Abstract
Recent reports suggest that the selective advantage of bioluminescence for bacteria is mediated by light-dependent stimulation of photolyase to repair DNA lesions. Despite evidence for this model, photolyase mutants have not been characterized in a naturally bioluminescent bacterium, nor has this hypothesis been tested in bioluminescent bacteria under natural conditions. We have now characterized the photolyase encoded by phr in the bioluminescent bacterium Vibrio fischeri ES114. Consistent with Phr possessing photolyase activity, phr conferred light-dependent resistance to UV light. However, upon comparing ES114 to a phr mutant and a dark ΔluxCDABEG mutant, we found that bioluminescence did not detectably affect photolyase-mediated resistance to UV light. Addition of the light-stimulating autoinducer N-3-oxo-hexanoyl homoserine lactone appeared to increase UV resistance, but this was independent of photolyase or bioluminescence. Moreover, although bioluminescence confers an advantage for V. fischeri during colonization of its natural host, Euprymna scolopes, the phr mutant colonized this host to the same level as the wild type. Taken together, our results indicate that at least in V. fischeri strain ES114, the benefits of bioluminescence during symbiotic colonization are not mediated by photolyase, and although some UV resistance mechanism may be coregulated with bioluminescence, we found no evidence that light production benefits cells by stimulating photolyase in this strain.
Bacterial bioluminescence is well understood biochemically and genetically (15, 27, 39); however, the functional significance of bioluminescence for bacteria remains uncertain (35). Generating luminescence presumably confers a selective advantage(s) under some circumstances, but this must be weighed against apparently significant energy costs (15, 21). Several hypotheses have been offered to explain how bioluminescence might be advantageous to the growth or survival of light-producing bacteria, and they propose that the value of luminescence might be due to the consumption of reducing equivalents, the reduction of oxygen, or an effect of light itself (29, 35). Recently, it was suggested that bioluminescence benefits bacteria by stimulating the light-dependent photolyase-mediated repair of DNA lesions (7-9, 23, 43). Photolyase is an enzyme that uses light to drive the “photoreactivation” repair of pyrimidine dimers, such as those caused by UV light, in adjacent base pairs on the same strand of DNA (42), and theoretically photolyase could be activated by bioluminescence. This led to the suggestion that bioluminescence may have evolved to stimulate photolyase in an otherwise dark environment (8).
Proponents of the hypothesis that luminescence benefits cells by stimulating photolyase have reported several experiments that support this model. Consistent with a role in repairing pyrimidine dimers, bioluminescence in Vibrio harveyi is stimulated by UV irradiation (7, 9). Furthermore, dark lux mutants of V. harveyi (9) and of other bacteria (23) were more sensitive to UV light than their respective wild-type parents, while Escherichia coli clones carrying V. harveyi bioluminescence-generating lux genes were more UV resistant. Also, in mixed cultures, a dark luxA mutant of V. harveyi was outcompeted by its bioluminescent parent when the cultures were exposed to UV light (8). Importantly, the relative sensitivity of dark strains in these experiments could be compensated for, at least in part, by exposure to external light, consistent with a benefit mediated by light itself (8, 9, 23). Finally, perhaps the most compelling evidence of a connection between bioluminescence and photolyase is the fact that the UV resistance conferred upon E. coli by the lux genes is dependent upon phr, the gene encoding photolyase (23).
Despite this evidence, it is not yet certain whether bioluminescence plays a significant role in photoreactivation. Some of the experiments described above are open to alternative interpretations. For example, bioluminescence could influence stress resistance and DNA repair through pathways that do not involve photolyase, and experiments such as those involving lux expression in E. coli may not be relevant to bioluminescent bacteria in the environment. Moreover, photolyase mutants have not been characterized in a naturally bioluminescent bacterium, nor has a connection between photolyase and bioluminescence been tested in bioluminescent bacteria under natural conditions where the bacteria produce light.
We tested the relationship between bioluminescence and photolyase in Vibrio fischeri ES114. The genome of this bioluminescent bacterium has been sequenced (34), allowing us to identify phr and generate a photolyase mutant, and we were also able to observe the bacterium under conditions where it is naturally luminescent, in the light organ of its host, Euprymna scolopes. We found that although bioluminescence enables V. fischeri to fully colonize its host (41), this effect is not mediated by photolyase. Taken together with analyses of cultured cells, our results cast doubt on the importance of bioluminescence in photoreactivation in this bacterium.
MATERIALS AND METHODS
Bacteria and culturing techniques.
The bacterial strains, plasmids, and oligonucleotides used in this study are described in Table 1. Plasmid constructs were transformed and maintained in E. coli strain DH5α or DH5αλpir, with the latter used specifically for plasmids containing the R6Kγ replication origin. The conjugative helper plasmid pEVS104 was maintained in E. coli strain CC118λpir. E. coli was grown in LB medium (28) or brain heart infusion medium, and V. fischeri was grown in either SWT medium, which contained 5 g of tryptone, 3 g of yeast extract, 3 ml of glycerol, and 700 ml of Instant Ocean (Aquarium Systems, Mentor, Ohio) per liter, or in LBS medium, which contained 10 g of tryptone, 5 g of yeast extract, 20 g of NaCl, and 20 mM Tris-hydrochloride (Tris, pH 7.5) per liter of water. Agar (15 mg ml−1) was added to solidify the media for plating experiments. Culture density was measured as the absorbance at 595 nm (A595) using a BioPhotometer (Brinkman Instruments, Westbury, N.Y.).
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides
| Strain, plasmid, or oligonucleotide | Relevant characteristicsa | Source or reference |
|---|---|---|
| V. fischeri | ||
| ES114 | Wild-type isolate from Euprymna scolopes | 3 |
| EW1 | ES114 Δphr (allele exchanged from pELW1) | This study |
| EW4 | ES114 Δphr ΔluxCDABEG (EW1 with lux allele exchanged from pEVS153) | This study |
| EVS102 | ES114 ΔluxCDABEG | J. L. Bose and E. V. Stabb, unpublished data |
| JRM100 | ES114 mini-Tn7-ermR (inserted at intergenic att site) | 26 |
| E. coli | ||
| CC118λpir | Δ(ara-leu) araD ΔlacX74 galE galK phoA20 thi-1 rpsE rpoB argE(Am) recA1; lysogenized with λpir | 20 |
| DH5α | F−F80dlacZΔM15 Δ(lacZYA-argF)U169 deoR supE44 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 14 |
| DH5αλpir | DH5α lysogenized with λpir | 10 |
| Plasmids | ||
| pCR-Blunt II-TOPO | PCR product cloning vector; ColE1 oriV kanR | Invitrogen |
| pALZ1 | PCR product (primers JBPHOTO1 and JBPHOTO2; ES114 template) in pCR-Blunt II-TOPO | This study |
| pALZ5 | SpeI-AvrII fragment of pALZ1 containing phr and flanking sequences cloned between SpeI-AvrII sites of pEVS118 | This study |
| pALZ6 | PCR product (primers JBPHOTO3 and JBPHOTO4; pALZ5 template) SmaI digested and self-ligated | This study |
| pBluescript KS(+) | ColE1 oriV ampR | Stratagene |
| pELW1 | SpeI-digested pALZ6 ligated to SpeI-digested pBluescript | This study |
| pELW2 | PCR product (primers JBPHOTO9 and JBPHOTO10; pALZ1 template) in pCR-Blunt II-TOPO | This study |
| pELW5 | phr-containing AvrII-KpnI fragment of pELW2 cloned between SpeI-KpnI sites of pVSV105 | This study |
| pEVS104 | R6Kγ oriV tra trb oriTRP4kanR; conjugative helper | 37 |
| pEVS118 | R6Kγ oriV oriTRP4chmR | 10 |
| pEVS153 | ColE1 oriV kanR chmR oriTRP4 with ΔluxCDABEG and flanking sequence from ES114 | Bose and Stabb, unpublished |
| pVSV105 | pES213 oriV chmR oriTRP4 | 11 |
| Oligonucleotides | ||
| JBPHOTO1 | 5′ GCT CTA GAG GTC TGG CAA GGT ACT TAA TAC GGC T 3′ | This study |
| JBPHOTO2 | 5′ CCG CCT AGG TGG GTG CCG CTG ACA CAA TAA TGA 3′ | This study |
| JBPHOTO3 | 5′ TCC CCC GGG CAT AGC GTA ATC TTA GCT CTT GTG GGT AAG 3′ | This study |
| JBPHOTO4 | 5′ TCC CCC GGG TAA CCA CGC TTG AGT ATG ATT AAT ACA TC 3′ | This study |
| JBPHOTO5 | 5′ CGC ATC GCC TTA GTT GAT TCT AAG GAT TC 3′ | This study |
| JBPHOTO6 | 5′ CAC CTC AGA AGC AGG CTT TAG CTG AGC TC 3′ | This study |
| JBPHOTO7 | 5′ GCC CCA CTC TGA AAT ACA AAA TTG CTC AG 3′ | This study |
| JBPHOTO8 | 5′ GTG TGG GTT GCG GCA CGA GCC ATG 3′ | This study |
| JBPHOTO9 | 5′ CCG CCC TAG GCA AGA GCT AAG ATT ACG CTA TGG AT 3′ | This study |
| JBPHOTO10 | 5′ GGG GTA CCG GAT GCC TTG CGA TGG GGT TAT ATC 3′ | This study |
ampR, ampicillin resistance (bla); chmR, chloramphenicol resistance (cat); ermR, erythromycin resistance; and kanR, kanamycin resistance (aph).
Chemicals were obtained from Sigma Chemical Co. (St. Louis, Mo.). When added to LB medium for selection of E. coli, ampicillin, chloramphenicol, and kanamycin were used at concentrations of 100, 20, and 40 μg ml−1, respectively. When added to brain heart infusion medium for selection of E. coli, erythromycin was used at a concentration of 150 μg ml−1. When added to LBS medium for selection of V. fischeri, chloramphenicol, erythromycin, and kanamycin were used at concentrations of 2, 5, and 100 μg ml−1, respectively. To add N-3-oxo-hexanoyl homoserine lactone (3-oxo-C6-HSL), a 200-μg ml−1 stock solution was prepared in ethyl acetate, an appropriate volume was dispensed into flasks or tubes, the ethyl acetate was allowed to evaporate, and then the medium was added so that the final concentration of 3-oxo-C6-HSL was 200 ng ml−1.
Genetic techniques and analyses.
We generated plasmids using standard cloning and DNA manipulation techniques. DNA ligase and restriction enzymes were obtained from New England Biolabs (Beverly, Mass.), and oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, Iowa). Plasmids were purified using QIAGEN Mini-prep kits (QIAGEN Inc., Valencia, Calif.), and DNA was recovered from restriction and ligation reactions with the DNA Clean and Concentrator-5 Kit (Zymo Research, Orange, Calif.). We used the ZeroBlunt-TOPO PCR Cloning Kit (Invitrogen, Carlsbad, Calif.) to clone PCR products into pCR-Blunt II-TOPO. PCR was performed with an iCycler (Bio-Rad Laboratories, Hercules, CA) using KOD HiFi DNA Polymerase (Novagen, Madison, Wis.). In addition to using a high-fidelity polymerase, we sequenced the cloned PCR products to ensure that unintended base pair alterations were not incorporated into our constructs. DNA sequencing was conducted at the University of Michigan DNA Sequencing Core Facility, and sequences were analyzed using Sequencher 4.1.2 (Gene Codes Corp., Ann Arbor, Mich.). Sequencing was completed using the primer oligonucleotides JBPHOTO5, JBPHOTO6, JBPHOTO7, and JBPHOTO8 (Table 1). Protein sequence comparisons were generated with BLAST (1), and similarities were calculated using the BLOSUM62 scoring matrix (19).
Details of plasmid construction are included in Table 1. To generate the Δphr mutant allele, we PCR amplified phr and flanking sequences from V. fischeri ES114 using primers JBPHOTO1 and JBPHOTO2, cloned this PCR product into pCR-Blunt II-TOPO, and then subcloned the phr-containing fragment into the small, mobilizable vector pEVS118 (10). We then used primers JBPHOTO3 and JBPHOTO4, which are directed outward from the start and stop codons of phr and contain SmaI recognition sites near their 5′ ends, to PCR amplify the vector and sequences flanking phr. The template was then destroyed by DpnI treatment, and the PCR product was digested with SmaI and self-ligated to generate pALZ6, which contains the genomic region surrounding phr but with the sequences between the phr start and stop codons replaced by 5′-CCCGGG-3′. pALZ6 was ligated to pBluescript KS(+) to generate pEW1, a mobilizable ColE1-containing vector for allelic exchange of the in-frame phr deletion. To generate the phr complementation vector pEW5, we specifically PCR amplified phr using primers JBPHOTO9 and JBPHOTO10, cloned this PCR product into pCR-Blunt II-TOPO, and subcloned a phr-containing fragment into shuttle vector pVSV105 (11).
Mutant alleles and the stable vectors for complementation were transferred to V. fischeri from E. coli by triparental mating using the conjugative helper plasmid pEVS104 (37). For mutant construction, recombinational marker exchange was scored by screening for the appropriate antibiotic resistance phenotype, and putative double recombinants were screened by PCR to determine which had reverted to wild type and which had incorporated the Δphr or ΔluxCDABEG mutant allele. The nonbioluminescent phenotype of the ΔluxCDABEG mutant also confirmed the replacement of the wild-type lux allele.
Measurement of UV resistance.
V. fischeri cells were grown in 3 ml of LBS in 18-mm tubes shaken at 28°C until the A595 was between 0.2 and 0.5. The cells were then pelleted by centrifugation, resuspended in filter-sterilized Instant Ocean, and diluted 10-fold in Instant Ocean, and 500-μl aliquots were placed into wells in two 24-well microtiter plates. This pelleting and resuspension did not detectably affect the bioluminescence (data not shown). One microtiter plate was exposed, with the lid removed, to a preset intensity (in μJ/cm2) of UV light in a UV-Stratalinker 1800 (Stratagene, La Jolla, Calif.), while the other plate was left unexposed. The cells were allowed to recover for 2 h before they were dilution plated onto LBS agar to determine the UV survival frequency, which was defined as the percentage of CFU ml−1 in the UV-treated sample relative to the number of CFU ml−1 in the untreated sample. To determine the influence of light on UV survival, cell suspensions were placed in four parallel microtiter plates, and two of these microtiter plates were exposed to UV light in a UV-Stratalinker that had been placed in a darkroom. Following exposure, one UV-treated plate and one untreated plate were allowed to recover for 2 h under white fluorescent laboratory lights, while the remaining plates for the “dark” treatment were kept in the darkroom during recovery and were dilution plated under a dim red light.
Euprymna scolopes colonization assays.
E. scolopes hatchlings were infected with V. fischeri using previously described inoculation procedures (32, 36) and overnight exposures of the squid to Instant Ocean containing V. fischeri. Within each experiment, similar numbers of mutant or wild-type cells were present in the respective inocula. Inoculant strains were pregrown unshaken in 5 ml of SWT in 50-ml conical tubes at 28°C so that the A595 was between 0.3 and 0.8, the cultures were diluted in Instant Ocean to between 1,000 and 3,000 total CFU ml−1, and E. scolopes juveniles were exposed to the inocula for between 12 and 14 h before being rinsed in V. fischeri-free Instant Ocean. The squid were homogenized at 48 h postinoculation, and the homogenates were serially diluted and plated onto LBS. Following overnight incubation, the colonies were counted to determine the number of CFU per animal. To generate mixed infections with both strains EW1 and JRM100, E. scolopes hatchlings were exposed to mixed inocula with roughly equal numbers of EW1 and JRM100 cells for 18 h before being rinsed in inoculum-free Instant Ocean. Ratios of EW1 to JRM100 were determined after dilution plating the cells on LBS by patching 50 colonies on LBS containing erythromycin. Relative competitive indices (RCIs) were calculated by dividing the ratio of EW1 to JRM100 in the squid after 48 h by the ratio of these strains in the inoculum.
RESULTS
The goal of this study was to characterize photolyase in Vibrio fischeri strain ES114, which is naturally bioluminescent and was originally isolated from the light organ of the Hawaiian bobtail squid, E. scolopes. Specifically, we sought to test whether there is a relationship between photolyase and bioluminescence both in culture and during symbiotic colonization.
Bioinformatic characterization of phr in V. fischeri ES114.
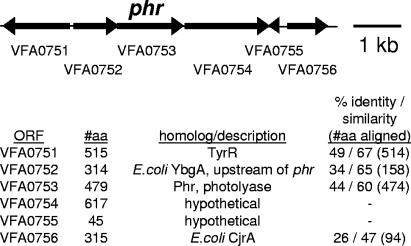
Analysis of the ES114 genome (34) revealed one putative photolyase (phr) gene. This open reading frame (ORF), which was designated VFA0753 by the genome project, is located on chromosome II and encodes a protein that is 44% identical and 60% similar to the photolyase of E. coli K12 (Fig. 1). The ORF was annotated by the genome project as photolyase, and we refer to it here as phr. The genetic context of VFA0753 further supports its phr designation, as the gene immediately upstream of it, VFA0752, encodes a protein that is homologous to YpgA, which is encoded by the ORF immediately upstream of phr in E. coli (Fig. 1). Moreover, ORFs encoding proteins similar to YbgA are also upstream of putative photolyase genes in Vibrio cholerae (17), Vibrio vulnificus (6), and Vibrio parahaemolyticus (25), although in each of these vibrios a putative transcriptional regulator is encoded in between ybgA and phr. In each of these four vibrios, the ORFs encoding proteins similar to Phr and YbgA are also found on chromosome II, although homologs of VFA0751 (TyrR), VFA0754, and VFA0756 (Fig. 1) are on chromosome I in these vibrios.
FIG. 1.
Schematic representation of the genetic organization near phr in V. fischeri ES114. ORF designations (e.g., VFA0753) are taken from the V. fischeri genome project (34). The arrows represent ORFs and indicate the direction of gene transcription, as well as the gene size, which is presented relative to the scale bar. “#aa” indicates the number of amino acids encoded by each ORF. Protein similarities were identified using BLAST (1) and the BLOSUM62 scoring matrix (19). The identity and similarity to E. coli K12 ORFs are presented, although closer matches with the same gene designation were found for less well-characterized ORFs (e.g., from vibrios). Several vibrios contain closer matches to ORF VFA0756, and most of these have been designated hypothetical.
The genetic organization near V. fischeri phr indicates that it may be expressed on a polycistronic transcript (Fig. 1). phr overlaps VFA0752 by 7 bp, and it is separated by only 59 bp from VFA0754, strongly suggesting that it is cotranscribed with the former and possibly with the latter. VFA0754 was annotated by the genome project as encoding methionine sulfoxide reductase, an enzyme that is similar to photolyase in that it repairs damaged macromolecules. However, we found no significant similarity between the protein encoded by VFA0754 and characterized methionine sulfoxide reductases, and we have annotated this ORF as encoding a hypothetical protein (Fig. 1).
The Δphr mutant lacks light-dependent UV resistance “photoreactivation” activity.
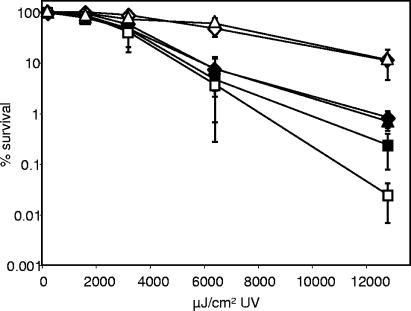
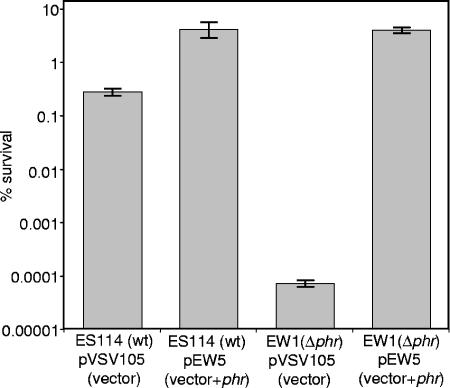
To test whether phr encodes a functional photolyase, we generated and characterized mutant EW1, which contains an in-frame deletion of the gene. Both EW1 and its wild-type parent, ES114, displayed a dose-dependent sensitivity to UV light; however, ES114 possessed a light-dependent mechanism of UV resistance that was absent in the mutant (Fig. 2). ES114 also appeared slightly more resistant to UV light than the mutant, even when recovery was in the dark, possibly due to minor photoreactivation stimulated by the darkroom red light or to Phr-assisted excision repair, which has been observed in E. coli (31). Restoring the phr gene in trans to the Δphr mutant complemented its UV sensitivity (Fig. 3), indicating that this phenotype was not due to effects on VFA072, VFA0754, or other genes. Taken together, our data indicate that V. fischeri phr encodes the bacterium's photoreactivating photolyase.
FIG. 2.
Effects of UV dose and subsequent recovery in the light (open symbols) or dark (filled symbols) on survival of V. fischeri ES114 (diamonds), the ΔluxCDABEG mutant EVS102 (triangles), or the Δphr mutant EW1 (squares). Percent survival was calculated relative to non-UV-exposed cells. The data represent means with standard errors for three independent cultures of each strain.
FIG. 3.
Complementation of UV sensivity in Δphr mutant EW1. Wild-type or EW1 (Δphr) cells carrying either pVSV105 or pEW1 (phr cloned in pVSV105) were exposed to 12,000 μJ/cm2 UV and allowed to recover under fluorescent white lights for 2 h prior to being plated. Percent survival was calculated relative to non-UV-exposed cells. The data represent means with standard errors for three independent cultures.
Bioluminescence does not appear to stimulate photolyase in V. fischeri ES114.
Over a range of UV doses, we saw no difference in the UV sensitivity, or the photoreactivation capacity, of ES114 or the dark ΔluxCDABEG mutant EVS102 (Fig. 2), suggesting that bioluminescence does not play a role in photoreactivation. On the other hand, the cells used in these experiments had been grown to low cell density and were relatively dim. It has been reported that exposure to UV light increases expression of the lux genes in some bacteria even at low cell density (7, 9); however, we considered the possibility that this was not the case in ES114 and that the similarity between ES114 and EVS102 might reflect the relatively dim luminescence ES114 generates in culture (3). We therefore amended the cultures with 200 ng ml−1 of the autoinducer 3-oxo-C6-HSL, an amount similar to that found in the light organ (4), which induced luminescence ∼500-fold to levels comparable to that seen in the symbiosis. Although addition of 3-oxo-C6-HSL stimulated luminescence in ES114, there was still no detectable difference between ES114 and the ΔluxCDABEG mutant EVS102 with regard to UV sensitivity or photoreactivation (Fig. 4).
FIG. 4.
Effects of pregrowth in 200 ng ml−1 3-oxo-C6-HSL (indicated by “+AI”) and outgrowth in the light (light bars) or dark (dark bars) on survival of V. fischeri ES114, the ΔluxCDABEG mutant EVS102, or the Δphr mutant EW1 following exposure to 12,000 μJ/cm2 UV. Percent survival was calculated relative to non-UV-exposed cells. The data represent means with standard errors for three independent cultures of each strain.
Interestingly, the addition of 3-oxo-C6-HSL appeared to increase resistance to UV light when the cells recovered from UV exposure in the dark; however, this trend was seen in the wild type, the dark mutant EVS102, and the Δphr mutant EW1 (Fig. 4), indicating that this effect is not directly related to bioluminescence or to photolyase. For each of the three strains, pregrowth in 3-oxo-C6-HSL significantly (P < 0.05) enhanced the survival of cells incubated in the dark following UV exposure in two out of three independent experiments, and when data for the three strains were pooled, the effect of 3-oxo-C6-HSL was significant (P < 0.05) in all three experiments.
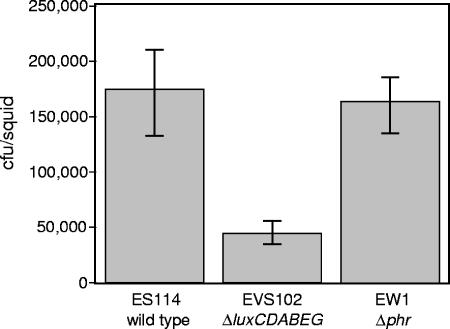
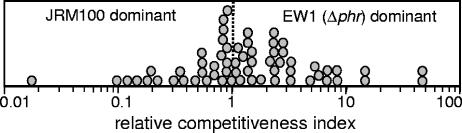
We also examined ES114, the dark ΔluxCDABEG mutant EVS102, and the phr mutant EW1 under conditions where bioluminescence is naturally induced and is beneficial to the bacteria, during symbiotic colonization of the E. scolopes light organ. Consistent with other studies (41; J. L. Bose, C. S. Rosenberg, and E. V. Stabb, unpublished data), we found that populations of the lux mutant in squid light organs 48 h after inoculation were three- to fourfold lower than the symbiont populations in ES114-infected animals (Fig. 5). However, colonization by the phr mutant EW1 was indistinguishable from that of ES114 (Fig. 5). In some instances, symbiotic defects can be detected only if strains are forced to compete for colonization; however, as shown in Fig. 6, EW1 was not outcompeted by JRM100, a marked derivative of ES114 with competitiveness similar to that of the wild type (26). Thus, in contrast to the benefit of bioluminescence in this environment, we found no evidence of an important role for photoreactivation during symbiotic colonization.
FIG. 5.
Symbiotic colonization of E. scolopes hatchlings by the wild type, ΔluxCDABEG mutant EVS102, or Δphr mutant EW1. Average colonization levels 48 h after inoculation with the indicated strain are shown, with standard errors (n = 13 or 14).
FIG. 6.
Competition during mixed infection between V. fischeri EW1 (Δphr) and JRM100. JRM100 is a derivative of ES114 marked with an intergenic mini-Tn7-ermR insertion that does not affect competitiveness (26). Juvenile squid were exposed to an ∼1:1 mixture of the strains at a total concentration of 2,500 CFU ml−1 for 14 h, and the RCI of EW1, defined as the ratio of EW1 to JRM100 in the squid divided by the ratio of these strains in the inoculum, was determined for each coinfected animal 48 h after inoculation. The RCI is plotted as a circle. A dotted line marks the RCI of 1, where the strain ratio in the squid matches that in the inoculum and there is no difference in strain competitiveness.
DISCUSSION
We have shown that the V. fischeri ES114 phr gene (ORF VFA0753) encodes the photolyase enzyme of ES114 and that the benefit of bioluminescence for this bacterium during host colonization is not mediated by stimulation of Phr. ORF VFA0753 encoded the only photolyase (Phr) homolog in the ES114 genome, an in-frame deletion mutant of phr lacked photoreactivation (Fig. 2 and 4), and this phenotype could be complemented with the phr gene in trans (Fig. 3). However, although bioluminescence enabled the bacteria to fully colonize the E. scolopes light organ, as reported elsewhere (41), the phr mutant was unaffected in host colonization (Fig. 5 and 6). Thus, in the light organ environment where the bacteria are naturally bioluminescent and where bioluminescence is advantageous for the bacteria, the benefits of bioluminescence are not mediated by photolyase. Moreover, bioluminescence did not detectably stimulate photoreactivation in culture, even when bioluminescence was stimulated by the addition of the autoinducer 3-oxo-C6-HSL (Fig. 4). Taken together, our data do not support the model that bioluminescence functions to stimulate photolyase-mediated DNA repair in V. fischeri ES114.
We cannot rule out the possibility that bioluminescence does benefit ES114 or other bacteria by stimulating photolyase-mediated DNA repair in some situations. It is worth noting that even with the addition of 3-oxo-C6-HSL, strain ES114 produces only about 1/10 the luminescence of V. fischeri strain MJ1 (Bose et al., submitted), and perhaps luminescence functions to stimulate photolyase only in very bright strains. Our characterization of photolyase in V. fischeri ES1114 should facilitate future studies of this enzyme in more highly luminescent strains, such as MJ1. It should also be noted that most of the evidence supporting a connection between photolyase and bioluminescence has been generated using V. harveyi or the V. harveyi lux genes cloned in E. coli (8, 9, 23), and our experiments do not address the possibility that luminescence in V. harveyi is tied to photolyase.
Nonetheless, we believe that several theoretical considerations make photoreactivation an unlikely natural role for bioluminescence. For example, photoreactivation in bacteria is optimally stimulated by 380-nm- to 440-nm-wavelength light (12, 22), but bioluminescent bacteria usually emit blue (∼500- nm-wavelength) light, with different strains emitting light with wavelengths ranging from 476 nm to 540 nm (33, 39). To our knowledge, photolyase has not been closely studied in a bioluminescent strain, but it has been examined in V. parahaemolyticus, a species that includes bioluminescent strains and is a close relative of V. harveyi and V. fischeri. In nonluminescent V. parahaemolyticus WP28, photolyase is maximally stimulated by light in the 375-nm to 425-nm range, 475-nm light is less than 10% as effective for photoreactivation, and light at >500 nm is ineffective (30). Thus, unless the photolyases of bioluminescent V. parahaemolyticus strains are highly diverged from that of WP28, there will be relatively little overlap in the bioluminescence and photoreactivation spectra in these strains.
The possibility that bioluminescence is not well attuned to the absorption requirements of photolyase raises the issues of efficiency and energetics. Hastings and Nealson calculated that the energy of each photon emitted by bioluminescence is equivalent to the hydrolysis of six ATP molecules (15). Even more energy would be required if the bioluminescence efficiency is <100%, as is likely the case, and this does not even take into account the considerable energy devoted to Lux protein synthesis (16, 21). Moreover, it seems likely that most of the photons generated will not be absorbed and utilized by photolyase, and this is supported by our observation that there was no detectable difference in the intensities of bioluminescence emitted from cultures of the wild type and the photolyase mutant (data not shown). Thus, energy devoted to bioluminescence-mediated photoreactivation could conceivably be equivalent to the hydrolysis of tens, hundreds, or possibly more ATP molecules per lesion repaired. This seems a remarkably inefficient process, although the energy cost might be outweighed by the importance of maintaining genetic integrity.
Perhaps more problematic is the question of whether there is a dark environment where bioluminescent bacteria would benefit from light-mediated activation of photolyase. Vibrio spp. are common near the ocean surface (13), where they are exposed to UV light. However, in this setting, the bacteria would also be exposed to light that could support photoreactivation, and at increased depths, UV light is filtered out to a greater degree than the visible light that would support photoreactivation (2, 18). Although photolyase also binds to DNA lesions other than pyrimidine dimers, and in E. coli it stimulates excision repair of such lesions (31), these examples do not constitute photoreactivation and do not require light. Until a natural condition is found under which photoreactivation is beneficial in an environment lacking light, the evidence suggesting that bioluminescence functions to activate photolyase arguably reflects artificial conditions.
One unexpected finding of our study was that phr is located on chromosome II in V. fischeri, V. cholerae, V. vulnificus, and V. parahaemolyticus (6, 17, 25, 34). In members of the family Vibrionaceae, chromosome II contains a higher relative representation of unusual and presumably niche-specific genes than does the larger chromosome I, which has a higher representation of housekeeping genes. Although photolyase might be considered one of the housekeeping genes found on chromosome II, it could also reflect the fact that the benefit of having phr is specific to certain environments, such as those exposed to UV light. In this regard, it is interesting to note that Photobacterium profundum strain SS9, a member of the Vibrionaceae that is adapted to deep, dark marine environments, lacks a photolyase gene (40).
Another interesting and unexpected finding was that the addition of 3-oxo-C6-HSL to stimulate bioluminescence also appeared to increase the survival of V. fischeri exposed to UV light and then kept in the dark but that this UV resistance was mediated by a mechanism independent of bioluminescence or photolyase (Fig. 4). This phenomenon was statistically significant (P < 0.05) for each strain in two out of three experiments, and it could provide an alternative explanation for the UV sensitivity of undefined (e.g., chemically induced) dark mutants. If bioluminescence and some mechanism(s) of UV resistance are coregulated, then some regulatory mutants might be both dark and UV sensitive without a direct causal relationship between bioluminescence and UV resistance.
The functional significance of bioluminescence for light-producing bacteria remains uncertain, and the observation that bioluminescence allows V. fischeri ES114 to fully colonize its host is equally mysterious. Several plausible explanations have been offered (35), suggesting that bioluminescence acts as a sink for excess reductant (5), as an antioxidant (24, 38, 41), or as a symbiotic signal to the host that is required for light organ tissue to develop into an environment receptive to colonization (M. McFall-Ngai and C. Whistler, personal communication). Given our results and the arguments above, we are skeptical of the idea that the driving force behind the evolution of bioluminescence was that it played a significant role in stimulating photolyase. There may not be a single, simple role for bioluminescence, and it seems likely that bioluminescence may serve different purposes depending on the bacterium and the environment. Our laboratory and others are actively investigating these multiple possible functions of bioluminescence.
Acknowledgments
We thank Andrea Zbell for technical assistance. Genome information was provided by the Vibrio fischeri Genome Project (http://ergo.integratedgenomics.com/Genomes/VFI), supported by the W. M. Keck Foundation.
This work was supported by a Graduate Recruitment Opportunities fellowship from the University of Georgia to E.L.W., by National Institutes of Health grant R01AI50661, and by a CAREER award to E.V.S. from the National Science Foundation (MCB-0347317).
Footnotes
Published ahead of print on 21 August 2006.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Atkins, W. R. G., and H. H. Poole. 1933. The photo-electric measurement of the penetration of light of various wave-lengths into the sea and the physiological bearing of the results. Phil. Trans. R. Soc. Lond. B 222:129-164. [Google Scholar]
- 3.Boettcher, K. J., and E. G. Ruby. 1990. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J. Bacteriol. 172:3701-3706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boettcher, K. J., and E. G. Ruby. 1995. Detection and quantification of Vibrio fischeri autoinducer from symbiotic squid light organs. J. Bacteriol. 177:1053-1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourgois, J.-J., F. E. Sluse, F. Baguet, and J. Mallefet. 2001. Kinetics of light emission and oxygen consumption by bioluminescent bacteria. J. Bioenerg. Biomembr. 33:353-363. [DOI] [PubMed] [Google Scholar]
- 6.Chen, C. Y., K. M. Wu, Y. C. Chang, C. H. Chang, H. C. Tsai, T. L. Liao, Y. M. Liu, H. J. Chen, A. B. Shen, J. C. Li, T. L. Su, C. P. Shao, C. T. Lee, L. I. Hor, and S. F. Tsai. 2003. Comparative genome analysis of Vibrio vulnificus, a marine pathogen. Genome Res. 13:2577-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Czyz, A., K. Plata, and G. Wegrzyn. 2002. Induction of light emission by luminescent bacteria treated with UV light and chemical mutagens. J. Appl. Genet. 43:377-389. [PubMed] [Google Scholar]
- 8.Czyz, A., K. Plata, and G. Wegrzyn. 2003. Stimulation of DNA repair as an evolutionary drive for bacterial bioluminescence. Luminescence 18:140-144. [DOI] [PubMed] [Google Scholar]
- 9.Czyz, A., B. Wrobel, and G. Wegrzyn. 2000. Vibrio harveyi bioluminescence plays a role in stimulation of DNA repair. Microbiology 146:283-288. [DOI] [PubMed] [Google Scholar]
- 10.Dunn, A. K., M. O. Martin, and E. V. Stabb. 2005. Characterization of pES213, a small mobilizable plasmid from Vibrio fischeri. Plasmid 54:114-134. [DOI] [PubMed] [Google Scholar]
- 11.Dunn, A. K., D. S. Millikan, D. M. Adin, J. L. Bose, and E. V. Stabb. 2006. New rfp- and pES213-derived tools for analyzing symbiotic Vibrio fischeri reveal patterns of infection and lux expression in situ. Appl. Environ. Microbiol. 72:802-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eker, A. P. M., P. Kooiman, J. K. C. Hessels, and A. Yasui. 1990. DNA photoreactivating enzyme from the cyanobacterium Anacystis nidulans. J. Biol. Chem. 265:8009-8015. [PubMed] [Google Scholar]
- 13.Franklin, M. P., I. R. McDonald, D. G. Bourne, N. J. Owens, R. C. Upstill-Goddard, and J. C. Murrell. 2005. Bacterial diversity in the bacterioneuston (sea surface microlayer): the bacterioneuston through the looking glass. Environ. Microbiol. 7:723-736. [DOI] [PubMed] [Google Scholar]
- 14.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 15.Hastings, J. W., and K. H. Nealson. 1977. Bacterial bioluminescence. Annu. Rev. Microbiol. 31:549-595. [DOI] [PubMed] [Google Scholar]
- 16.Hastings, J. W., W. H. Riley, and J. Massa. 1965. The purification, properties, and chemiluminescent quantum yield of bacterial luciferase. J. Biol. Chem. 240:1473-1481. [PubMed] [Google Scholar]
- 17.Heidelberg, J. F., J. A. Eisen, W. C. Nelson, R. A. Clayton, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, L. Umayam, S. R. Gill, K. E. Nelson, T. D. Read, H. Tettelin, D. Richardson, M. D. Ermolaeva, J. Vamathevan, S. Bass, H. Qin, I. Dragoi, P. Sellers, L. McDonald, T. Utterback, R. D. Fleishmann, W. C. Nierman, O. White, S. L. Salzberg, H. O. Smith, R. R. Colwell, J. J. Mekalanos, J. C. Venter, and C. M. Fraser. 2000. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 406:477-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helbling, E. W., E. S. Barbieri, M. A. Marcoval, R. J. Goncalves, and V. E. Villafane. 2005. Impact of solar ultraviolet radiation on marine phytoplankton of Patagonia, Argentina. Photochem. Photobiol. 81:807-818. [DOI] [PubMed] [Google Scholar]
- 19.Henikoff, S., and J. G. Henikoff. 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89:10915-10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Herrero, M., V. De Lorenzo, and K. N. Timmis. 1990. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172:6557-6567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karl, D., and K. H. Nealson. 1980. Regulation of cellular metabolism during synthesis and expression of the luminous system in Beneckea and Photobacterium. J. Gen. Microbiol. 117:357-368. [Google Scholar]
- 22.Kelner, A. 1951. Action spectra for photoreactivation of ultraviolet-irradiated Escherichia coli and Streptomyces griseus. J. Gen. Physiol. 34:835-852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozakiewicz, J., M. Gajewska, R. Lyzen, A. Czyz, and G. Wegrzyn. 2005. Bioluminescence-mediated stimulation of photoreactivation in bacteria. FEMS Microbiol. Lett. 250:105-110. [DOI] [PubMed] [Google Scholar]
- 24.Lyzen, R., and G. Wegrzyn. 2005. Sensitivity of dark mutants of various strains of luminescent bacteria to reactive oxygen species. Arch. Microbiol. 183:203-208. [DOI] [PubMed] [Google Scholar]
- 25.Makino, K., K. Oshima, K. Kurokawa, K. Yokoyama, T. Uda, K. Tagomori, Y. Iijima, M. Najima, M. Nakano, A. Yamashita, Y. Kubota, S. Kimura, T. Yasunaga, T. Honda, H. Shinagawa, M. Hattori, and T. Iida. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet 361:743-749. [DOI] [PubMed] [Google Scholar]
- 26.McCann, J., E. V. Stabb, D. S. Millikan, and E. G. Ruby. 2003. Population dynamics of Vibrio fischeri during infection of Euprymna scolopes. Appl. Environ. Microbiol. 69:5928-5934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meighen, E. A. 1994. Genetics of bacterial bioluminescence. Annu. Rev. Genet. 28:117-139. [DOI] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Nealson, K. H., and J. W. Hastings. 1979. Bacterial bioluminescence: its control and ecological significance. Microbiol. Rev. 43:496-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okaichi, K., T. Ohnishi, and K. Nozu. 1984. Photoreactivation of UV-killing in Vibrio parahaemolyticus WP28. Photochem. Photobiol. 39:771-773. [DOI] [PubMed] [Google Scholar]
- 31.Ozer, Z., J. T. Reardon, D. S. Hsu, K. Malhotra, and A. Sancar. 1995. The other function of DNA photolyase: stimulation of excision repair of chemical damage to DNA. Biochemistry 34:15886-15889. [DOI] [PubMed] [Google Scholar]
- 32.Ruby, E. G., and L. M. Asato. 1993. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch. Microbiol. 159:160-167. [DOI] [PubMed] [Google Scholar]
- 33.Ruby, E. G., and K. Nealson. 1977. A luminous bacterium that emits yellow light. Science 196:432-434. [DOI] [PubMed] [Google Scholar]
- 34.Ruby, E. G., M. Urbanowski, J. Campbell, A. Dunn, M. Faini, R. Gunsalus, P. Lohstroh, C. Lupp, J. McCann, D. Millikan, A. Schaefer, E. Stabb, A. Stevens, K. Visick, C. Whistler, and E. P. Greenberg. 2005. Complete genome sequence of Vibrio fischeri: a symbiotic bacterium with pathogenic congeners. Proc. Natl. Acad. Sci. USA 102:3004-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stabb, E. V. 2005. Shedding light on the bioluminescence “paradox.” ASM News 71:223-229. [Google Scholar]
- 36.Stabb, E. V., and E. G. Ruby. 2003. Contribution of pilA to competitive colonization of Euprymna scolopes by Vibrio fischeri. Appl. Environ. Microbiol. 69:820-826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stabb, E. V., and E. G. Ruby. 2002. RP4-based plasmids for conjugation between Escherichia coli and members of the Vibrionaceae. Methods Enzymol. 358:413-426. [DOI] [PubMed] [Google Scholar]
- 38.Timmins, G. S., S. K. Jackson, and H. M. Swartz. 2001. The evolution of bioluminescent oxygen consumption as an ancient oxygen detoxification mechanism. J. Mol. Evol. 52:321-332. [DOI] [PubMed] [Google Scholar]
- 39.Tu, S.-C., and H. I. X. Mager. 1995. Biochemistry of bacterial bioluminescence. Photochem. Photobiol. 62:615-624. [DOI] [PubMed] [Google Scholar]
- 40.Vezzi, A., S. Campanaro, M. D'Angelo, F. Simonato, N. Vitulo, F. M. Lauro, A. Cestaro, G. Malacrida, B. Simionati, N. Cannata, C. Romualdi, D. H. Bartlett, and G. Valle. 2005. Life at depth: Photobacterium profundum genome sequence and expression analysis. Science 307:1459-1461. [DOI] [PubMed] [Google Scholar]
- 41.Visick, K. L., J. Foster, J. Doino, M. McFall-Ngai, and E. G. Ruby. 2000. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J. Bacteriol. 182:4578-4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weber, S. 2005. Light-driven enzymatic catalysis of DNA repair: a review of recent biophysical studies on photolyase. Biochim. Biophys. Acta 1707:1-23. [DOI] [PubMed] [Google Scholar]
- 43.Wegrzyn, G., and A. Czyz. 2002. How do marine bacteria produce light, why they are luminescent, and can we employ bacterial bioluminescence in aquatic biotechnology. Oceanologia 44:291-305. [Google Scholar]