Abstract
Recently we discovered two novel, deeply branching lineages in the domain Bacteria from termite guts by PCR-based analyses of 16S rRNA (Y. Hongoh, P. Deevong, T. Inoue, S. Moriya, S. Trakulnaleamsai, M. Ohkuma, C. Vongkaluang, N. Noparatnaraporn, and T. Kudo, Appl. Environ. Microbiol. 71:6590-6599, 2005). Here, we report on the specific detection of these bacteria, the candidate phylum TG3 (Termite Group 3) and a subphylum in the phylum Fibrobacteres, by fluorescence in situ hybridization in the guts of the wood-feeding termites Microcerotermes sp. and Nasutitermes takasagoensis. Both bacterial groups were detected almost exclusively from the luminal fluid of the dilated portion in the hindgut. Each accounted for approximately 10% of the total prokaryotic cells, constituting the second-most dominant groups in the whole-gut microbiota. The detected cells of both groups were in undulate or vibroid forms and apparently resembled small spirochetes. The cell sizes were 0.2 to 0.4 by 1.3 to 6.0 μm and 0.2 to 0.3 by 1.3 to 4.9 μm in the TG3 and Fibrobacteres, respectively. Using PCR screenings with specific primers, we found that both groups are distributed among various termites. The obtained clones formed monophyletic clusters that were delineated by the host genus rather than by the geographic distance, implying a robust association between these bacteria and host termites. TG3 clones were also obtained from a cockroach gut, lake sediment, rice paddy soil, and deep-sea sediments. Our results suggest that the TG3 and Fibrobacteres bacteria are autochthonous gut symbionts of various termites and that the TG3 members are also widely distributed among various other environments.
Termites harbor an abundance and diversity of gut bacteria, which are thought to play essential roles in the carbon and nitrogen metabolism of their host termites (4, 26). Recent culture-independent analyses have revealed that the bacterial gut microbiota comprises many termite-specific lineages that are as yet uncultured (11-13, 27, 33, 35, 39). Among them, the candidate phylum Termite Group I (TG1) was first recognized in our previous study as a novel, deeply branching lineage specific to termites (27) and later found to constitute a new phylum, together with clones from various environments (13, 15). Now, the termite-specific cluster in this candidate phylum has been partly characterized as endosymbionts of gut protists in various lower termites (28, 38), whereas no isolate exists so far from this phylum. In higher termites, which generally lack gut symbiotic protists and harbor only prokaryotes (in contrast to lower termites that harbor both), there have been found other novel, deeply branching lineages in the domain Bacteria. Using clonal analyses of 16S rRNA, we recently discovered a novel phylum-level cluster, temporarily named TG3 (Termite Group 3), and a novel subphylum-level cluster in the phylum Fibrobacteres (designated Fibrobacteres subphylum 2 in this study) from the guts of the wood-feeding higher termites Microcerotermes spp. (11). Each group accounted for approximately 10% of the analyzed clones, constituting the second-most dominant groups, together with the orders Bacteroidales and Clostridiales, following the predominant genus, Treponema. The candidate phylum TG3 can be divided into two subphyla. Subphylum 1 was abundantly found in the analyzed clones from the Microcerotermes termites and related to several clones from other environments, such as rice paddy soil and salt marsh sediment (11). Subphylum 2 contains a few phylotypes that were found only rarely in Microcerotermes spp. and the fungus-growing termite Macrotermes gilvus (12) and that were related to a few marine clones. No isolate has been obtained from these groups, and no information other than the 16S rRNA sequences is available to date, as with other many-candidate (sub)phyla that have no cultured representatives. In fact, while the number of phylum-level clusters has been increasing as 16S sequence data accumulate, most of them have never been investigated, even for their localization and diversity.
In the present study, we attempted to detect TG3 and Fibrobacteres bacteria in the guts of Microcerotermes sp. and another wood-feeding higher termite, Nasutitermes takasagoensis, by fluorescence in situ hybridization (FISH) with specific probes. Moreover, we designed specific PCR primers in order to detect them from various environments, including termite and cockroach guts, lake sediment, sea sediments, rice paddy soil, and others. Our study reveals the in situ localization, morphology, diversity, and broad distribution of these novel bacterial lineages.
MATERIALS AND METHODS
Sample collection and DNA extraction.
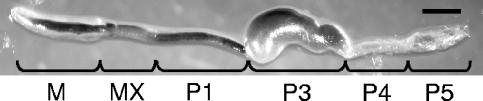
Termites and wood-feeding cockroaches were collected with their nest or nest log and carefully transported to our laboratory without heating or sunlight exposure. Five to 20 worker termites were randomly chosen from each colony immediately after collection or after being kept with their nest log for several months in the laboratory. Whole guts were isolated from these individuals by using sterile forceps. For cockroaches, one adult individual was randomly chosen and the whole gut was removed by dissection on ice with sterile scissors and forceps. To investigate the in situ localization of bacteria, 10 guts of adult workers of the termite Nasutitermes takasagoensis, collected in Iriomote Island, Japan, were cut into five pieces, i.e., midgut, mixed segment, proctodeal segment part 1 (P1), P3, and P4 and P5 combined (Fig. 1), as described previously (39). A colony of Microcerotermes sp. used for FISH analyses was collected in Bangkok, Thailand.
FIG. 1.
The gut of Nasutitermes takasagoensis. M, midgut; MX, mixed segment; P1 to P5, hindgut sections. Scale bar = 1 mm.
DNA was extracted from the gut homogenates by using an Isoplant II kit (Nippon Gene Co.), which chemically lyses bacterial cell walls and membranes with benzyl chloride. The extracts were further purified using a DNeasy tissue kit (QIAGEN) as described previously (40). Soil samples were subjected to an additional extraction step using cetyl trimethyl ammonium bromide between the Isoplant and DNeasy steps as described previously (12).
PCR amplification.
PCR was performed with the Bacteria-specific primer pair 27F (5′-AGAGTTTGATYMTGGCTCAG) and 1390R (5′-ACGGGCGGTGTGTACAA) (39) to amplify the near-full-length 16S rRNA gene. For the construction of a clone library from N. takasagoensis, the PCR was conducted, as described previously (11), with the following program: an initial 2-min denaturation at 95°C, 12 cycles of denaturation (30 s at 95°C), annealing (1 min at 50°C) and extension (4 min at 72°C), and a final 10-min extension at 72°C.
For the detection of specific bacterial groups, the PCR products after amplification as described above underwent 20 to 24 cycles instead of 12, were diluted to approximately the same concentration among samples, and were used as the template for the nested PCR with the taxon-specific primers listed in Table 1. The annealing temperature for each pair of primers was optimized using the gradient program of a PTC-200 thermal cycler (MJ Research). For the detection of TG3 subphylum 1 or 2, 65°C was chosen as the annealing temperature and 72°C was chosen for the detection of Fibrobacteres subphylum 2. After checking the amplifications with 25 cycles, 10 to 20 cycles of PCR were performed for cloning.
TABLE 1.
FISH probes and PCR primers designed in this study
| Probe or primer | Sequence (5′→3′) | Target |
|---|---|---|
| FISH probes | ||
| TG3S1-168 | GCCCCGCGTTGGCAAGGT | TG3 subphylum 1 |
| TG3S2-35 | ATTAAGCACTCCGCTAGC | TG3 subphylum 2 |
| FibS2-416 | GTTTACACGCCTAGGCGC | Fibrobacteres subphylum 2 |
| Spiro-36 | CTTAAGACGCGCCGCCAG | Spirochaetes |
| Bactd-937 | CCACATGTTCCTCCGCTT | Bacteroidales |
| PCR primers | ||
| TG3S1-164F | GGGATAACCTTGCCAACGC | TG3 subphylum 1 |
| TG3S2-44F | AGTGAACGCTRGCGGAG | TG3 subphylum 2 |
| TG3-1225R | RCCATTGTAGCACGTGTC | TG3 |
| FibS2-53F | GCTGGYGGCGTGTYTKATG | Fibrobacteres subphylum 2 |
| FibS2-1186R | ACCTTCCTCCGGGTTGTCC | Fibrobacteres subphylum 2 |
Cloning and sequencing.
The PCR products were purified using a MonoFas DNA purification kit (GL Sciences), and TA cloning was performed using a TOPO TA cloning kit for sequencing (Invitrogen). Clones were randomly chosen from the constructed libraries, and sequencing was performed using a BigDye Terminator cycle sequencing kit (PerkinElmer) and an ABI 3700 genetic analyzer as described previously (13). All sequenced clones were subjected to the identification of chimeric sequences by using the online programs RDP II Chimera Check (21) and Bellerophon (14) as described previously (11). The detected chimeras were eliminated from the following analyses. The remaining clones were sorted into phylotypes with a criterion of 97.0 or 99.0% sequence identity by using the program DOTUR, version 1.5 (31). The statistical comparisons of clone libraries were conducted using the program ∫-LIBSHUFF, version 1.21 (32), as described previously (11).
Phylogenetic analysis.
Alignment and preliminary phylogenetic affiliation of the clones were performed using ARB software (20). The sequences of clones were incorporated into the ARB database ssujun02, which was modified in our previous study (12), and the alignment was corrected manually. Closely related sequences, found by a BLAST search (2), and all termite gut clones available in the public databases DDBJ, GenBank, and EMBL (accessed in March 2006), were also added to the ARB database. A clone with the least PCR errors, as judged with the definition by Acinas et al. (1), was chosen as the representative of a phylotype and used for the construction of phylogenetic trees. Maximum likelihood (ML) trees were constructed using the PHYML, version 2.4.4, program (9) with the general time-reversible (GTR) nucleotide substitution model. The heterogeneity of nucleotide substitution rates among sites was approximated by a gamma distribution (G) and an assumption of invariable sites (I). Minimum evolution (ME) trees were constructed by the tree bisection-reconnection of a neighbor-joining tree using PAUP* (version 4.0b10; D. Swofford, Sinauer Associates, Sunderland, MA). The inferred trees were depicted by using the tree-drawing function of MEGA, version 3.1 (18).
FISH.
We designed oligonucleotide probes targeting 16S rRNA specific to each of TG3 subphyla 1 and 2 and Fibrobacteres subphylum 2 (Table 1) by using the probe-designing function of ARB (20). To elucidate the taxonomic composition of bacteria in the guts of Microcerotermes sp. and N. takasagoensis, probes specific to each of the order Bacteroidales and the phylum Spirochaetes were also designed (Table 1). These probes were labeled at the 5′ end with either Texas Red or 6-carboxyfluorescein (FAM) and used for FISH, basically as described previously (24, 25). The sequence specificity of these probes was checked in the probe match program in RDP II (21), and the optimal condition for specific hybridization was determined by Clone-FISH as described by Schramm et al. (34). Briefly, plasmids carrying a target sequence (the positive control) or a nontarget sequence (the negative control) were introduced into the λDE3 lysogen of NovaBlue (Novagen) and the insert was transcribed by T7 RNA polymerase induced by the addition of 1 mM isopropyl-β-d-thiogalactoside (IPTG). After the transcripts were accumulated by an addition of 170 μg/ml chloramphenicol, the host cells were collected and subjected to FISH. The clones and a cultured isolate, used as the controls, and detailed information on the specificity of the probes are described in Table S1 in the supplemental material. The hybridization temperature was set to 60°C for all probes, and for just the probe specific to the Bacteroidales, 20% formamide and 0.05 pmol/μl of the competitors comp-Bact1 (5′-CCACATGCTCCTCCGCTT) and comp-Bact2 (5′-CCACATGTTCCACCGCTT) were added. No cross-hybridization was observed under these conditions between any pair of specific probes. The mixture of probes EUB338 (3) and EUB338II and -III (7) was used to detect most cells in the domain Bacteria, with the hybridization temperature at 60°C. The specimens were observed with an Olympus epifluorescence microscope (BX-60).
Enumeration of cells.
The total number of prokaryotic cells in the whole guts of termites was estimated by using 4′,6′-diamidino-2-phenylindole HCl (DAPI) as described previously (11). The taxonomic composition of bacteria in termite guts was determined from the proportion of cells identified by FISH with specific probes against DAPI-stained cells, which were mounted on silane-coated slide glasses in a density of approximately 400 to 1,200 cells per 5.3 × 10−3 mm2 for Microcerotermes sp. and 100 to 300 cells per 5.3 × 10−3 mm2 for N. takasagoensis. In Microcerotermes sp., the proportion of cells detected by FISH against a total of approximately 2,000 to 3,400 DAPI-stained cells was calculated from three or four microscopic fields. In N. takasagoensis, 10 microscopic fields per sample were observed and the proportion against 1,200 to 3,000 DAPI-stained cells was calculated. The FISHs were conducted with the combinations of probes TG3S1-168 and FibS2-416, TG3S2-35 and Bactd-937, and Spiro-36 and the EUB338 mixture. Each of a pair was labeled with a different dye (Texas Red or FAM), and captured fluorescence microscopic images were overlaid so as to distinguish signals visualized with a red or green color from insect tissues and debris emitting autofluorescence with a yellowish color. Since we could not design an appropriate probe for another dominant bacterial group in termite guts, the order Clostridiales, cells with an endospore and/or that were gram positive were counted as gram-positive bacteria, including the Clostridiales. Gram staining was performed using a ViaGram Red+ bacterial Gram stain kit (Molecular Probes). In this kit, gram-positive cells were bound by Texas Red-labeled wheat germ agglutinin and detected by fluorescence microscopy (36). Since some spirochetes were found to be Gram stained with this method, we excluded the spirochete form cells from the count for gram-positive cells. The enumeration was performed as described for the FISH analyses. Significant differences in frequency among samples were detected using chi-square tests, and a sample that caused a difference was specified by confirming insignificancy when excluding the sample from a comparison. Cell sizes were shown as width times wavelength. Data are expressed throughout this paper as the means ± standard deviations unless otherwise stated.
Nucleotide sequence accession numbers.
The 16S rRNA sequences generated in this study have been deposited with DDBJ under accession numbers AB255887 to AB256016.
RESULTS
Clonal analysis of gut bacteria in N. takasagoensis.
We sequenced 170 clones of 16S rRNA, amplified by PCR of the gut homogenate of N. takasagoensis, and found 51 phylotypes defined with ≥97.0% sequence identity. Phylogenetic analysis revealed that the phylotypes were affiliated with seven phyla, including the candidate phylum TG3 and the phylum Fibrobacteres (data not shown). The taxonomic composition based on the clone frequency was very similar to those of Microcerotermes spp. at the higher taxonomic level (Table 2), although only one phylotype overlapped and the clone libraries were statistically different (∫-LIBSHUFF test; P < 0.0001).
TABLE 2.
Taxonomic composition of bacteria in the gut of Microcerotermes sp. and Nasutitermes takasagoensis, shown by clonal and FISH analyses of 16S rRNAe
| Taxon of bacteria | Clone frequency (%)
|
Frequency of FISH-detected cells against DAPI count (%)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Mspa | Nt | Msp | Nt-avb | Nt-1c | Nt-2c | Nt-3c | Nt-4c | Nt-5c | |
| TG3S1 | 6.4 | 9.4 | 10.4 | 10.9 | 4.3 | 4.9 | 23.7 | 11.2 | 10.4 |
| Fibrobacteres S2 | 7.6 | 14.1 | 12.6 | 13.5 | 14.0 | 16.0 | 10.8 | 12.4 | 14.2 |
| Spirochaetes | 58.7 | 57.1 | 55.2 | 59.3 | 64.3 | 56.8 | 53.6 | 62.4 | 59.3 |
| Bacteroidales | 6.3 | 4.7 | 3.7 | 2.2 | 1.8 | 3.6 | 2.2 | 1.8 | 1.5 |
| Gram positives | 12.2 | 5.3 | 11.8d | 5.7d | 8.0d | 7.1d | 5.1d | 4.6d | 3.6d |
| Others | 8.9 | 9.4 | 6.3 | 8.5 | 7.6 | 11.6 | 4.6 | 7.6 | 11.0 |
Results are averaged data obtained in our previous study (11).
Results are averaged data from five individuals, Nt-1 to Nt-5.
Data are from individual workers Nt-1 to Nt-5.
Data were obtained using Gram staining instead of FISH.
Msp, Microcerotermes sp.; Nt, Nasutitermes takasagoensis; S2, subphylum 2.
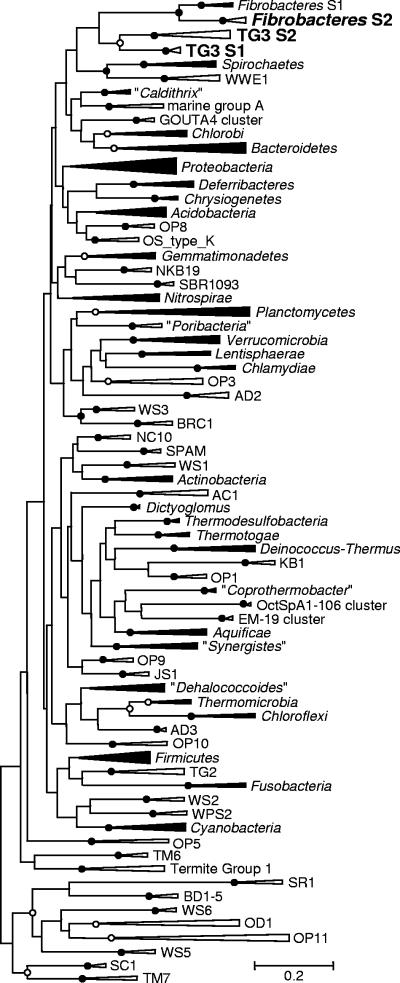
The distinctness of the candidate phylum TG3 from other bacterial phyla was confirmed by phylogenetic analysis with 67 other known phyla or phylum-level clusters (Fig. 2). The monophyly between TG3 subphyla 1 and 2 as well as that between Fibrobacteres subphyla 1 and 2 was also confirmed in this analysis. These monophylies were consistent when different nucleotide substitution models, tree-inferring methods, and sets of reference sequences were used, while the branching order among the phylum-level clusters, including that between TG3 and the Fibrobacteres, was unstable (data not shown).
FIG. 2.
Phylogenetic tree showing the phylum-level clusters in the domain Bacteria based on 16S rRNA sequences. Two or more publicly available sequences were chosen for each phylum-level cluster, and a maximum likelihood tree was constructed with the GTR+G+I model. A total of 1,180 unambiguously aligned nucleotides were used, corresponding to positions 28 to 1388 in Escherichia coli (J01695). Bootstrap tests were performed with 100 resamplings. Open and closed circles at the nodes indicate the bootstrap confidence values 70 to 94 and 95 to 100, respectively. Clusters that have cultured representatives are shown next to closed wedges; clusters represented by only environmental clones are shown next to open wedges. Asterisks indicate the phylum-level clusters recognized in this study in addition to the TG3 phylum. S1 and S2 indicate subphyla 1 and 2, respectively. The alignment is available upon request.
In situ detection by FISH.
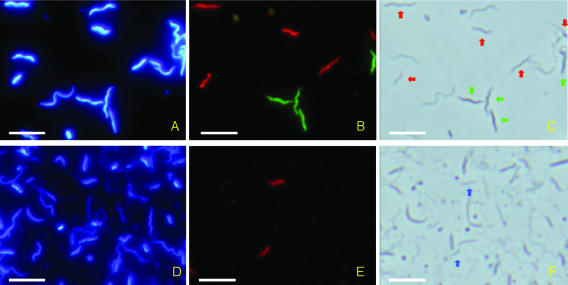
We successfully detected cells of TG3 subphyla 1 and 2 and Fibrobacteres subphylum 2 in the gut homogenates of Microcerotermes sp. and N. takasagoensis by using FISH with specific probes (Fig. 3). Both TG3 subphylum 1 and Fibrobacteres subphylum 2 were in undulate forms with tapered ends (Fig. 3C). The cell sizes were 0.2 to 0.4 by 1.3 to 6.0 μm, with amplitudes of 0.3 to 0.8 μm (n = 200), and 0.2 to 0.3 by 1.3 to 4.9 μm, with amplitudes of 0.3 to 0.6 μm (n = 200), in TG3 subphylum 1 and Fibrobacteres subphylum 2, respectively. Smaller types of cells of both groups appeared as vibroid forms and were similar to the cells of TG3 subphylum 2 shown in Fig. 3F. The cell size of TG3 subphylum 2 was 0.2 to 0.3 by 1.4 to 3.2 μm (n = 38). Although these detected cells were apparently similar to small spirochetes under phase-contrast microscopy (Fig. 3C), we confirmed that they were not spirochetes by counterstaining with probe Spiro-36 that was designed specific to almost all of the spirochete phylotypes found in Microcerotermes spp. and N. takasagoensis (see Table S1 in the supplemental material).
FIG. 3.
Detection of TG3 subphylum 1 and Fibrobacteres subphylum 2 in the P3 section of the hindgut of Nasutitermes takasagoensis (A to C) and TG3 subphylum 2 in the whole-gut homogenate of Microcerotermes sp. (D to F). (A) DAPI image. (B) Cells of TG3 subphylum 1 and Fibrobacteres subphylum 2 were simultaneously detected by FISH with FAM (green) and Texas Red (red), respectively. (C) Phase-contrast image. Cells of TG3 subphylum 1 are indicated by green arrows; cells of Fibrobacteres subphylum 2 are indicated by red arrows. The other undulate or helical cells are presumably spirochetes. (D) DAPI image. (E) Cells of TG3 subphylum 2 detected by FISH with Texas Red (red). (F) Phase-contrast image. Cells of TG3 subphylum 2 are indicated by blue arrows. Bars = 5 μm.
The cell sizes of TG3 subphylum 1 and Fibrobacteres subphylum 2 were significantly different between the host termites. The cells of TG3 subphylum 1 were significantly longer in N. takasagoensis (3.9 ± 0.8 μm) than in Microcerotermes sp. (2.1 ± 0.5 μm) (t test; P < 0.00001). This was also the case with Fibrobacteres subphylum 2; cells were 3.0 ± 0.7 μm in N. takasagoensis and 2.3 ± 0.5 μm in Microcerotermes sp. (t test; P < 0.00001). Since only a few cells of TG3 subphylum 2 were found from N. takasagoensis, it was impossible to compare statistically for this group.
Localization in the gut of N. takasagoensis.
Nested PCRs using taxon-specific primers detected TG3 subphyla 1 and 2 and Fibrobacteres subphylum 2 from only the hindgut sections (P1, P3, and P4 and P5) of N. takasagoensis and not from the midgut or the mixed segment (data not shown). Using FISH analyses, we detected abundant cells of TG3 subphylum 1 and Fibrobacteres subphylum 2 from the P3 section (Fig. 3A to C), rarely so from P4 and P5, and no signal was detected from the midgut, mixed segment and P1 section. No attachment of these cells to the gut wall fragments was observed, while numerous bacterial cells colonized the surface of the fragments, as visualized with DAPI or the EUB338 probe mixture (data not shown). An attachment to wood particles was also not observed. Thus, it is likely that these bacteria are free swimming or only loosely attached to gut wall or food particles. Since cells of TG3 subphylum 2 were found only rarely, we were unable to determine their localizations by FISH.
Enumeration of cells.
We enumerated cells of the bacterial groups dominant in the clone library by FISH with taxon-specific probes or Gram staining. Each whole gut of five adult workers of N. takasagoensis was tested. Most DAPI-stained cells were hybridized with the EUB338 mixture, up to 98.4% ± 0.9%. The taxonomic composition based on FISH was basically similar among individual workers (Table 2). Only the frequency of TG3 subphylum 1 in sample Nt-3 was significantly higher among the individuals (chi-square test; P < 0.0001). The taxonomic composition averaged among the individuals was well congruent with that based on the clonal analysis (Table 2). TG3 subphylum 1 and Fibrobacteres subphylum 2 occupied, on average, 10.9% ± 7.8% and 13.5% ± 2.0% of the DAPI count, respectively. Thus, one gut contained an average of 1.1 × 107 ± 0.8 × 107 and 1.3 × 107 ± 0.2 × 107 cells, based on the total number of prokaryotic cells detected with DAPI (9.8 × 107 ± 0.1 × 107 per gut).
In Microcerotermes sp., the enumeration was conducted for the mixture of the whole gut from 40 worker termites. The cells hybridized with the EUB338 mixture accounted for up to 99.6% of DAPI-stained cells. As with N. takasagoensis, the taxonomic composition obtained here coincided with the results from the clonal analyses in our previous study (11) (Table 2). Thus, the taxonomic compositions based on both clone and FISH analyses were very similar for Microcerotermes sp. and N. takasagoensis at the higher taxonomic level. TG3 subphylum 1 and Fibrobacteres subphylum 2 occupied 10.2% and 12.6% of the DAPI count, corresponding to 6.5 × 105 and 7.9 × 105 cells per gut, respectively, based on the total number of prokaryotic cells in a gut of Microcerotermes sp., 6.2 × 106 ± 2.4 × 106, which was estimated in our previous study (11). The cells of TG3 subphylum 2 were relatively rare, accounting for 1.2% of the DAPI count. This corresponds to 7.5 × 104 cells per gut.
Distribution among various environments.
We conducted PCR screenings with the taxon-specific primers listed in Table 1. PCR products were successfully obtained from the guts of various termites (Table 3) and some other environments (Table 4). We confirmed the amplification of the targets by sequencing eight clones per sample. We obtained TG3 clones from all or most of the termites in the family Termitidae, i.e., higher termites which lack gut protists, but from none or only two species in the other families, i.e., lower termites which harbor gut protists (Table 3). Clones of Fibrobacteres subphylum 2 were obtained from most of the higher termites and four lower termite species. Clones of TG3 subphyla 1 and 2 were also obtained from other environments, including the gut of the wood-feeding cockroach Panesthia angustipennis in the family Panesthiidae, rice paddy soil from three distinct locations, lake sediment, and deep-sea sediments, whereas Fibrobacteres subphylum 2 was never detected from these other environments (Table 4).
TABLE 3.
Detection of TG3 subphylum 1, TG3 subphylum 2, and Fibrobacteres subphylum 2 from termite gut samples by specific amplification of the 16S rRNA genee
| Termite (sub)family | Termite species | Food | TG3S1 | TG3S2 | FibS2 | Collection site |
|---|---|---|---|---|---|---|
| Mastotermitidae | Mastotermes darwiniensis | Wood | − | − | − | Darwin, Australiab |
| Termopsidae | Hodotermopsis sjoestedti | Wood | − | + | + | Yakushima, Japan |
| Termopsidae | Archotermopsis sp. | Wood | − | − | − | Nan, Thailandc |
| Kalotermitidae | Neotermes koshunensis | Wood | − | − | + | Okinawa, Japan |
| Rhinotermitidae | Coptotermes formosanus | Wood | − | + | + | Okinawa, Japan |
| Rhinotermitidae | Reticulitermes speratus | Wood | − | − | − | Tanzawa, Japanc |
| Rhinotermitidae | Reticulitermes amamianus | Wood | − | − | + | Amami, Japanc |
| Rhinotermitidae | Reticulitermes okinawanus | Wood | − | − | − | Okinawa, Japanc |
| Rhinotermitidae | Reticulitermes sp. | Wood | − | − | − | Nan, Thailandc |
| Termitidae | ||||||
| Macrotermitinae | Macrotermes gilvus | Litter | + | + | − | Pathum Thani, Thailandc |
| Apicotermitinae | Speculitermes sp. | Grass | + | −a | − | Pathum Thani, Thailand |
| Termitinae | Termes comis | w/sd | + | + | + | Pathum Thani, Thailandc |
| Termitinae | Pericapritermes nitobei | Soil | + | −a | + | Iriomote, Japan |
| Termitinae | P. latignathus | Soil | + | + | + | Pathum Thani, Thailand |
| Termitinae | Microcerotermes sp. 1 | Wood | + | + | + | Pathum Thani, Thailandc |
| Termitinae | Microcerotermes sp. 2 | Wood | + | + | + | Prachinburi, Thailandc |
| Nasutitermitinae | Nasutitermes dimorphus | Wood | + | + | + | Bangkok, Thailand |
| Nasutitermitinae | N. takasagoensis | Wood | + | + | + | Iriomote, Japan |
Although a faint signal was obtained, the sequences were not of TG3S2, but TG3S1.
The live termites were provided by Michael Lenz in CSIRO, Australia.
w/s, interface between dead wood and soil.
FibS1, Fibrobacteres subphylum 1; TG3S1, TG3 subphylum 1; TG3S2, TG3 subphylum 2; +, detected; −, not detected.
TABLE 4.
Detection of TG3 subphylum 1, TG3 subphylum 2, and Fibrobacteres subphylum 2 from various environments by specific amplification of the 16S rRNA geneh
| Environment | Host and/or location | TG3S1 | TG3S2 | FibS2 |
|---|---|---|---|---|
| Cockroach gut | Panesthia angustipennis, Yakushima, Japan | − | + | − |
| Cockroach gut | Cryptocercus punctulatus, Appalachian region, United Statesb | − | − | − |
| Bovine rumen | Tsukuba, Japanc | − | − | − |
| Activated sludge | Sewage disposal plant, Kyushu, Japand | − | − | − |
| Anaerobic digester | Sewage disposal plant, Japand | − | − | − |
| Orchard soil | Pathum Thani, Thailand | − | − | − |
| Rice paddy soil | Niigata, Japane | + | −a | − |
| Rice paddy soil | Nagano, Japane | + | −a | − |
| Rice paddy soil | Tainan, Taiwane | + | −a | − |
| Lake sediment | Kasumi-ga-ura, Japand | + | − | − |
| Sea sediment | 24 m and 42 m depth, Setouchi, Japanf | − | − | − |
| Deep-sea sediment | 700 m depth, Toyama, Japang | + | + | − |
| Deep-sea sediment | 1,000 m depth, Toyama, Japang | + | −a | − |
| Deep-sea sediment | 4,700 m depth, the Chishima Trench, Japang | − | − | − |
Although a faint or clear signal was obtained, the sequences were not of TG3S2, but TG3S1.
The live insects were provided by Christine A. Nalepa in North Carolina State University.
The purified DNA sample was provided by Akio Takenaka in NILGS, Japan.
The purified DNA samples were provided by Hideyuki Tamaki and Yoichi Kamagata in AIST, Japan.
The purified DNA samples were provided by Sanae Sakai and Hiroyuki Imachi in Nagaoka University of Technology, Japan.
The purified DNA samples were provided by Ikuo Yoshinaga in Kyoto University, Japan.
The purified DNA samples were provided by Shizuka Arakawa and Chiaki Kato in JAMSTEC, Japan.
FibS1, Fibrobacteres subphylum 1; TG3S1, TG3 subphylum 1; TG3S2, TG3 subphylum 2; +, detected; −, not detected.
Phylogenetic diversity.
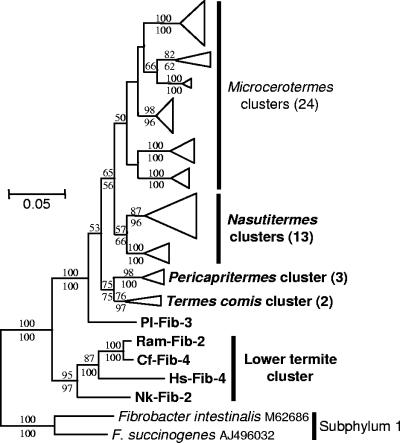
The clones obtained with the taxon-specific amplifications were sorted into phylotypes with a criterion of 99.0% sequence identity for detailed phylogenetic analyses. The TG3 and Fibrobacteres clones obtained with the Bacteria-specific primers in this and the previous studies (11, 12) were also reclassified with the same criterion. In Fibrobacteres subphylum 2, the phylotypes from the lower termites constituted a monophyletic cluster distinct from that of the higher termites (Fig. 4). The latter was further divided into subclusters specific to the genus of the termite host. No overlap of phylotypes among the host termite species was found except between Microcerotermes species 1 and 2. The sequence similarity was more than 85.3% within subphylum 2 and 81.3 to 84.3% between subphyla 1 and 2. The full tree of Fig. 4 is shown in Fig. S1 in the supplemental material.
FIG. 4.
Phylogenetic tree showing the relationship of the 16S rRNA phylotypes affiliated with Fibrobacteres subphylum 2. An ML tree was constructed with the GTR model. A tree obtained with the GTR+G+I model was basically congruent with this tree. An ME tree was also constructed with the GTR+G+I model. A total of 1,054 unambiguously aligned nucleotides were used, corresponding to positions 54 to 1164 in E. coli (J01695). Bootstrap tests were performed with 100 resamplings for both the ML tree and ME trees, and the confidence values are indicated above (ML) and below (ME) the branches. The phylotypes and clusters obtained in this study are shown in bold letters. The number of contained phylotypes in the compressed clusters are shown in parentheses. The host termites are indicated in the clone codes as abbreviations listed in Table 2. The full tree is published as Fig. S1 in the supplemental material; Pl, P. latignathus; Ram, R. amamianus; CF, C. formosanus; Hs, H. sjoestedti; NK, N. koshunensis.
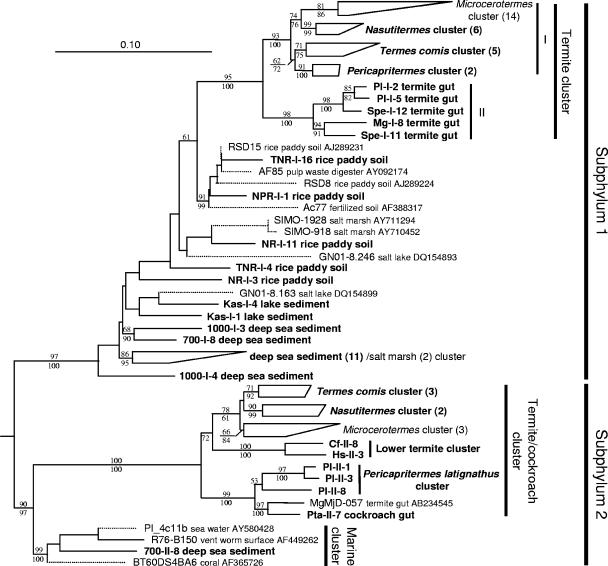
In TG3 subphylum 1, the phylotypes derived from termites formed a monophyletic cluster (Fig. 5). This termite-specific cluster comprised two major subclusters, I and II, as shown in the phylogenetic tree. Among them, subcluster I was further divided into clusters specific to the genus of the termite host as in Fibrobacteres subphylum 2. In both bacterial groups, phylotypes obtained from Nasutitermes dimorphus from Thailand and N. takasagoensis from Japan formed a monophyletic cluster as did those from Pericapritermes latignathus from Thailand and Pericapritermes nitobei from Japan (see Fig. S1 and S2 in the supplemental material). In TG3 subphylum 2, the phylotypes derived from termites and a cockroach formed a monophyletic cluster which also contained subclusters specific to host termite genera. No overlap of phylotypes among the host species was found, except between Microcerotermes species 1 and 2.
FIG. 5.
Phylogenetic tree showing the relationship of the 16S rRNA phylotypes affiliated with the candidate phylum TG3. An ML tree was constructed as the framework using the fastDNAml program implemented in ARB. The tree topology was basically congruent with the ML trees constructed using the PHYML program with the GTR or GTR+G+I model. An ME tree was also constructed with the GTR+G+I model. A total of 1,007 unambiguously aligned nucleotides were used, corresponding to positions 165 to 1225 in E. coli (J01695). Some phylotypes of the subphylum 1 were obtained by PCR using the primer set designed specific to the subphylum 2. This was caused by unexpected matching of the forward primer for subphylum 2 to some of the subphylum 1 phylotypes that had not been obtained before this study. Bootstrap tests were performed with 100 resamplings for both the ML tree with the GTR model and the ME tree, and the confidence values are indicated above (ML) and below (ME) the branches. The phylotypes and clusters obtained in this study are shown in bold letters. The short sequences (connected by dotted lines) found in public databases were added later by means of the ARB parsimony tool without changing the overall topology. The number of contained phylotypes in the compressed clusters are shown in parentheses. The origin of phylotypes are indicated in the clone codes as abbreviations listed in Table 2 for termites and Table 3 for other environments. Chlorobium limicola (Y10640) and Prosthecochloris vibrioforme (Y10649) were used as the outgroups. The full tree before the short sequences were added is published as Fig. S2 in the supplemental material. Pl, P. latignathus; Spe, Speculitermes sp.; Mg, M. gilvus; Cf, C. formosanus; Hs, H. sjoestedti; TNR, Tainan, Taiwan; NPR, Niigata, Japan; NR, Nagano, Japaan; Kas, Kasumi-ga-ura, Japan; 700, 700 m depth, Toyama, Japan; 1,000, 1,000 m depth, Toyama, Japan.
Most phylotypes from the deep-sea sediments constituted a monophyletic cluster in TG3 subphylum 1, together with two short sequences from salt marsh sediments found only in the public databases (AY710950 and AY711286) (Fig. 5). Two phylotypes were shared between the samples from distinct depths, 700 m and 1,000 m (see Fig. S2 in the supplemental material). Only one phylotype from deep-sea sediment was affiliated with TG3 subphylum 2, and it formed a monophyletic cluster, together with three marine clones in the public databases, including one from the surface of the vent worm Riftia pachyptila (19) (Fig. 5). The sequence similarity was more than 82.9% within subphylum 1 and more than 80.4% within subphylum 2. The similarity between subphyla 1 and 2 was 77.5 to 86.2%. The TG3 clones shared only below 80% sequence identities with any other known sequences, including those of the Fibrobacteres. This low similarity to other phyla and the consistent monophyly of subphyla 1 and 2 completely fulfill the definition of new candidate phylum (division) for uncultured bacteria with only 16S sequences proposed by Hugenholtz et al. (16) and Rappe and Giovannoni (30), whereas we should wait for further diagnostic information on morphology and physiology for describing this group in authentic nomenclature.
DISCUSSION
Over the last decade, clonal analyses of environmental 16S rRNA have disclosed many phylum-level clusters in the domain Bacteria. However, while more than 30 candidate phyla without cultured representatives have been recognized, most have never been characterized, even by 16S rRNA-based analyses, such as FISH and selective PCR amplification. This is disappointing because the information on their diversity, in situ localization, and favored habitats may enhance the possibility of further characterization of these uncultured bacteria. In the present study, we successfully detected cells of the candidate phylum TG3 and Fibrobacteres subphylum 2 by FISH with specific probes and obtained diverse phylotypes from various environments by PCR screenings. Using the FISH analyses, we demonstrated that these bacteria were the second-most dominant groups in the whole-gut microbiota of both the termites Microcerotermes sp. and N. takasagoensis. Since we revealed in our previous study that the bacterial community structures in guts are similar within a genus of termites (11), it is likely that the dominance of these bacteria is consistent through the genera Microcerotermes and Nasutitermes. Although the gut bacterial communities of Nasutitermes termites, including N. takasagoensis, have often been studied using microscopy, no reports have referred to these bacteria (6, 8, 29, 41). This might be due to the apparent similarity of their morphologies with those of small spirochetes.
Since these bacteria were detected from all or most of the diverse higher termite samples by PCR screenings, it is likely that they are distributed commonly among higher termites. However, their abundance seems dependent on the taxonomic and/or feeding group of the termite host. In the comprehensive 16S clonal analyses using Bacteria-specific primers, either TG3 or Fibrobacteres subphylum 2 was never or rarely found from other higher termites, i.e., the fungus grower Macrotermes gilvus (12), the interface feeder Termes comis (39), and the soil feeder Cubitermes orthognathus (33). In lower termites, while TG3 subphylum 2 and/or Fibrobacteres subphylum 2 members were detected from a few species by PCR screenings, they have never been found by comprehensive 16S clonal analyses (11, 13, 35; Y. Hongoh, unpublished data). Therefore, the dominance of these bacteria could be unique to some wood-feeding higher termites. Nevertheless, the termite-specific clusters delineated by the host genus and not by the geographic distance suggest that these bacteria are autochthonous gut symbionts that have a robust association with termite hosts. On the other hand, the shared phylotypes between Microcerotermes species 1 and 2 that inhabit the same locations imply that cospeciation is not strict, as discussed in our previous study (11).
The localization in the gut is a clue to the physiology of these as-yet-uncultured bacteria. The physicochemical condition in the highly compartmentalized guts of higher termites has been investigated at a fine scale in some species of Microcerotermes and Nasutitermes (5). The dilated proctodeal segment (P3), where both TG3 subphylum 1 and Fibrobacteres subphylum 2 were found abundantly by FISH, had a pH of around 7 and was completely anoxic around the central region, while a microoxic region existed toward the gut epithelium. Since both bacterial groups were found only from the luminal fluid, these bacteria appear to favor an anoxic condition and moderate pH. In the P3 section of Nasutitermes walkeri, acetate was the predominant short-chain fatty acid (37), which is a typical product of microbial fermentation. Since the described species of the Fibrobacteres, Fibrobacter intestinalis and F. succinogenes, characteristically ferment cellulose and produce acetate as a major component in the rumen or cecum of mammals (22), one may expect a similar function for the Fibrobacteres bacteria in termite guts. However, only traces of cellulase have been detected from the hindguts of N. takasagoensis (42) and other Nasutitermes termites (10).
While Fibrobacteres subphylum 2 was detected from only termite guts, we successfully detected TG3 subphyla 1 and 2 from other environments. It is remarkable that as many as 15 phylotypes affiliated with TG3 subphylum 1 were recovered from deep-sea sediments. Since TG3 subphylum 1 clones were also obtained from lake sediment, rice paddy soil, and salt marsh sediment (found in the public databases), soil or sediments submerged in water may be favored habitats for this bacterial group. In TG3 subphylum 2, clones from marine environments as well as from termite and cockroach guts were recovered. These raise the possibility that the TG3 bacteria prevail widely among water-associated environments and the guts of various insects feeding on dead plant matters, although their ecological functions remain unknown. Fortunately, we found that the TG3 and Fibrobacteres bacteria are abundantly and consistently harbored in a specific region of the gut by termites. This will enable us to further investigate these bacteria for their detailed morphologies and possible functions with reproducibility, such as by rRNA-based scanning electron microscopy (17), microautoradiography-FISH (23), and metagenomic analysis.
Supplementary Material
Acknowledgments
We sincerely thank the following people who generously provided their precious samples: S. Arakawa, H. Imachi, Y. Kamagata, C. Kato, M. Lenz, S. Moriya, C. A. Nalepa, S. Sakai, A. Takenaka, H. Tamaki, and I. Yoshinaga. We also thank C. Vongkaluang and S. Savitr for helping with our study in Thailand. We thank A. Yamada and T. Sato for assisting with experiments.
Y.H. and S.H. are recipients of a Special Postdoctoral Research Fellowship from RIKEN. This work was partially supported by the International Cooperative Research Project of Japan Science and Technology Agency (JST-ICORP), grants for the Bioarchitect Research Program, and the Eco Molecular Science Research Program from RIKEN.
Footnotes
Published ahead of print on 21 August 2006.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Acinas, S. G., V. Klepac-Ceraj, D. E. Hunt, C. Pharino, I. Ceraj, D. L. Distel, and M. F. Polz. 2004. Fine-scale phylogenetic architecture of a complex bacterial community. Nature 430:551-554. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madssen, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breznak, J. A. 2000. Ecology of prokaryotic microbes in the guts of wood- and litter-feeding termites, p. 209-232. In T. Abe, D. E. Bignell, and M. Higashi (ed.), Termites: evolution, sociality, symbioses, ecology. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 5.Brune, A., D. Emerson, and J. A. Breznak. 1995. The termite gut microflora as an oxygen sink: microelectrode determination of oxygen and pH gradients in guts of lower and higher termites. Appl. Environ. Microbiol. 61:2681-2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Czolij, R. T., M. Slaytor, and R. W. O'Brien. 1985. Bacterial flora of the mixed segment and the hindgut of the higher termite Nasutitermes exitiosus Hill (Termitidae, Nasutitermitinae). Appl. Environ. Microbiol. 49:1226-1236. [Google Scholar]
- 7.Daims, H., A. Bruhl, R. Amann, K. H. Schleifer, and M. Wagner. 1999. The domain-specific probe EUB338 is insufficient for the detection of all Bacteria: development and evaluation of a more comprehensive probe set. Syst. Appl. Microbiol. 22:434-444. [DOI] [PubMed] [Google Scholar]
- 8.Eutick, M. L., P. C. Veivers, R. W. O'Brien, and M. Slaytor. 1978. Dependence of the higher termite, Nasutitermes exitiosus and the lower termite, Coptotermes lacteus on their gut flora. J. Insect Physiol. 24:363-368. [Google Scholar]
- 9.Guindon, S., and O. Gascuel. 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 10.Hogan, M. E., P. C. Veivers, M. Slaytor, and R. T. Czolij. 1988. The site of cellulose breakdown in higher termites (Nasutitermes walkeri and Nasutitermes exitiosus). J. Insect Physiol. 34:891-899. [Google Scholar]
- 11.Hongoh, Y., P. Deevong, T. Inoue, S. Moriya, S. Trakulnaleamsai, M. Ohkuma, C. Vongkaluang, N. Noparatnaraporn, and T. Kudo. 2005. Intra- and interspecific comparisons of bacterial diversity and community structure support coevolution of gut microbiota and termite host. Appl. Environ. Microbiol. 71:6590-6599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hongoh, Y., L. Ekpornprasit, T. Inoue, S. Moriya, S. Trakulnaleamsai, M. Ohkuma, N. Noparatnaraporn, and T. Kudo. 2006. Intracolony variation of bacterial gut microbiota among castes and ages in the fungus-growing termite Macrotermes gilvus. Mol. Ecol. 15:505-516. [DOI] [PubMed] [Google Scholar]
- 13.Hongoh, Y., M. Ohkuma, and T. Kudo. 2003. Molecular analysis of bacterial microbiota in the gut of the termite Reticulitermes speratus (Isoptera; Rhinotermitidae). FEMS Microbiol. Ecol. 44:231-242. [DOI] [PubMed] [Google Scholar]
- 14.Huber, T., G. Faulkner, and P. Hugenholtz. 2004. Bellerophon: a program to detect chimeric sequences in multiple sequence alignments. Bioinformatics 20:2317-2319. [DOI] [PubMed] [Google Scholar]
- 15.Hugenholtz, P., B. M. Goebel, and N. R. Pace. 1998. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 180:4765-4774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hugenholtz, P., C. Pitulle, K. L. Hershberger, and N. R. Pace. 1998. Novel division level bacterial diversity in a Yellowstone hot spring. J. Bacteriol. 180:366-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kenzaka, T., A. Ishidoshiro, N. Yamaguchi, K. Tani, and M. Nasu. 2005. rRNA sequence-based scanning electron microscopic detection of bacteria. Appl. Environ. Microbiol. 71:5523-5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5:150-163. [DOI] [PubMed] [Google Scholar]
- 19.Lopez-Garcia, P., F. Gaill, and D. Moreira. 2002. Wide bacterial diversity associated with tubes of the vent worm Riftia pachyptila. Environ. Microbiol. 4:204-215. [DOI] [PubMed] [Google Scholar]
- 20.Ludwig, W., O. Strunk, R. Westram, L. Richter, H. Meier, Yadhukumar, A. Buchner, T. Lai, S. Steppi, G. Jobb, W. Förster, I. Brettske, S. Gerber, A. W. Ginhart, O. Gross, S. Grumann, S. Hermann, R. Jost, A. König, T. Liss, R. Lüssmann, M. May, B. Nonhoff, B. Reichel, R. Strehlow, A. Stamatakis, N. Stuckmann, A. Vilbig, M. Lenke, T. Ludwig, A. Bode, and K. H. Schleifer. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32:1363-1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. J. Parker, P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Montgomery, L., B. Flesher, and D. Stahl. 1988. Transfer of Bacteroides succinogenes (Hungate) to Fibrobacter gen. nov. as Fibrobacter succinogenes comb. nov. and description of Fibrobacter intestinalis sp. nov. Int. J. Syst. Evol. Microbiol. 38:430-435. [Google Scholar]
- 23.Nielsen, J. L., and P. H. Nielsen. 2005. Advances in microscopy: microautoradiography of single cells. Methods Enzymol. 397:237-256. [DOI] [PubMed] [Google Scholar]
- 24.Noda, S., T. Inoue, Y. Hongoh, M. Kawai, C. A. Nalepa, C. Vongkaluang, T. Kudo, and M. Ohkuma. 2006. Identification and characterization of ectosymbionts of distinct lineages in Bacteroidales attached to flagellated protists in the gut of termites and a wood-feeding cockroach. Environ. Microbiol. 8:11-20. [DOI] [PubMed] [Google Scholar]
- 25.Noda, S., M. Ohkuma, A. Yamada, Y. Hongoh, and T. Kudo. 2003. Phylogenetic position and in situ identification of ectosymbiotic spirochetes on protists in the termite gut. Appl. Environ. Microbiol. 69:625-633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ohkuma, M. 2003. Termite symbiotic systems: efficient bio-recycling of lignocellulose. Appl. Microbiol. Biotechnol. 61:1-9. [DOI] [PubMed] [Google Scholar]
- 27.Ohkuma, M., and T. Kudo. 1996. Phylogenetic diversity of the intestinal bacterial community in the termite Reticulitermes speratus. Appl. Environ. Microbiol. 62:461-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohkuma, M., S. Noda, T. Iida, and T. Kudo. 2001. Phylogenetic identification of endosymbionts of the flagellated protists in the gut of termites. Presented at the the 9th International Symposium on Microbial Ecology (ISME-9), Amsterdam, The Netherlands.
- 29.Paster, B. J., F. E. Dewhirst, S. M. Cooke, V. Fussing, L. K. Poulsen, and J. A. Breznak. 1996. Phylogeny of not-yet-cultured spirochetes from termite guts. Appl. Environ. Microbiol. 62:347-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rappe, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 31.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schloss, P. D., B. R. Larget, and J. Handelsman. 2004. Integration of microbial ecology and statistics: a test to compare gene libraries. Appl. Environ. Microbiol. 70:5485-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmitt-Wagner, D., M. W. Friedrich, B. Wagner, and A. Brune. 2003. Phylogenetic diversity, abundance, and axial distribution of bacteria in the intestinal tract of two soil-feeding termites (Cubitermes spp.). Appl. Environ. Microbiol. 69:6007-6017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schramm, A., B. M. Fuchs, J. L. Nielsen, M. Tonolla, and D. A. Stahl. 2002. Fluorescence in situ hybridization of 16S rRNA gene clones (Clone-FISH) for probe validation and screening of clone libraries. Environ. Microbiol. 4:713-720. [DOI] [PubMed] [Google Scholar]
- 35.Shinzato, N., M. Muramatsu, T. Matsui, and Y. Watanabe. 2005. Molecular phylogenetic diversity of the bacterial community in the gut of the termite Coptotermes formosanus. Biosci. Biotechnol. Biochem. 69:1145-1155. [DOI] [PubMed] [Google Scholar]
- 36.Sizemore, R. K., J. J. Caldwell, and A. S. Kendrick. 1990. Alternate Gram staining technique using a fluorescent lectin. Appl. Environ. Microbiol. 56:2245-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Slaytor, M., P. C. Veivers, and N. Lo. 1997. Aerobic and anaerobic metabolism in the higher termite Nasutitermes walkeri (Hill). Insect Biochem. Mol. Biol. 27:291-303. [Google Scholar]
- 38.Stingl, U., R. Radek, H. Yang, and A. Brune. 2005. “Endomicrobia”: cytoplasmic symbionts of termite gut protozoa form a separate phylum of prokaryotes. Appl. Environ. Microbiol. 71:1473-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thongaram, T., Y. Hongoh, S. Kosono, M. Ohkuma, S. Trakulnaleamsai, N. Noparatnaraporn, and T. Kudo. 2005. Comparison of bacterial communities in the alkaline gut segment among various species of higher termites. Extremophiles 9:229-238. [DOI] [PubMed] [Google Scholar]
- 40.Thongaram, T., S. Kosono, M. Ohkuma, Y. Hongoh, M. Kitada, T. Yoshinaka, S. Trakulnaleamsai, N. Noparatnaraporn, and T. Kudo. 2003. Gut of higher termites as a niche for alkaliphiles as shown by culture-based and culture-independent studies. Microbes Environ. 18:152-159. [Google Scholar]
- 41.Tokuda, G., T. Nakamura, R. Murakami, and I. Yamaoka. 2001. Morphology of the digestive system in the wood-feeding termite Nasutitermes takasagoensis (Shiraki) (Isoptera: Termitidae). Zool. Sci. 18:869-877. [Google Scholar]
- 42.Tokuda, G., H. Watanabe, T. Matsumoto, and H. Noda. 1997. Cellulose digestion in the wood-eating higher termite, Nasutitermes takasagoensis (Shiraki): distribution of cellulases and properties of endo-beta-1,4-glucanase. Zool. Sci. 14:83-93. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.