTaauR (Fig. 2D). Likewise, no significant differences in growth rates between the wild type and the mutant were seen with most of the other amino acids tested (see Table S1 in the supplemental material) (Fig. 2E). One interesting exception was serine. When serine was supplied as the sole source of carbon, the KTaauR mutant grew markedly better than the wild type (Fig. 2F). Succinate as an additional carbon source had effects similar to those of glucose, i.e., it significantly improved the growth rate of the KTaauR mutant when Glu or Asp was provided as a nitrogen source (data not shown). Comparable results were obtained with the KTaauS mutant, although this strain exhibited longer doubling times on amino acids (data not shown).
FIG. 2.
Growth of Pseudomonas putida KT2440 (open symbols) and the KTaauR deletion mutant (closed symbols) on amino acids (circles) or amino acids supplemented with glucose (squares). Cells were pregrown overnight on M9+ medium (see Materials and Methods) and then transferred to M9 minimal medium supplemented with 10 mM amino acids as the sole carbon and nitrogen sources or amino acids with 22 mM glucose. Relative growth rates were estimated from absorption measurements at 595 nm between 0 and 26 h after transfer to fresh medium. The solid lines are fits of a modified logistic model to the data (26).
Identification of aau-regulated proteins.
In order to identify proteins that depend on the aau system for expression, the proteomes of P. putida KT2440 and the KTaauR mutant strain were compared by 2-D polyacrylamide gel electrophoresis. Wild-type and mutant cells grown on NH4+ as a nitrogen source exhibited very similar protein patterns (data not shown). During growth of the KTaauR mutant on Glu, a number of proteins were downregulated compared to the wild-type strain (see Fig. S1a and b, spots R1 to R4, in the supplemental material). In another experiment, the protein expression profiles of KTaauR grown on Glu on one hand or glucose-NH4+ on the other were examined. In that case, three major spots (see Fig. S1b and c, spots D1 to D3, in the supplemental material) were downregulated on glucose-NH4+. All of these proteins could be identified and assigned to proteins deduced from the P. putida KT2440 genome. Table 2 lists their names, the encoding genes, and their known or putative functions, as well as their isoelectric points and masses as calculated from the genome data.
TABLE 2.
Characteristics of differentially expressed proteins in P. putida KT2440 and an isogenic aauR mutant during growth on glutamate and glucose-NH4+
| Growth conditionsc | Spot | Identification | No. of matching peptides | Locus | pIcalca | Masscalc (kDa)b | Function |
|---|---|---|---|---|---|---|---|
| Glutamate | R1 | Carboxyphosphonoenolpyruvate phosphonomutase | 13 | PP1389 | 5.26 | 31.9 | Unknown |
| R2 | Conserved hypothetical protein | 11 | PP3089 | 5.42 | 19.5 | Unknown | |
| R3 | Amino acid ABC transporter, periplasmic amino acid binding protein | 20 | PP1071 | 8.61 | 33.5 | Amino acid transport | |
| R4 | PGA | 22 | PP2453 ansB | 6.9 | 38.6 | Hydrolysis of Gln and Asn | |
| Glucose-NH4+ | D1 | Branched-chain amino acid ABC transporter, periplasmic amino acid binding protein | 15 | PP4867 | 5.93 | 40.3 | Amino acid transport |
| D2 | Branched-chain amino acid ABC transporter, periplasmic amino acid binding protein | 15 | braC (PP1141) | 6.02 | 39.4 | Amino acid transport | |
| D3 | General amino acid ABC transporter, periplasmic binding protein | 17 | aapJ (PP1297) | 5.84 | 36.4 | Amino acid transport |
pIcalc, calculated isoelectric point.
Masscalc, calculated mass.
Spots were downregulated during growth of the KTaauR mutant.
Two of the protein spots no longer expressed in the mutant were identified as glutaminase/asparaginase (R4; PP2453) and the periplasmic solute-binding protein associated with the Glu/Asp transporter encoded by PP1070 to PP1068 (R3). As mentioned above, both proteins are strongly induced by glutamate and repressed by glucose (17). Spot R1 was identified as the protein product of PP1389 (annotated as carboxyphosphonoenolpyruvate phosphonomutase), while spot R2 corresponds to a conserved protein of unknown function. The spots expressed during growth of the mutant on Glu but downregulated on glucose-NH4+ (spots D1 to D3) all correspond to solute-binding proteins of amino acid transporters of the ABC type, including BraC and AapJ. The putative functions of these transporters in the KTaauR mutant are discussed below.
Role of the aau system in amino acid uptake.
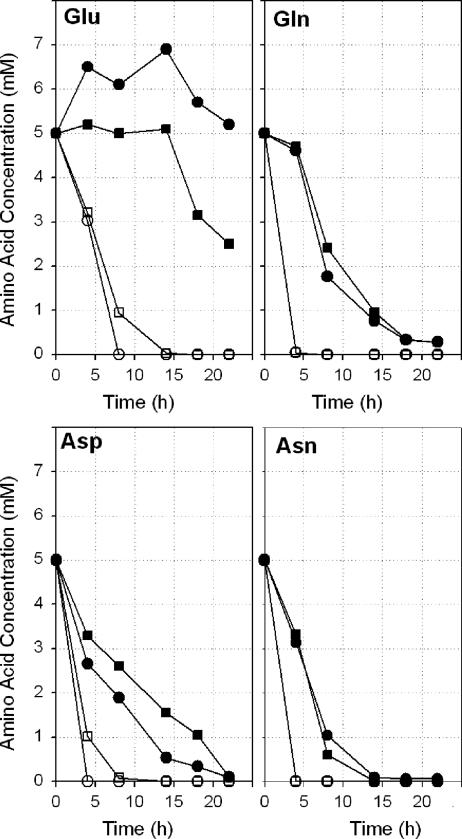
In another experiment, the uptake of acidic amino acids and their amides by wild-type P. putida KT2440 and the KTaauR mutant was examined. After cells pregrown on M9+ were transferred to amino acid-containing medium, the concentrations of the respective amino acids in the medium were monitored as a function of time. As shown in Fig. 3, the wild-type strain consumed all of the amino acids tested within 5 to 8 h after transfer (note that in these experiments the initial cell densities were markedly higher than those in Fig. 2). In the KTaauR deletion mutant, the consumption of Gln, Asp, and Asn was moderately delayed, whereas Glu was not taken up at all until growth of the mutant commenced after 15 to 20 h. In the absence of glucose, the mutant cells even secreted glutamate into the medium, as indicated by an increase of the initial Glu concentration by about 30% (Fig. 3).
FIG. 3.
Amino acid uptake by P. putida KT2440 (open symbols) and the KTaauR mutant (closed symbols) during growth on 5 mM amino acids (circles) and on 22 mM glucose with amino acids (squares). The cells were pregrown on M9+ medium (see Materials and Methods) overnight and then transferred to M9 minimal medium supplemented with 5 mM amino acids as the sole carbon and nitrogen sources and amino acids in combination with 22 mM glucose. High initial cell densities (A600 = 0.5 to 0.6) were used for these experiments. The concentrations of the amino acids remaining in the medium are shown as a function of incubation time (see Materials and Methods).
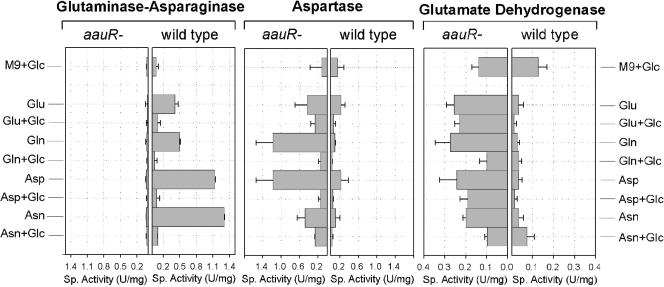
Effects of the aauR deletion on key enzymes of acidic amino acid metabolism.
In order to identify other targets of the aau system, the specific activities of key enzymes of Glu and Asp metabolism were assayed during growth of wild-type cells and KTaauR on various sources of carbon and nitrogen (Fig. 4). The enzymes examined included PGA, glutamate dehydrogenase, glutamine synthetase, GOGAT, and aspartase. As described previously (16), the PGA activity of wild-type cells is enhanced by Asn, Asp, Gln, and Glu and repressed by glucose. By contrast the KTaauR mutant failed to express significant amounts of PGA. The activities of aspartase and glutamate dehydrogenase, on the other hand, were elevated up to 10-fold in the mutant, regardless of the nitrogen and carbon sources supplied. Note that aspartase, like PGA, was subject to catabolite repression by glucose. This effect has been observed before with E. coli aspartase (11). The activities of glutamine synthetase and glutamate synthase were almost the same in wild-type cells and KTaauR and did not significantly respond to glucose (data not shown).
FIG. 4.
Specific activities of key enzymes of glutamate and aspartate metabolism in P. putida KT2440 (wild type) and KTaauR mutant cells. The cells were grown to late exponential phase (A600 = 0.6 to 0.8) in M9 medium supplemented with the indicated amino acids (10 mM each) as the sole carbon and nitrogen sources and 22 mM glucose plus the amino acids as nitrogen sources. Crude cell extracts were assayed for the specific activities (U/mg protein) of PGA, aspartase, and GDH, as described in Materials and Methods. The error bars indicate standard deviations. The means ± standard deviations were determined from at least three independent experiments.
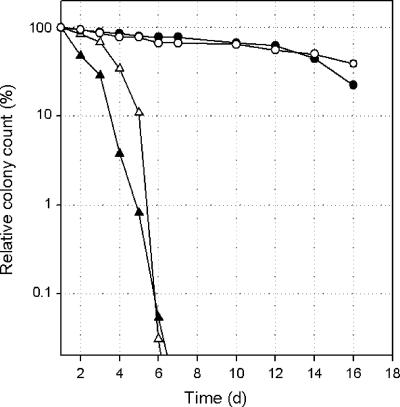
aauR and gltB knockout mutations result in similar phenotypes.
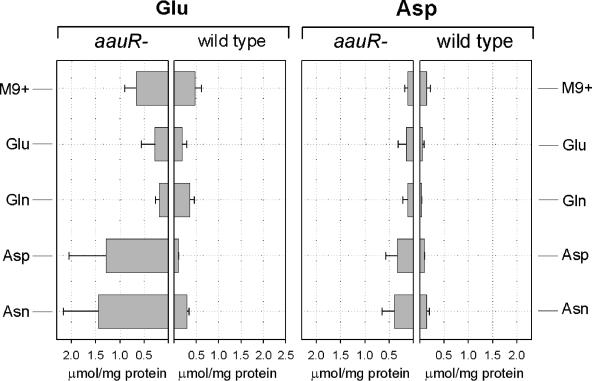
As shown previously, P. putida transposon mutants defective in gltB (a gene encoding one of the subunits of GOGAT) exhibit greatly reduced survival times under conditions of nitrogen starvation (18). A similar effect was seen with strain KTaauR, which rapidly lost viability after a few days of starvation, while more than 50% of the wild-type cells survived for several weeks under these conditions (Fig. 5). A second, unexpected feature of our gltB deletion mutants was the accumulation of intracellular Glu. As shown in Fig. 6, a similar effect was seen with KTaauR. The intracellular concentrations of Glu and Asp were both markedly enhanced during growth on Asp or Asn, whereas no such accumulation was seen with wild-type cells, regardless of the nitrogen source supplied. While the intracellular Glu concentration reached values of 2 μmol/mg of cellular protein, the concentrations of the amides Asn and Gln remained much lower (0.01 to 0.05 μmol/mg) in both the wild type and the mutant and did not significantly vary with the type of nitrogen source provided (data not shown).
FIG. 5.
Survival of P. putida KT2440 and the KTaauR deletion mutant in M9 medium containing 5 mM aspartate (open symbols) or glutamate (closed symbols). The numbers of viable cells (as a fraction of original cell number) are shown as a function of incubation time for P. putida KT2440 (circles) and the KTaauR mutant (triangles).
FIG. 6.
Intracellular concentrations of glutamate and aspartate (expressed as μmol per mg of cellular protein) as a function of time after transfer to M9+ medium or to M9 minimal medium containing 10 mM each of Glu, Gln, Asp, and Asn. The error bars indicate standard deviations. The means ± standard deviations were determined from at least three independent experiments.
AauR binds to the putative promoter elements of two target genes.
In order to confirm that the observed defects of KTaauR with respect to the utilization of Glu, Asp, and Gln were indeed the consequences of aauR inactivation, an intact copy of aauR was cloned as a BamHI/HindIII fragment into plasmid pMMB67EH. The resulting plasmid was electroporated into KTaauR to generate strain KTaauR+. In addition, reporter lacZ gene fusions that contained the putative promoter regions of PP2453 (ansB) and PP1071 were generated. As described above, both genes are inducible by Glu and also are targets of the aau system. The β-galactosidase activities of exponentially growing cultures in Glu-containing media were measured (Miller units). As shown in Table 3, the galactosidase activities of KTansB::lacZ and KTABC::lacZ were reduced by about 35% compared to the wild type. In the complemented aauR+ strain, these values were higher, reaching about 80% of the wild-type activities. Like wild-type KT2440, the complemented aauR+ strain efficiently utilized Asp and Glu as sole sources of carbon and nitrogen, indicating that the inactivation of aauR was the cause of the observed metabolic defect (data not shown).
TABLE 3.
Activities of the ansB and ABC transporter promoters in P. putida KT2440, its isogenic KTaauR mutant, and the KTaauR+ strain
| Promoter-lacZ fusion | β-Galactosidase activitya
|
||
|---|---|---|---|
| P. putida KT2440 | KTaauR mutant | KTaauR+ mutant | |
| KTansB::lacZ | 6,284 ± 120 | 4,156 ± 80 | 5,051 ± 89 |
| KTABC::lacZ | 4,887 ± 114 | 2,986 ± 90 | 3,886 ± 105 |
| pDNlacΩ (control) | 280 ± 70 | 214 ± 87 | 260 ± 83 |
Values are means ± standard deviations.
Structural relationship between AauS-AauR and other two-component systems.
Searches in the Comprehensive Microbial Resource database using AauS and AauR as query sequences revealed the existence of a number of orthologous systems in related organisms and at least two paralogues in P. putida KT2440. Closely related orthologues in other pseudomonads include the two-component systems encoded by PA1337-PA1336 in P. aeruginosa PAO1 (Fig. 1), the system PFL4875-PFL4876 of Pseudomonas fluorescens Pf5, and PSPTO4175-PSPTO4176 of Pseudomonas syringae DC3000. Fig. S2 in the supplemental material shows alignments of some of the conserved “boxes” regularly found in two-component systems to illustrate the high degree of sequence similarity between these systems. The closest paralogues of the aau system in P. putida KT2240 are the two-component systems encoded by PP1402-PP1401 and PP0264-PP0263. While the function of the latter is unknown, the system encoded by PP1402-PP1401 was annotated as dct (for dicarboxylate transport). The dct regulatory system controls the uptake of dicarboxylic acids, such as succinate, fumarate, malate, and aspartate, and has been most thoroughly studied in rhizobia (for a review, see reference 25). From Fig. S2 in the supplemental material, it is evident that, as judged by sequence similarity, both aau paralogues are more distantly related to P. putida KT2440 aauS-aauR than to the putative aau systems of P. aeruginosa, P. fluorescens, and P. syringae.
DISCUSSION
The P. putida KT2440 genome encodes more than 130 sensor kinases and response regulators that make up at least 50 different two-component systems. At present, most of these putative regulatory systems have not been associated with any defined function. Here, we show that the two-component system encoded by PP1067-PP1066 (aauS-aauR) is required for efficient growth of P. putida KT2440 on acidic amino acids (Glu and Asp) and their amides (Gln and Asn). According to our present results, the system has at least two targets: it controls the expression of PGA, and thus the hydrolysis of exogenous Asn and Gln in the periplasmic space. In addition, aau stimulates the uptake of periplasmic glutamate produced by PGA by enhancing the expression of an ABC transporter encoded by PP1071 to PP1068. Finally, the aau system directly or indirectly affects the activities of key enzymes of acidic amino acid metabolism, such as aspartase and glutamate dehydrogenase. As far as P. putida KT2440 is concerned, AauS-AauR is the first two-component system shown to participate in acidic amino acid utilization.
The aau system is required for glutamate utilization.
The AauR-AauS system seems to be selective for the metabolism of acidic amino acids and their amides, as the growth of KTaauR and KTaauS on other amino acids was indistinguishable from that of wild-type cells. The main effect of aau inactivation on growth was a very extended lag phase that lasted for 15 h or more before growth started, with reduced rates and yields. We assume that during this lag another yet-unidentified regulatory system is activated that partially compensates for the loss of aau function, enabling less-efficient growth on acidic amino acids.
The present data further suggest that aau mutant strains are impaired mainly in the ability to utilize acidic amino acids as a source of carbon, since additional carbon sources, like glucose or succinate, partially restored growth, whereas a supply of NH4+ did not improve growth rates. The fact that succinate is still efficiently utilized by KTaauR and KTaauS indicates that aau is functionally distinct from the dctBD system, which regulates dicarboxylate uptake and metabolism in various bacteria. In rhizobia, for instance, mutations in any of the dct genes result in a loss of the ability to transport and utilize C4-dicarboxylates (3).
Target genes.
The proteomics data presented here, together with the results of reporter gene fusions, indicate that aauR acts as a σ54-dependent transcriptional activator of at least two genes or operons, i.e., ansB, encoding PGA, and PP1071 to PP1068, which encodes a glutamate transporter of the ABC type. As shown before, the expression of these two genes was also diminished in an rpoN-deficient mutant of strain KT2440 (17), indicating that the alternate sigma factor σ54 is required for their expression, as well. Additional evidence for this assumption is provided by the finding that the ansB (PP2543) promoter contains the −12/−24 sequence motif characteristic of σ54-responsive genes (5). A similar motif was detected by analysis of the putative promoter sequence of PP1071 using PromScan (17). While our proteomics data (see Fig. S1 in the supplemental material) and the activity assay results shown in Fig. 4 indicate that inactivation of aauR largely abolishes ansB expression, the activities of the lacZ reporter gene fusions were only moderately reduced (Table 3). Comparative experiments with KT2440 genome arrays could resolve this discrepancy and may also help to identify further aau-dependent genes.
In contrast to the proteins already described, a third Glu-inducible and aau-dependent protein (spot R1 in Fig. S1 in the supplemental material) has no recognizable role in amino acid metabolism. It was annotated as putative carboxyphosphonoenolpyruvate phosphonomutase, an enzyme involved in the biosynthesis of the antibiotic bialaphos in Streptomyces hygroscopicus (6). As there is no evidence that pseudomonads synthesize bialaphos, this annotation remains doubtful.
Effects of AauR-AauS on glutamate and aspartate metabolism.
At present, very little is known about the transporters involved in the uptake of acidic amino acids by P. putida. Here, we show that the ABC transporter encoded by PP1071 to PP1068 is essential for the uptake of glutamate. Figure 3 suggests that the initial failure of KTaauR to grow on Glu as a sole source of carbon and nitrogen is due to its inability to take up glutamate. Mutants in which the periplasmic glutamate-binding protein encoded by PP1071 was inactivated failed to grow on both Glu and Gln for up to 24 h, while growth on Asp and Asn was largely unaffected (B. Singh, unpublished data).
The intracellular accumulation of glutamate (Fig. 6) and the secretion of Glu into the medium (Fig. 3) were unexpected features of KTaauR. However, similar effects have been described for Rhizobium leguminosarum by Hosie and coworkers and Walshaw and coworkers (4, 22). They found that mutants with defects in tricarboxylic acid (TCA) cycle enzymes exhibited greatly elevated intracellular glutamate levels and also excreted glutamate. These steps are thought to constitute an overflow pathway with the function of removing excess carbon and reducing equivalents. Interestingly, in R. leguminosarum, the highest intracellular Glu accumulation was observed with aspartate as the nitrogen source (13). As shown by Fig. 6, the same holds for P. putida KT2440. This finding and the fact that the levels of both aspartase and GDH are elevated in KTaauR indicate that a similar overflow pathway operates in that mutant. Aspartase may participate in such a pathway by converting aspartate to the TCA cycle intermediate fumarate, while GDH, the equilibrium state of which favors amino acid synthesis (8), forms Glu from α-ketoglutarate. The finding that, in P. putida, significant excretion of glutamate occurred only in the presence of glucose (Fig. 3) may be rationalized by the fact that glucose degradation replenishes the TCA cycle to create additional 2-oxoglutarate.
A final piece of evidence for the existence of an overflow pathway in P. putida KT2440 aau mutants is provided by the observation that KTaauR growing on Glu (but not on glucose-NH4+) strongly expresses solute-binding proteins associated with amino acid ABC transporters (see Fig. S1, spots D1 to D3, in the supplemental material). Two of these (BraDEFG, encoded by PP1140 to PP1137, and a transporter encoded by PP4866 to PP4863) are thought to transport branched-chain amino acids, while the third (AapQMP, encoded by PP1298 to PP1300) is annotated as a general amino acid transporter. As shown by Walshaw and Poole (21), it is transporters of the Aap and Bra types that mediate the selective efflux of glutamate from R. leguminosarum. Experiments to identify additional target genes of the aau system in intermediary metabolism that may explain the observed phenotype are being conducted in our laboratory.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Deutsche Forschungsgemeinschaft via SFB 395.
We thank Andrej Hasilik, Marburg, Germany, for providing laboratory facilities to carry out 2-D experiments. The cooperation of Rajesh Chandramohandas in the 2-D studies is gratefully acknowledged.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bagdasarian, M., and K. N. Timmis. 1982. Host-vector systems for gene cloning in Pseudomonas. Curr. Top. Microbiol. Immunol. 96:47-67. [DOI] [PubMed] [Google Scholar]
- 2.Derst, C., A. Wehner, V. Specht, and K. H. Röhm. 1994. States and functions of tyrosine residues in Escherichia coli asparaginase II. Eur. J. Biochem. 224:533-540. [DOI] [PubMed] [Google Scholar]
- 3.Engelke, T., D. Jording, D. Kapp, and A. Puhler. 1989. Identification and sequence analysis of the Rhizobium meliloti dctA gene encoding the C4-dicarboxylate carrier. J. Bacteriol. 171:5551-5560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hosie, A. H., D. Allaway, M. A. Jones, D. L. Walshaw, A. W. Johnston, and P. S. Poole. 2001. Solute-binding protein-dependent ABC transporters are responsible for solute efflux in addition to solute uptake. Mol. Microbiol. 40:1449-1459. [DOI] [PubMed] [Google Scholar]
- 5.Hüser, A., U. Klöppner, and K. H. Röhm. 1999. Cloning, sequence analysis, and expression of ansB from Pseudomonas fluorescens, encoding periplasmic glutaminase/asparaginase. FEMS Microbiol. Lett. 178:327-335. [DOI] [PubMed] [Google Scholar]
- 6.Lee, S. H., T. Hidaka, H. Nakashita, and H. Seto. 1995. The carboxyphosphonoenolpyruvate synthase-encoding gene from the bialaphos-producing organism Streptomyces hygroscopicus. Gene 153:143-144. [DOI] [PubMed] [Google Scholar]
- 7.Meister, A. 1985. Glutamate synthase from E. coli, Klebsiella aerogenes and Saccharomyces cerevisiae. Methods Enzymol. 113:327-337. [DOI] [PubMed] [Google Scholar]
- 8.Merrick, M. J., and R. A. Edwards. 1995. Nitrogen control in bacteria. Microbiol. Rev. 59:604-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meyer, J. M., and E. R. Stadtman. 1981. Glutamine synthetase of pseudomonads: some biochemical and physicochemical properties. J. Bacteriol. 146:705-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 11.Nishimura, N., and M. Kisumi. 1984. Aspartase-hyperproducing mutants of Escherichia coli B. Appl. Environ. Microbiol. 48:1072-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pridmore, R. D. 1987. New and versatile cloning vectors with kanamycin-resistance marker. Gene 56:309-312. [DOI] [PubMed] [Google Scholar]
- 13.Reid, C. J., D. L. Walshaw, and P. S. Poole. 1996. Aspartate transport by the dct system in Rhizobium leguminosarum negatively affects nitrogen-regulated operons. Microbiology 142:2603-2612. [DOI] [PubMed] [Google Scholar]
- 14.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Biotechnology 1:784-791. [Google Scholar]
- 16.Sonawane, A., U. Klöppner, C. Derst, and K. H. Röhm. 2003. Utilization of acidic amino acids and their amides by pseudomonads: role of periplasmic glutaminase-asparaginase. Arch. Microbiol. 179:151-159. [DOI] [PubMed] [Google Scholar]
- 17.Sonawane, A., U. Klöppner, S. Hövel, U. Völker, and K. H. Röhm. 2003. Identification of Pseudomonas proteins coordinately induced by acidic amino acids and their amides: a two-dimensional electrophoresis study. Microbiology 149:2909-2918. [DOI] [PubMed] [Google Scholar]
- 18.Sonawane, A. M., and K. H. Röhm. 2004. A functional gltB gene is essential for utilization of acidic amino acids and expression of periplasmic glutaminase/asparaginase (PGA) by Pseudomonas putida KT2440. Mol. Genet. Genomics 271:33-39. [DOI] [PubMed] [Google Scholar]
- 19.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 20.Suresh, S. V., M. M. Shareef, A. P. V. Shetty, and K. T. Shetty. 2002. HPLC method for amino acids profile in biological fluids and inborn metabolic disorders of aminoacidopathies. Indian J. Clin. Biochem. 17:7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walshaw, D. L., and P. S. Poole. 1996. The general l-amino acid permease of Rhizobium leguminosarum is an ABC uptake system that also influences efflux of solutes. Mol. Microbiol. 21:1239-1252. [DOI] [PubMed] [Google Scholar]
- 22.Walshaw, D. L., A. Wilkinson, M. Mundy, M. Smith, and P. S. Poole. 1997. Regulation of the TCA cycle and the general amino acid permease by overflow metabolism in Rhizobium leguminosarum. Microbiology 143:2209-2221. [DOI] [PubMed] [Google Scholar]
- 23.West, A. H., and A. M. Stock. 2001. Histidine kinases and response regulator proteins in two-component signaling systems. Trends Biochem. Sci. 26:369-376. [DOI] [PubMed] [Google Scholar]
- 24.Williams, V. R., and D. J. Lartigue. 1969. Aspartase. Methods Enzymol. 13:354-361. [Google Scholar]
- 25.Yurgel, S. N., and M. L. Kahn. 2004. Dicarboxylate transport by rhizobia. FEMS Microbiol. Rev. 28:489-501. [DOI] [PubMed] [Google Scholar]
- 26.Zwietering, M. H., I. Jongenburger, F. M. Rombouts, and K. van't Riet. 1990. Modeling of the bacterial growth curve. Appl. Environ. Microbiol. 56:1875-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.