Abstract
Previously available primer sets for detecting anaerobic ammonium-oxidizing (anammox) bacteria are inefficient, resulting in a very limited database of such sequences, which limits knowledge of their ecology. To overcome this limitation, we designed a new primer set that was 100% specific in the recovery of ∼700-bp 16S rRNA gene sequences with >96% homology to the “Candidatus Scalindua” group of anammox bacteria, and we detected this group at all sites studied, including a variety of freshwater and marine sediments and permafrost soil. A second primer set was designed that exhibited greater efficiency than previous primers in recovering full-length (1,380-bp) sequences related to “Ca. Scalindua,” “Candidatus Brocadia,” and “Candidatus Kuenenia.” This study provides evidence for the widespread distribution of anammox bacteria in that it detected closely related anammox 16S rRNA gene sequences in 11 geographically and biogeochemically diverse freshwater and marine sediments.
Anaerobic ammonium oxidation (anammox) involves the direct autotrophic conversion of ammonium and nitrite to N2 under anoxic conditions. First confirmed in a fluidized wastewater bed reactor in 1994 (23), anammox bacteria are now thought to form a deep-branching lineage of the Planctomycetales with high genus level diversity (7, 18). The anammox reaction occurs within a specialized cell compartment, the anammoxosome, in which ammonium is oxidized via hydrazine (N2H4) and hydroxylamine (NH2OH) intermediates (16). This process produces twice the amount of N2 per molecule of nitrite consumed as denitrification and does not require an external reducing agent (9, 20).
The role of anammox bacteria in the nitrogen cycle has been investigated for only a few environments. Anammox has been proposed to be responsible for the consumption of more than 40% of the fixed N in the anoxic water in the Black Sea (12), 19 to 35% of the total N2 formation in the anoxic waters of a coastal bay in Costa Rica (3), 24 to 67% of total N2 production in continental shelf sediments (4), and 1 to 24% of the N lost in estuarine sediments (15, 22). In a global context, the anammox reaction may account for 30 to 70% of oceanic N2 production (5), representing a major N sink.
While isotope-pairing studies have been used as indicators of anammox metabolism, anammox bacteria in environmental samples have been detected primarily by fluorescence in situ hybridization. Few environmentally derived anammox 16S rRNA gene sequences have been obtained by the use of “universal” primers, primers targeted to the Planctomycetales, or anammox-specific primers. Sequences that fall within the documented anammox group have been identified in relatively few, mostly marine environments (1, 8, 11, 12, 15, 21). The primer sets previously available for detecting anammox bacteria have resulted in a very limited database of such sequences and may perhaps be limiting the knowledge concerning the distribution of these important N-cycling bacteria.
In this study we explored the distribution and diversity of anammox-related bacteria in sediment samples from geographically and biogeochemically distinct environments. Eleven sites were analyzed, representing subtropical freshwater wetlands, a northern fen wetland, a hypereutrophic lake, deep ocean, continental margin, a shallow marine environment, and Siberian permafrost. We designed a new primer set that was 100% specific in the recovery of 700-bp 16S rRNA gene sequences of the “Candidatus Scalindua” group of anammox bacteria, and we detected this group of bacteria in all habitats sampled. We also developed a new primer set that was more efficient than any previously used for the recovery of full-length 1,380-bp sequences. This study provides evidence for the wide distribution of anammox bacteria within aquatic environments, greatly increases the number of anammox-related sequences deposited, and represents a rapid and efficient method for determining the presence of anammox bacteria before undertaking more exhaustive N isotope studies to determine in situ activity.
Our sampling sites were chosen to represent contrasting geographical, biogeochemical, and temporal attributes. Freshwater sediment samples were collected from beneath shallow (<1-m) waters at two long-term monitoring sites at Kellogg Biological Station, Mich.: Wintergreen Lake, a small, groundwater-fed, hypereutrophic lake (42°23′56"N, 85°22′59"W), and Sheriff's Marsh (42°25′19"N, 85°30′58"W), a fen wetland that receives groundwater and stream flow. Sediments from subtropical wetlands were collected using a piston corer in Florida Everglades Water Conservation Area 2A at two sites: a phosphorus (P)-impacted eutrophic site, F1 (0.4-m depth; 26°21′59"N, 80°22′23"W), and a reference P level oligotrophic site, U3 (0.5-m depth; 26°17′24"N, 80°25′08"W). Marine samples were collected at six sites using a multicorer: East Hanna Shoal (160-m depth; 72°38′13"N, 158°40′01"W) and East Hanna Shoal Deep (1,450-m depth; 72°51′07"N, 158°12′25"W), both from the Alaskan maritime environment in the Chuckchi Sea; Turning Basin (12-m depth; 47°03′08"N, 122°54′27"W) and Shallow Bud Inlet (3-m depth; 47°04′59"N, 122°54′07"W), both in Puget Sound; and Washington Margin (1,138-m depth; 46°25′34"N, 124°41′30"W) and an area west of the Juan de Fuca Ridge (3,869-m depth; 46°47′00"N, 133°40′00"W), both in the Pacific Ocean. Siberian frozen alluvial sandy loam, deposited in the Middle Pleistocene Epoch, 300,000 to 400,000 years ago, was collected from Cape Svyatoi Nos in the tundra zone on the Laptev Sea coast (72°55′01"N, 140°10′12"E) as described previously (24).
Ten primers targeting the 16S rRNA genes of “Ca. Scalindua,” “Candidatus Brocadia,” and “Candidatus Kuenenia” were initially designed, screened against GenBank for target organisms, and tested in various sediments by PCR amplification and sequencing. Brod541F (5′-GAGCACGTAGGTGGGTTTGT-3′)-Brod1260R (5′-GGATTCGCTTCACCTCTCGG-3′) was the only primer set to consistently produce single bands and specifically target anammox bacteria (720-bp amplicons). Specifically, Brod541F exhibited 100% homology to four groups in GenBank: “Candidatus Scalindua brodae” (18), uncultured ammonia-oxidizing bacteria (15), unidentified bacteria and planctomycetes from Antarctica (1), and uncultured planctomycetes from the Black Sea (8). Primers An7F (5′-GGCATGCAAGTCGAACGAGG-3′) and An1388R (5′-GCTTGACGGGCGGTGTG-3′) were designed for the capture of greater phylogenetic variability with 1,380-bp amplicons. DNA was isolated using the Ultraclean soil DNA kit (MoBio Laboratories Inc.), and PCR amplifications of 16S rRNA genes targeting the anammox group were performed in a model 9600 thermal cycler (Perkin-Elmer Cetus) from 30 to 80 ng μl−1 environmental DNA extracts. Reaction conditions for the primer pair Brod541F-Brod1260R were as follows: 10 pmol of each primer, 200 ng μl−1 of bovine serum albumin (Roche), 2.5 U of Taq polymerase (Promega), 200 μM each deoxyribonucleoside triphosphate (Invitrogen), 150 mM MgCl2 (Promega), and 1/10 volume of 10× PCR buffer provided with the enzyme. The PCR program was as follows: 95°C for 3 min; 30 cycles of 95°C for 45 s, 60°C (for Brod541F-Brod1260R) or 63°C (for An7F-An1388R) for 1 min, and 72°C for 1 min; and 72°C for 7 min. Clone libraries were created using TOPO TA cloning (Invitrogen), and clones were sequenced using an ABI 3730 genetic analyzer or an ABI Prism 3700 DNA analyzer (Applied Biosystems). Sequences were analyzed against GenBank with BLAST and Ribosomal Database Project-II (RDP) (2). The presence of chimeric sequences was determined by using the CHIMERA_CHECK program from RDP. Sequences were aligned with ClustalW, and phylogenetic trees were constructed using MEGA, version 3.1 (10), using a neighbor-joining method with bootstrapping.
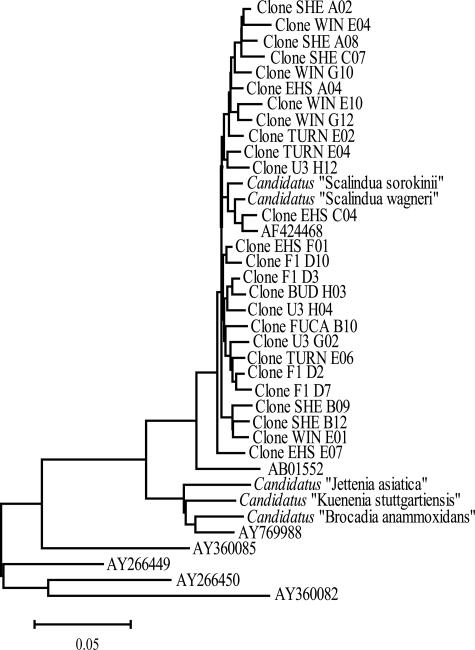
The screening primer set Brod541F-Brod1260R resulted in consistent amplification from all sediments tested and was subsequently used to create clone libraries of each site. The resulting 475 sequences exhibited >96% nucleotide identity to either “Candidatus Scalindua wagneri” or “Ca. Scalindua brodae” and represented 100% specificity for the anammox group. Partial and redundant sequences were removed and 698-bp fragments were aligned, resulting in 287 unique sequences. These were compared against the anammox group consisting of “Ca. Scalindua,” “Ca. Brocadia,” “Ca. Kuenenia,” and “Candidatus Jettenia asiatica,” in addition to environmental and wastewater-obtained sequences that fall within the anammox group. A neighbor-joining tree was constructed using the 25 sequences that remained after 98% redundancy was removed (Fig. 1), which completely excluded the East Hanna Shoal Deep, Siberian permafrost, and Washington Margin sequences. Clones generated from Sheriff's Marsh and Siberian permafrost exhibited the highest (88%) and the lowest (28%) proportions of unique sequences, respectively. There was no discernible grouping either between freshwater and marine samples or among sites with different nutrient loadings. Thus, Brod541F-Brod1260R-generated 16S rRNA gene sequences from the 11 different sites cluster independently, although sequences from individual sites cluster together when 99% redundancy is included, reflecting a certain level of endemism.
FIG. 1.
16S rRNA gene-based bootstrapped phylogenetic tree reflecting the relationships of Brod541F-Brod1260R clones from East Hanna Shoal (EHS), East Hanna Shoal Deep, Washington margin, Turning Basin (TURN), Shallow Bud Inlet (BUD), Juan de Fuca (FUCA), Wintergreen Lake (WIN), Sheriff's Marsh (SHE), Everglades F1 (F1), Everglades U3 (U3), and Siberian permafrost sediments, with 98% redundancy removed and with the anammox group and related bacteria given as GenBank accession numbers. Bar, 5% sequence divergence.
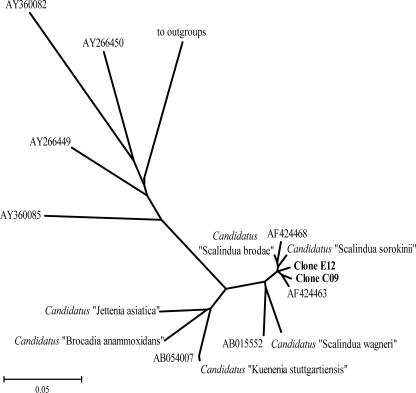
Additional clone libraries were created to confirm that the 698-bp Brod541F-Brod1260R 16S rRNA gene sequences obtained did indeed represent full-length sequences of known anammox bacteria. Primer An7F, targeting “Ca. Scalindua,” “Ca. Brocadia,” and “Ca. Kuenenia,” was first combined with the universal primer 1392R. The resulting 68 clones lacked any relationship to the anammox group, as was the case with the Brod541F-1392R primer set, despite the increased specificity of the forward primers. To resolve this problem, An7F was combined with An1388R, and two 1,380-bp sequences were obtained out of 21 passing sequences. Clone FLE12 (positions 7 to 1388) was 97% homologous to “Ca. Scalindua brodae” and 92% homologous to “Ca. Scalindua wagneri.” Positions 541 to 560 and 1241 to 1260 were identical to the Brod541F and Brod1260R primer target sites, respectively. Clone FLC09 (positions 5 to 1388) was 96% homologous to “Ca. Scalindua brodae” and 92% homologous to “Ca. Scalindua wagneri.” There was one mismatch within the Brod541F site and 100% homology in the Brod1260R site. A lack of homology to the planctomycete-specific primer S-P-Planc-0046-a-A-18 (Pla46F) (13) was observed for both clones. Sequencing error was estimated at 4 bases in An1388R, and the anammox sequence-targeting efficiency of this primer was estimated to be 10% of the total retrieved passing sequences. A neighbor-joining tree was constructed using these full-length sequences for comparison to the anammox-related clones derived from GenBank (Fig. 2).
FIG. 2.
16S rRNA gene-based bootstrapped neighbor-joining tree reflecting the relationships of full-length An7F-An1388R-generated amplicons, with the anammox group as well as other sequences derived from environmental samples given as GenBank accession numbers. Bar, 5% sequence divergence.
Specific 16S rRNA sequences belonging to the anammox group have historically been difficult to recover from environmental samples. The planctomycete-specific forward primer Pla46F, utilized as a “capture” primer for all anammox bacteria, has been used to construct clone libraries, generally from samples which show evidence of anammox activity via 15N isotope-pairing studies. However, Pla46F underrepresents anammox bacterial abundance in various studies (6, 17, 18). For example, in an enriched sample where anammox bacteria represented 40% of the bacterial population, only 36% of Pla46F-derived 16S rRNA clones sequenced belonged to the anammox group (17). In environmental samples, this method results in very low target sequence retrieval efficiencies, possibly due to inherent PCR template-to-product ratio bias (14) or the lack of Pla46F specificity. For example, the lack of Pla46 primer annealing site homology with the 1,380-bp clone FLE12 and clone FLC09 sequences in this study would have resulted in the omission of these clones from a Pla46F-generated library.
In the context of this study, the novel primer set Brod541F-Brod1260R provides an efficient method of screening environmental samples for the presence of “Ca. Scalindua”-like anammox bacteria. A previously suggested primer, S-*-BS-820-a-A-22 (19), which targets “Ca. Scalindua wagneri” and “Candidatus Scalindua sorokinii,” appears to be specific for anammox bacteria with up to three mismatches but is homologous only with approximately 50% of the sequences recovered in this study. Conversely, primer S-*-Amx-0820-a-A-22, specific for “Ca. Brocadia” and “Ca. Kuenenia” (17), exhibited at least three mismatches in approximately 80% and four mismatches in 50% of the sequences retrieved with the Brod541F-Brod1260R primer set, which was expected due to our targeting of “Ca. Scalindua” bacteria. Lastly, primer S-*-Scabr-1114-a-A-22, specific for “Ca. Scalindua brodae” and recommended for use as a reverse primer with Pla46F (18), exhibited at least one mismatch in 50% of the sequences retrieved. This suggests that use of these primers could result in the underrepresentation of anammox bacteria in environmental samples and illustrates the difficulty of retrieving specifically anammox sequences.
The identification of sequences with >96% nucleotide identity to “Ca. Scalindua” anammox bacteria in all freshwater, marine, and permafrost samples tested constitutes the first broad-range 16S rRNA gene-based molecular investigation of anammox bacterial distribution. The presence of anammox 16S rRNA gene sequences in anoxic deep-sea sediments, shallow perturbed marine sediments, shallow organic rich freshwater sediments, periphyton-dominated aerobic sediments, and ancient frozen permafrost sediments suggests that anammox bacteria in sediments are not constrained by conditions that appear to be unfavorable to their metabolism and thus may be more widely distributed than previously thought. While the presence of anammox sequences does not equate with in situ anammox activity, these sequences do serve to identify habitats for more intensive study and serve as molecular markers for better tracking of candidate populations for anaerobic ammonium oxidation. The broad, perhaps universal distribution of anammox sequences in sediments suggests that the process may be not only a significant oceanic nitrogen sink but also an important method of nitrogen removal from freshwater wetlands and lakes.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the sequences reported here are DQ869865 to DQ870184, DQ869384, and DQ869385.
Acknowledgments
We thank Susan Newman and the staff of the South Florida Water Management District for Everglades sampling support, David Gilichinsky of the Russian Academy of Sciences for providing the permafrost sample and information, Stephen Hamilton for providing the information for the Kellogg Biological Station sites, and Amy Burgin for additional sampling assistance.
This research was supported by the Biotechnology Investigations-Ocean Margins Program, Office of Biological and Environmental Research, Office of Science, U.S. Department of Energy.
REFERENCES
- 1.Bowman, J. P., and R. D. McCuaig. 2003. Biodiversity, community structural shifts, and biogeography of prokaryotes within Antarctic continental shelf sediment. Appl. Environ. Microbiol. 69:2463-2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cole, J. R., B. Chai, R. J. Farris, Q. Wang, S. A. Kulam, D. M. McGarrell, G. M. Garrity, and J. M. Tiedje. 2005. The Ribosomal Database Project (RDP-II): sequences and tools for high-throughput rRNA analysis. Nucleic Acids Res. 33(Database Issue):D294-D296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dalsgaard, T., D. E. Canfield, J. Petersen, B. Thamdrup, and J. Acuña-González. 2003. N2 production by the anammox reaction in the anoxic water column of Golfo Dulce, Costa Rica. Nature 422:606-608. [DOI] [PubMed] [Google Scholar]
- 4.Dalsgaard, T., and B. Thamdrup. 2002. Factors controlling anaerobic ammonium oxidation with nitrite in marine sediments. Appl. Environ. Microbiol. 68:3802-3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devol, A. H. 2003. Nitrogen cycle—solution to a marine mystery. Nature 422:575-576. [DOI] [PubMed] [Google Scholar]
- 6.Egli, K., U. Franger, P. J. J. Alvarez, H. R. Siegrist, J. R. Van der Meer, and A. J. B. Zehnder. 2001. Enrichment and characterization of an anammox bacterium from a rotating biological contractor treating ammonium-rich leachate. Arch. Microbiol. 175:198-207. [DOI] [PubMed] [Google Scholar]
- 7.Freitag, T. E., and J. I. Prosser. 2003. Community structure of ammonia-oxidizing bacteria within anoxic marine sediments. Appl. Environ. Microbiol. 69:1359-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kirkpatrick, J., B. Oakley, C. Fuchsman, S. Srinivasan, J. T. Staley, and J. W. Murray. 2006. Diversity and distribution of planctomycetes and related bacteria in the suboxic zone of the Black Sea. Appl. Environ. Microbiol. 72:3079-3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuai, L., and W. Verstraete. 1998. Ammonium removal by the oxygen-limited autotrophic nitrification-denitrification system. Appl. Environ. Microbiol. 64:4500-4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 11.Kuypers, M. M. M., G. Lavik, D. Woebken, M. Schmid, B. M. Fuchs, R. Amann, B. B. Jørgensen, and M. S. M. Jetten. 2005. Massive nitrogen loss from the Benguela upwelling system through anaerobic ammonium oxidation. Proc. Natl. Acad. Sci. USA 102:6478-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuypers, M. M. M., A. O. Sliekers, G. Lavik, M. Schmid, B. B. Jørgensen, J. G. Kuenen, J. S. Sinninghe Damsté, M. Strous, and M. S. M. Jetten. 2003. Anaerobic ammonium oxidation by anammox bacteria in the Black Sea. Nature 422:608-611. [DOI] [PubMed] [Google Scholar]
- 13.Neef, A., R. I. Amann, H. Schlesner, and K.-H. Schleifer. 1998. Monitoring a widespread bacterial group: in situ detection of Planctomycetes with 16S rRNA-targeted probes. Microbiology 144:3257-3266. [DOI] [PubMed] [Google Scholar]
- 14.Polz, M. F., and C. M. Cavanaugh. 1998. Bias in template-to-product ratios in multitemplate PCR. Appl. Environ. Microbiol. 64:3724-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Risgaard-Peterson, N., R. L. Meyer, M. Schmid, M. S. M. Jetten, A. Enrich-Prast, S. Rysgaard, and N. P. Revsbech. 2004. Anaerobic ammonium oxidation in an estuarine sediment. Aquat. Microb. Ecol. 36:293-304. [Google Scholar]
- 16.Schalk, J., H. Oustad, J. G. Kuenen, and M. S. M. Jetten. 1998. The anaerobic oxidation of hydrazine: a novel reaction in microbial nitrogen metabolism. FEMS Microbiol. Lett. 158:61-67. [DOI] [PubMed] [Google Scholar]
- 17.Schmid, M., U. Twachtmann, M. Klein, M. Strous, S. Juretschko, M. S. M. Jetten, J. Metzger, K.-H. Schleifer, and M. Wagner. 2000. Molecular evidence for genus level diversity of bacteria capable of catalyzing anaerobic ammonium oxidation. Syst. Appl. Microbiol. 23:93-106. [DOI] [PubMed] [Google Scholar]
- 18.Schmid, M., K. Walsh, R. Webb, W. I. C. Rijpstra, K. T. van de Pas-Schoonen, M. J. Verbruggen, T. Hill, B. Moffett, J. Fuerst, S. Schouten, J. S. Sinninghe Damsté, J. Harris, P. Shaw, M. S. M. Jetten, and M. Strous. 2003. Candidatus “Scalindua brodae,” sp. nov., Candidatus “Scalindua wagneri”, sp. nov., two new species of anaerobic ammonium oxidizing bacteria. Syst. Appl. Microbiol. 26:529-538. [DOI] [PubMed] [Google Scholar]
- 19.Schmid, M. C., B. Maas, A. Dapena, K. van de Pas-Schoonen, J. van de Vossenberg, B. Kartal, L. van Niftrik, I. Schmidt, I. Cirpus, J. G. Kuenen, M. Wagner, J. S. Sinninghe Damsté, M. Kuypers, N. P. Revsbech, R. Mendez, M. S. M. Jetten, and M. Strous. 2005. Biomarkers for in situ detection of anaerobic ammonium-oxidizing (anammox) bacteria. Appl. Environ. Microbiol. 71:1677-1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Strous, M., J. G. Kuenen, and M. S. M. Jetten. 1999. Key physiology of anaerobic ammonium oxidation. Appl. Environ. Microbiol. 65:3248-3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tal, Y., J. E. M. Watts, and H. J. Schreier. 2005. Anaerobic ammonia-oxidizing bacteria and related activity in Baltimore inner harbor sediment. Appl. Environ. Microbiol. 71:1816-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trimmer, M., J. C. Nicholls, and B. Deflandre. 2003. Anaerobic ammonium oxidation measured in sediments along the Thames estuary, United Kingdom. Appl. Environ. Microbiol. 69:6447-6454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van de Graaf, A., A. Mulder, P. de Bruijn, M. S. M. Jetten, L. Robertson, and J. Kuenen. 1995. Anaerobic oxidation of ammonium is a biologically mediated process. Appl. Environ. Microbiol. 61:1246-1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vishnivetskaya, T., S. Kathariou, J. McGrath, D. Gilichinsky, and J. M. Tiedje. 2000. Low-temperature recovery strategies for the isolation of bacteria from ancient permafrost sediments. Extremophiles 4:165-173. [DOI] [PubMed] [Google Scholar]