Abstract
A polyphasic, culture-independent study was conducted to investigate the abundance and population structure of ammonia-oxidizing bacteria (AOB) in canal sediments receiving wastewater discharge. The abundance of AOB ranged from 0.2 to 1.9% and 1.6 to 5.7% of the total bacterial fraction by real-time PCR and immunofluorescence staining, respectively. Clone analysis and restriction endonuclease analysis revealed that the AOB communities influenced by the wastewater discharge were dominated by Nitrosomonas, were similar to each other, and were less diverse than the communities outside of the immediate discharge zone.
Tokyo Bay is a representative enclosed eutrophic bay in the southern Kanto region of Japan. More than 86% of the shoreline is reclaimed, and artificial structures, such as canals and landfill islands, occupy the inner bay area (29). These artificial structures often hamper water exchange and accelerate precipitation of sludge containing excess organic matter, heavy metals, and toxic compounds (16, 17, 29). These human activities negatively affect the sea-bottom environment and limit natural remediation in the coastal ecosystem (19).
Nitrification is essential to the nitrogen cycle in aquatic environments. When coupled with denitrification and/or anaerobic ammonium oxidation, it relieves the negative impacts of eutrophication through removal of nitrogen to the atmosphere as nitrous oxide or molecular nitrogen (5, 9, 38). However, nitrification is sensitive to environmental stress and contaminants (18, 23, 37). Ammonia-oxidizing bacteria (AOB) carry out the first, rate-limiting step of nitrification: conversion of ammonia to nitrite. The ecology and physiology of AOB are particularly difficult to study by conventional cultivation techniques because of their long generation times and low growth rates, which can result in underestimations of their numbers in the environment (21). Compared to the studies of AOB diversity in nature (1, 4, 6, 12, 13, 22), quantitative studies have been limited, especially for marine environments (11). Therefore, a rapid, culture-independent detection technique for AOB would be useful for the study of marine AOB. Through this study, we developed and combined molecular and novel immunofluorescence staining approaches to investigate the spatial distribution, abundance, and population structure of AOB in a canal area of Tokyo Bay that is heavily polluted by excess organic and nutrient loading from wastewater treatment plant discharges. Particularly important for this study was quantification of AOB because of the deficiency of quantitative studies of AOB populations in marine environments (11).
Sampling sites.
Water and sediment samples were collected on 28 May 2003 in Tokyo Bay (see Fig. S1A in the supplemental material). Six sampling sites were chosen to represent a range of environments both inside and outside of the Keihin Canal area in order to investigate the distribution and population structure of ammonia oxidizers (see Fig. S1B in the supplemental material). A large-scale wastewater treatment plant, Morigasaki Water Reclamation Center, discharges 1,130,000 tons day−1 of effluent through two drains located in the canal area. The designations of the sampling sites, such as canal points (C21, C23, and C24), bay points (B18 and B23), and Tokyo long-term beacon (TLB), correspond to some of the sampling stations employed by the Environmental Bureau of the Tokyo Metropolitan Government to regularly monitor water quality. Sampling was conducted at low tide (see Fig. S1C in the supplemental material).
Physicochemical profiles and nutrient concentrations in water and sediments.
Vertical profiles of physicochemical data in the water column were obtained using a sounder (Hydrolab Sonde 4a; Hach Environmental, Loveland, CO) (see Table S1 in the supplemental material). A simultaneous multidepth water sampler was used for collecting water samples for nutrient analysis (39). Water samples were filtered through cellulose-acetate syringe filters (pore size, 0.8 μm). Nutrient samples were analyzed using an autoanalyzer (TRAACS-800; Bran+Luebbe, Tokyo, Japan). Wastewater effluent was the main source of freshwater draining into the canal. Wastewater runoff dramatically influenced water chemistry near C23 and C24, in which sharp increases in NH4+ and NO3− concentrations, respectively, were observed in the surface water. Ammonium was discharged mainly at C23 (318.5 μM NH4+-N), whereas NO3− was discharged mainly at C24 (251.1 μM NO3−-N) because of the difference of wastewater treatment systems. Moreover, the precipitation of particulate organic matter is probably accelerated at these sites as a result of the tidal flow. Consequently, heavy loading of organic matter was recorded in sediment samples collected from these sites (see Table S2 in the supplemental material).
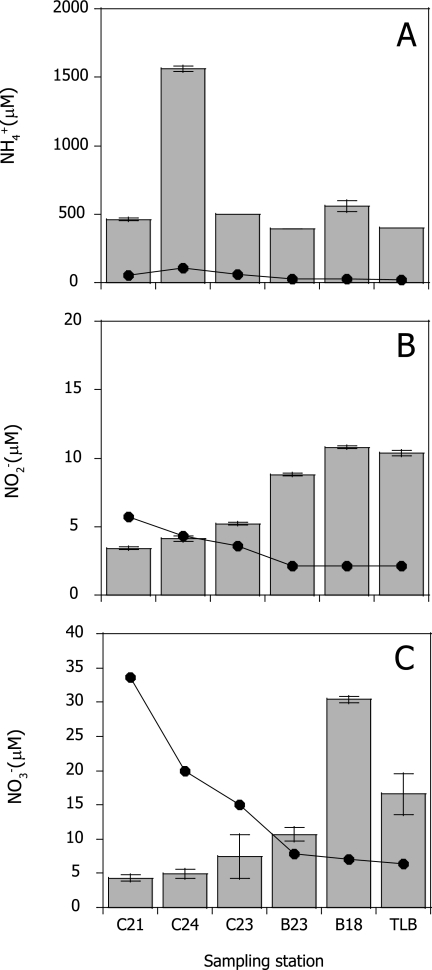
Sediment samples were collected using a core sampler (110-mm inner diameter); triplicate subcores (40-mm inner diameter) were taken from each site. The top 2-cm section of sediment was sliced from each core and used for nucleic acid extractions and determination of sediment characteristics (39) (see Table S2 in the supplemental material). The dominant nitrogen compound in porewater was NH4+ (see Fig. S1 in the supplemental material). The highest NH4+ concentration was detected in the interstitial water sample collected at C24 (1,559 μM), where the oxygen saturation level of bottom water was 5.1% during our sampling period (see Table S1 in the supplemental material). The concentration of NH4+ in the porewater was greater than the NH4+ concentration in the bottom water (Fig. 1A). The bottom water NH4+ concentration correlated with that of NH4+ in porewater (r2 = 0.75; P < 0.05) due to the diffusion of NH4+ in porewater to the bottom water (Fig. 1A). The concentrations of NO2− and NO3− in the bottom water were high inside the canal and decreased toward the outside of the canal due to the diffusion of river and wastewaters (Fig. 1B and C). Both were correlated strongly (r2 = 0.98; P < 0.01). On the contrary, the concentrations of NO2− and NO3− in the porewater were low inside the canal and increased toward the outside of the canal, possibly due to the nitrification activity. The highest NO2− and NO3− concentrations in the porewater were detected at B18, where we observed significantly lower percentages of total organic carbon and nitrogen and a large sand content (41.2%) compared to neighboring sites (P < 0.01) due to an unimpeded water flow from the Sumida River (see Fig. S1 and Table S2 in the supplemental material).
FIG. 1.
Concentrations of ammonium (A), nitrite (B), and nitrate (C) in porewater (bars) and bottom water (lines). Error bars indicate standard deviations of triplicate measurements.
RFLP analyses.
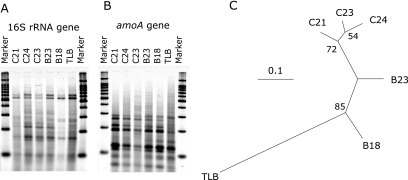
To investigate the heterogeneity and spatial variation of AOB community structures in marine sediments, restriction fragment length polymorphisms (RFLP) analysis was carried out for PCR-amplified 16S rRNA (Fig. 2A) and amoA genes (Fig. 2B) using the method of Urakawa and colleagues (39). A total of 23 informative sites, 12 from 16S rRNA genes and 11 from amoA genes, were used to generate a distance matrix based on the presence and absence of DNA bands on the gel. A dendrogram was constructed from the distance matrix by the neighbor-joining (NJ) method using the PAUP* software package (version 4.0b10) (Fig. 2C). Results show that the AOB communities receiving the canal discharge were similar to each other and were less diverse than the communities outside of the immediate discharge zone. The RFLP patterns among the canal sites were similar (87.0 to 91.3% similarity) and were clustered together with a 72% bootstrap value (Fig. 2C). This result showed clear contrast with the previous report by Urakawa and colleagues (39), in which the change of AOB communities was apparent along an environmental gradient created by river outflow.
FIG. 2.
RFLP patterns of PCR-amplified 16S rRNA (A) and amoA (B) genes and the neighbor-joining dendrogram deduced from RFLP data (C). HhaI and HaeIII were mixed and used for digestion of 16S rRNA genes, and MseI and RsaI were mixed and used for amoA genes. The bar represents 10% divergence. Numbers at the branch nodes are bootstrap values obtained from neighbor-joining analysis; only values greater than 50% are indicated.
In general, oxygen concentration, ammonium concentration, and salinity are thought to be extremely important environmental parameters that affect the nitrification rate and determine the nitrifier community (10, 31). Recent reports revealed that the population shift of dominant AOB occurred in the estuarine region with the sharpest gradients in salinity (4, 10, 14). The range of salinity values detected in this study (see Table S1 in the supplemental material) was smaller than that found in other studies (4, 10, 12, 14, 39). Here we investigated the effect of low oxygen and high NH4+ concentrations and found no major difference in the AOB population at C24, where anomalous low oxygen and high NH4+ concentrations were observed (see Table S1 in the supplemental material) due to active microbial degradation of fresh organic matter (Fig. 1A). These results implied that even if the ammonium gradient we observed was large among AOB habitats, it did not strongly influence the AOB population in canal sediments. Our results were similar to those found by Prinčič and colleagues (31), who reported that an extremely high concentration of NH4+ (214 mM) selected a novel nitrifier population, whereas moderate concentrations (3.6 to 71 mM) did not engender a population change.
Culture-independent identification of dominant phylogenetic groups.
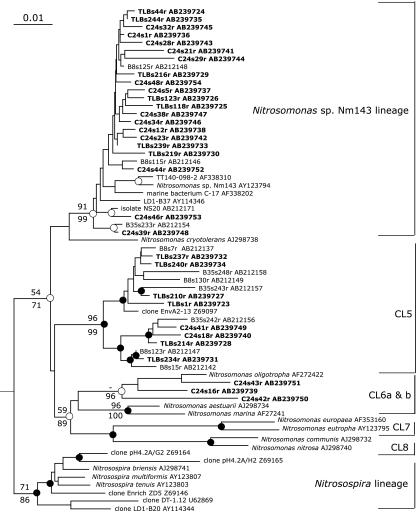
Two 16S rRNA gene libraries were constructed with a primer set of β-AMOf (25) and β-AOB1236r (39) using the reverse transcription-PCR method to determine the major AOB groups that potentially maintain a high cellular rRNA content, as described previously (39). A total of 121 clones from the two clone libraries were analyzed: 49 and 72 clones, respectively, from sites C24 and TLB. First, partial (ca. 500 bp) sequences were determined and compared to available published sequences using nucleotide-nucleotide BLAST to determine tentative phylogenetic affiliation of clones. Second, 32 representative sequences (19 and 13 clones, respectively, from sites C24 and TLB) were determined (ca. 1.1 kb) and used for phylogenetic analyses (maximum-likelihood [ML], maximum parsimony, and NJ). Phylogenetic analyses revealed that all sequences belonged to the Nitrosomonas lineage, but no Nitrosospira sequences were detected (Fig. 3). The clone composition of the offshore site, TLB, was similar to clone compositions obtained from a previous survey of Tokyo Bay sediments (39). The majority of clone sequences were grouped into the Nitrosomonas sp. strain Nm143 lineage, which has been observed exclusively from estuarine or coastal marine habitats and has adapted to conditions ranging from high oxygen concentrations to low oxygen concentrations (33). The second-largest group belongs to cluster 5, which was proposed by Stephen and colleagues (36) and has been found exclusively in marine environments, especially organic material-rich environments (1, 10, 22, 26, 39). Our results were consistent with the results of recent stable isotope probing analysis, in which Freitag and colleagues (14) first confirmed the activity of the Nitrosomonas sp. strain Nm143 group within an estuarine sediment ecosystem, while Nitrosospira cluster 1 was found in DNA libraries but was undetectable by stable isotope probing analysis.
FIG. 3.
Neighbor-joining tree of the 16S rRNA genes of ammonia-oxidizing bacteria belonging to the Betaproteobacteria. The clone sequences reported in this study are depicted in bold. For outgroups, two Betaproteobacteria, Herbaspirillum sp. isolate G8A1 and Dechloromonas sp. strain HZ, are used (not shown). The total alignment length for the analysis was 1,103 bp. The bar represents 0.01 estimated substitutions per site. The branch nodes supported by three phylogenetic analyses (ML, maximum parsimony, and NJ) are indicated as closed circles. The white circle at the branch node indicates the support of topology by at least two phylogenetic analyses. Numbers at branch nodes are quarter puzzling support values obtained from ML analysis (above the branch) and bootstrap values obtained from NJ analysis (below the branch); only values greater than 50% and important to define the phylogenetically major clusters (CL) are indicated. Designations of clusters are adapted from the work of Stephen and colleagues (36) and Purkhold and colleagues (32). The Nitrosomonas sp. strain Nm143 lineage was proposed by Purkhold and colleagues (32).
We anticipated obtaining many Nitrosomonas sequences that are common in wastewater treatment plants, because a considerable amount of wastewater effluent containing nitrifying bacteria has been discharged into the Keihin Canal area. Limpiyakorn and colleagues (24) reported that Nitrosomonas oligotropha and Nitrosomonas communis were major AOB species in the wastewater treatment plants in the Tokyo area. This result resembled those of previous reports that specifically examined other wastewater treatment plants and their effluents (7, 8, 15). Moreover, it has been reported that N. oligotropha, rather than N. communis or Nitrosomonas ureae, was the major detectable AOB in the wastewater treatment plant linked to the Keihin Canal area (24). As expected, we obtained three clones clustered with N. oligotropha at a high bootstrap value (96%) (Fig. 3). These clones were found only at C24 but not at TLB or two other offshore sampling points (39), suggesting that discharged wastewater is a possible source of these N. oligotropha-like clones. However, these clones comprised only a minor part of the libraries. These results suggest that the Nitrosomonas groups that are common in wastewater do not predominate as members of active AOB communities in marine sediment, even though they have been detected from river water and estuaries via DNA-based techniques (4, 7, 8, 10).
Quantitative PCR.
A real-time PCR assay, based on the quenching-primer PCR method, was developed for 16S rRNA gene analysis to estimate the spatial distribution and numbers of AOB in marine sediments in our previous study (39). The copy number of β-AOB ranged from 1.9 × 102 to 2.0 × 103 per ng of DNA. The gene's copy number was converted to cell numbers using a formula based on the total bacterial counts and the total amount of extracted DNA (39). The population sizes of β-AOB were between 8.0 × 107 ± 1.3 × 107 (mean ± standard error; n = 4) and 5.7 × 108 ± 0.6 × 108 cells per g of dry sediments (Table 1). Estimated numbers of AOB corresponded to 0.2 to 1.9% of 4′,6-diamidino-2-phenylindole counts. These are greater than estimates for soils (27) but comparable to the percentages reported previously in sewage treatment systems in Tokyo, 0.01 to 2.8% (1.0 × 109 to 9.2 × 1010 cells liter−1) (24), and a municipal wastewater treatment system, 2.9% (1.2 × 1010 ± 0.9 × 1010 cells liter−1), by real-time PCR quantification based on 16S rRNA genes of β-AOB (15). Our finding supports the idea that coastal marine sediment provides a crucial habitat for AOB and maintains numerous AOB cells. This finding recalls that of a previous report that the interface between water and sediment is a major habitat for nitrifying bacteria in aquatic environments (2).
TABLE 1.
Quantitative analyses of AOB by real-time PCR and immunofluorescence staining
| Sampling site | Real-time PCR resulta
|
Immunofluorescence staining resulta
|
||
|---|---|---|---|---|
| No. of AOB (SE) | % of AOB (SE) | No. of AOB (SE) | % of AOB (SE) | |
| C21 | 3.2 × 108 (3.0 × 107) | 1.1 (0.11) | 1.1 × 109 (3.3 × 108) | 3.6 (1.10) |
| C24 | 8.0 × 107 (1.3 × 107) | 0.2 (0.03) | 6.4 × 108 (1.5 × 107) | 1.6 (0.04) |
| C23 | 5.7 × 108 (5.7 × 107) | 1.9 (0.19) | 1.6 × 109 (6.5 × 108) | 5.2 (2.17) |
| B23 | 1.6 × 108 (2.9 × 107) | 0.7 (0.13) | 1.0 × 109 (2.9 × 108) | 4.4 (1.26) |
| B18 | 2.9 × 108 (4.4 × 107) | 1.0 (0.15) | 1.7 × 109 (2.5 × 108) | 5.7 (0.86) |
| TLB | 5.2 × 108 (5.9 × 107) | 1.5 (0.18) | 1.8 × 109 (2.5 × 108) | 5.1 (0.74) |
Data are means ± standard errors(n = 4 for real-time PCR; n = 2 for immunofluorescence staining). AOB are counted as cells g−1 (dry weight) of sediment.
AOB quantification by immunofluorescence staining.
In addition to nucleic acid-based techniques, we also used a novel method of AOB immunofluorescence detection based on anti-hydroxylamine oxidoreductase (HAO) antibodies, which could detect both beta- and gammaproteobacterial AOB (39). Compared to molecular studies, which can provide phylogenetic information, antibody studies are limited to quantification, but antibody techniques are an effective way to detect natural bacterial communities without cultivation (40). In particular, antibodies targeting proteins such as AmoB (ammonia monooxygenase subunit B) (30) and HAO offer a great opportunity to estimate AOB populations in nature with high specificity and sensitivity.
After examination of anti-HAO antibodies with pure cultures, marine sediment samples were analyzed for AOB. We observed numerous AOB cells associated with detritus-like materials along with other bacteria, as previously reported (35). The population size of AOB ranged from 6.4 × 108 ± 0.2 × 108 (mean ± standard error; n = 2) to 1.8 × 109 ± 0.3 × 109 cells per g of dry sediment (Table 1). The cell numbers corresponded to between 1.6% (C24) and 5.7% (B18) of total cell numbers in sediment samples. The number of AOB determined by immunofluorescence staining was larger than that found by real-time PCR (Table 1), but the numbers obtained with these two methods were correlated (r2 = 0.68; P < 0.01). This finding was in accordance with that of our previous study in which AOB in Tokyo Bay sediments were examined quantitatively: 0.1 to 1.1% for real-time PCR and 1.2 to 4.3% for immunofluorescence staining (39).
The difference in quantification results between the two methods is likely explainable by the presence of γ-AOB (28, 40, 41) and anaerobic ammonia oxidizers (anammox), which are potential targets of anti-HAO antibodies (34). Moreover, the existence of ammonia-oxidizing archaea in seawater environments was recently reported (20). Bergmann and colleagues (3) indicated that the HAO-like protein-encoding gene is present in the genome of non-AOB, although the induction of HAO-like protein in the bacterial cells has not been completely understood. For comprehensive understanding of the global nitrogen cycle, further efforts are necessary to identify, quantify, and isolate novel and uncultured nitrifiers that play important roles in the environment. The sequences reported in this study are available from DDBJ/EMBL/GenBank under accession numbers AB239723 to AB239754. A figure showing the reproducibility of RFLP analysis can be requested from the corresponding author.
Supplementary Material
Acknowledgments
We thank M. Kato and S. Uchidaya for excellent technical assistance and H. Sekiguchi for helping with collection of samples. We also thank S. Kurata and H. Sogou at Kankyo Engineering Co., Ltd., for helping with optimization of real-time PCR assays. We gratefully acknowledge O. Yagi and F. Kurisu at the University of Tokyo for providing additional wastewater data. We also appreciate J. J. Kelly at Loyola University, Chicago, for his critical reading of our manuscript. Helpful comments and suggestions by three anonymous reviewers are gratefully acknowledged.
This research was partially supported by the Environmental Technology Development Fund of the Ministry of the Environment of Japan and a Grant-in-Aid for Young Scientists (no. 17688009) from the Ministry of Education, Culture, Sports, Science and Technology of Japan to H.U.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bano, N., and J. T. Hollibaugh. 2000. Diversity and distribution of DNA sequences with affinity to ammonia-oxidizing bacteria of the beta subdivision of the class Proteobacteria in the Arctic Ocean. Appl. Environ. Microbiol. 66:1960-1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belser, L. W. 1979. Population ecology of nitrifying bacteria. Annu. Rev. Microbiol. 33:309-333. [DOI] [PubMed] [Google Scholar]
- 3.Bergmann, D. J., A. B. Hooper, and M. G. Klotz. 2005. Structure and sequence conservation of hao cluster genes of autotrophic ammonia-oxidizing bacteria: evidence for their evolutionary history. Appl. Environ. Microbiol. 71:5371-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernhard, A. E., T. Donn, A. E. Giblin, and D. A. Stahl. 2005. Loss of diversity of ammonia-oxidizing bacteria correlates with increasing salinity in an estuary system. Environ. Microbiol. 7:1289-1297. [DOI] [PubMed] [Google Scholar]
- 5.Blackburn, T. H., and N. D. Blackburn. 1992. Model of nitrification and denitrification in marine sediments. FEMS Microbiol. Lett. 100:517-521. [Google Scholar]
- 6.Caffrey, J. M., N. Harrington, I. Solem, and B. B. Ward. 2003. Biogeochemical processes in a small California estuary. 2. Nitrification activity, community structure and role in nitrogen budgets. Mar. Ecol. Prog. Ser. 248:27-40. [Google Scholar]
- 7.Cébron, A., T. Berthe, and J. Garnier. 2003. Nitrification and nitrifying bacteria in the lower Seine River and estuary (France). Appl. Environ. Microbiol. 69:7091-7100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cébron, A., M. Coci, J. Garnier, and H. J. Laanbroek. 2004. Denaturing gradient gel electrophoretic analysis of ammonia-oxidizing bacterial community structure in the lower Seine River: impact of Paris wastewater effluents. Appl. Environ. Microbiol. 70:6726-6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalsgaard, T., and B. Thamdrup. 2002. Factors controlling anaerobic ammonium oxidation with nitrite in marine sediments. Appl. Environ. Microbiol. 68:3802-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Bie, M. J. M., A. G. C. L. Speksnijder, G. A. Kowalchuk, T. Schuurman, G. Zwart, J. R. Stephen, O. E. Diekmann, and H. J. Laanbroek. 2001. Shifts in the dominant populations of ammonia-oxidizing beta-subclass Proteobacteria along the eutrophic Schelde estuary. Aquat. Microb. Ecol. 23:225-236. [Google Scholar]
- 11.Dollhopf, S. L., J. H. Hyun, A. C. Smith, H. J. Adams, S. O'Brien, and J. E. Kostka. 2005. Quantification of ammonia-oxidizing bacteria and factors controlling nitrification in salt marsh sediments. Appl. Environ. Microbiol. 71:240-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francis, C. A., G. D. O'Mullan, and B. B. Ward. 2003. Diversity of ammonia monooxygenase (amoA) genes across environmental gradients in Chesapeake Bay sediments. Geobiology 1:129-140. [Google Scholar]
- 13.Freitag, T. E., and J. I. Prosser. 2003. Community structure of ammonia-oxidizing bacteria within anoxic marine sediments. Appl. Environ. Microbiol. 69:1359-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freitag, T. E., L. Chang, and J. I. Prosser. 2006. Changes in the community structure and activity of betaproteobacterial ammonia-oxidizing sediment bacteria along a freshwater-marine gradient. Environ. Microbiol. 8:684-696. [DOI] [PubMed] [Google Scholar]
- 15.Harms, G., A. C. Layton, H. M. Dionisi, I. R. Gregory, V. M. Garrett, S. A. Hawkins, K. G. Robinson, and G. S. Sayler. 2003. Real-time PCR quantification of nitrifying bacteria in a municipal wastewater treatment plant. Environ. Sci. Technol. 37:343-351. [DOI] [PubMed] [Google Scholar]
- 16.Hosokawa, Y., M. Yasui, K. Yoshikawa, Y. Tanaka, and M. Suzuki. 2003. The nationwide investigation of endocrine disruptors in sediment of harbours. Mar. Pollut. Bull. 47:132-138. [DOI] [PubMed] [Google Scholar]
- 17.Hosomi, M., T. Matsuo, S. Dobashi, S. Katou, and H. Abe. 2003. Survey of dioxins in Tokyo Bay bottom sediment. Mar. Pollut. Bull. 47:68-73. [DOI] [PubMed] [Google Scholar]
- 18.Juliette, L. Y., M. R. Hyman, and D. J. Arp. 1993. Inhibition of ammonia oxidation in Nitrosomonas europaea by sulfur compounds: thioethers are oxidized to sulfoxides by ammonia monooxygenase. Appl. Environ. Microbiol. 59:3718-3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kemp, W. M., P. Sampou, J. Caffrey, M. Mayer, K. Henriksen, and W. R. Boynton. 1990. Ammonium recycling versus denitrification in Chesapeake Bay sediments. Limnol. Oceanogr. 35:1545-1563. [Google Scholar]
- 20.Konneke, M., A. E. Bernhard, J. R. de la Torre, C. B. Walker, J. B. Waterbury, and D. A. Stahl. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543-546. [DOI] [PubMed] [Google Scholar]
- 21.Konuma, S., H. Satoh, T. Mino, and T. Matsuo. 2001. Comparison of enumeration methods for ammonia-oxidizing bacteria. Water Sci. Technol. 43:107-114. [PubMed] [Google Scholar]
- 22.Kowalchuk, G. A., and J. R. Stephen. 2001. Ammonia-oxidizing bacteria: a model for molecular microbial ecology. Annu. Rev. Microbiol. 55:485-529. [DOI] [PubMed] [Google Scholar]
- 23.Laanbroek, H. J., P. L. E. Bodelier, and S. Gerards. 1994. Oxygen-consumption kinetics of Nitrosomonas europaea and Nitrobacter hamburgensis grown in mixed continuous cultures at different oxygen concentrations. Arch. Microbiol. 161:156-162. [Google Scholar]
- 24.Limpiyakorn, T., Y. Shinohara, F. Kurisu, and O. Yagi. 2005. Communities of ammonia-oxidizing bacteria in activated sludge of various sewage treatment plants in Tokyo. FEMS Microbiol. Ecol. 54:205-217. [DOI] [PubMed] [Google Scholar]
- 25.McCaig, A. E., T. M. Embley, and J. I. Prosser. 1994. Molecular analysis of enrichment cultures of marine ammonia oxidisers. FEMS Microbiol. Lett. 120:363-367. [DOI] [PubMed] [Google Scholar]
- 26.McCaig, A. E., C. J. Phillips, J. R. Stephen, G. A. Kowalchuk, S. M. Harvey, R. A. Herbert, T. M. Embley, and J. I. Prosser. 1999. Nitrogen cycling and community structure of proteobacterial beta-subgroup ammonia-oxidizing bacteria within polluted marine fish farm sediments. Appl. Environ. Microbiol. 65:213-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendum, T. A., R. E. Sockett, and P. R. Hirsch. 1999. Use of molecular and isotopic techniques to monitor the response of autotrophic ammonia-oxidizing populations of the beta subdivision of the class Proteobacteria in arable soils to nitrogen fertilizer. Appl. Environ. Microbiol. 65:4155-4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nold, S. C., J. Zhou, A. H. Devol, and J. M. Tiedje. 2000. Pacific Northwest marine sediments contain ammonia-oxidizing bacteria in the beta subdivision of the Proteobacteria. Appl. Environ. Microbiol. 66:4532-4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ogura, N. 1990. Tokyo Bay—its environmental changes. Koseisha Koseikaku, Tokyo, Japan.
- 30.Pinck, C., C. Coeur, P. Potier, and E. Bock. 2001. Polyclonal antibodies recognizing the AmoB protein of ammonia oxidizers of the β-subclass of the class Proteobacteria. Appl. Environ. Microbiol. 67:118-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prinčič, A., I. I. Mahne, F. Megušar, E. A. Paul, and J. M. Tiedje. 1998. Effects of pH and oxygen and ammonium concentrations on the community structure of nitrifying bacteria from wastewater. Appl. Environ. Microbiol. 64:3584-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Purkhold, U., A. Pommerening-Röser, S. Juretschko, M. C. Schmid, H. P. Koops, and M. Wagner. 2000. Phylogeny of all recognized species of ammonia oxidizers based on comparative 16S rRNA and amoA sequence analysis: implications for molecular diversity surveys. Appl. Environ. Microbiol. 66:5368-5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Purkhold, U., M. Wagner, G. Timmermann, A. Pommerening-Röser, and H. P. Koops. 2003. 16S rRNA and amoA-based phylogeny of 12 novel betaproteobacterial ammonia-oxidizing isolates: extension of the dataset and proposal of a new lineage within the nitrosomonads. Int. J. Syst. Evol. Microbiol. 53:1485-1494. [DOI] [PubMed] [Google Scholar]
- 34.Schalk, J., S. de Vries, J. G. Kuenen, and M. S. M. Jetten. 2000. Involvement of a novel hydroxylamine oxidoreductase in anaerobic ammonium oxidation. Biochemistry 39:5405-5412. [DOI] [PubMed] [Google Scholar]
- 35.Stehr, G., B. Böttcher, R. Dittberner, G. Rath, and H. P. Koops. 1995. The ammonia-oxidizing nitrifying population of the River Elbe estuary. FEMS Microbiol. Ecol. 17:177-186. [Google Scholar]
- 36.Stephen, J. R., A. E. McCaig, Z. Smith, J. I. Prosser, and T. M. Embley. 1996. Molecular diversity of soil and marine 16S rRNA gene sequences related to β-subgroup ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 62:4147-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephen, J. R., Y. J. Chang, S. J. Macnaughton, G. A. Kowalchuk, K. T. Leung, C. A. Flemming, and D. C. White. 1999. Effect of toxic metals on indigenous soil β-subgroup proteobacterium ammonia oxidizer community structure and protection against toxicity by inoculated metal-resistant bacteria. Appl. Environ. Microbiol. 65:95-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thamdrup, B., and T. Dalsgaard. 2002. Production of N2 through anaerobic ammonium oxidation coupled to nitrate reduction in marine sediments. Appl. Environ. Microbiol. 68:1312-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Urakawa, H., S. Kurata, T. Fujiwara, D. Kuroiwa, H. Maki, S. Kawabata, T. Hiwatari, H. Ando, T. Kawai, M. Watanabe, and K. Kohata. 2006. Characterization and quantification of ammonia-oxidizing bacteria in eutrophic coastal marine sediments using polyphasic molecular approaches and immunofluorescence staining. Environ. Microbiol. 8:787-803. [DOI] [PubMed] [Google Scholar]
- 40.Ward, B. B., and A. F. Carlucci. 1985. Marine ammonia-oxidizing and nitrite-oxidizing bacteria—serological diversity determined by immunofluorescence in culture and in the environment. Appl. Environ. Microbiol. 50:194-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ward, B. B., and G. D. O'Mullan. 2002. Worldwide distribution of Nitrosococcus oceani, a marine ammonia-oxidizing gamma-proteobacterium, detected by PCR and sequencing of 16S rRNA and amoA genes. Appl. Environ. Microbiol. 68:4153-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.