Abstract
The biodegradation of polychlorinated biphenyls (PCBs) relies on the ability of aerobic microorganisms such as Burkholderia xenovorans sp. LB400 to tolerate two potential modes of toxicity presented by PCB degradation: passive toxicity, as hydrophobic PCBs potentially disrupt membrane and protein function, and degradation-dependent toxicity from intermediates of incomplete degradation. We monitored the physiological characteristics and genome-wide expression patterns of LB400 in response to the presence of Aroclor 1242 (500 ppm) under low expression of the structural biphenyl pathway (succinate and benzoate growth) and under induction by biphenyl. We found no inhibition of growth or change in fatty acid profile due to PCBs under nondegrading conditions. Moreover, we observed no differential gene expression due to PCBs themselves. However, PCBs did have a slight effect on the biosurface area of LB400 cells and caused slight membrane separation. Upon activation of the biphenyl pathway, we found growth inhibition from PCBs beginning after exponential-phase growth suggestive of the accumulation of toxic compounds. Genome-wide expression profiling revealed 47 differentially expressed genes (0.56% of all genes) under these conditions. The biphenyl and catechol pathways were induced as expected, but the quinoprotein methanol metabolic pathway and a putative chloroacetaldehyde dehydrogenase were also highly expressed. As the latter protein is essential to conversion of toxic metabolites in dichloroethane degradation, it may play a similar role in the degradation of chlorinated aliphatic compounds resulting from PCB degradation.
Polychlorinated biphenyls (PCBs) are organic chemicals belonging to a class of hydrocarbons that are among the most important environmental contaminants, as their physicochemical properties allow them to persist in the environment and bioaccumulate (34). PCBs accumulate in lipid-rich tissues of all organisms, including humans, where they alter immune functions (43) and cause neurological (40), developmental (31), respiratory (45), and reproductive problems as well as cancer due to estrogenic activity (12). These health concerns prioritize PCBs as major targets for environmental cleanup.
The bioremediation strategy with the highest potential for destruction of PCB mixtures is a sequential anaerobic-aerobic process (1). Anaerobic bacteria can reductively dechlorinate highly chlorinated congeners but cannot complete the process, and they accumulate congeners with fewer chlorines (4, 22, 35). The burden of PCB degradation lies with aerobic cometabolism of the products of reductive dechlorination by biphenyl- and chlorobenzoate-degrading organisms (22, 38). Recently a sequential anaerobic-aerobic process was shown at the laboratory scale to remove over 50% of Aroclor 1242 from river sediments (39).
Two of the most effective aerobic PCB-degrading bacteria, Burkholderia xenovorans strain LB400 and Rhodococcus sp. strain RHA1, vary greatly in their levels of PCB toxicity resistance (41), yet the sources of this difference are unknown. Indeed recent studies of PCB-tolerant LB400 (13-15) suggest that cell systems other than the biphenyl pathway contribute to the efficient degradation of biphenyl and PCBs. Although the biphenyl pathway has been extensively studied, the role of auxiliary mechanisms, which may provide tolerance to PCBs, has been lightly investigated, and information on the biological effects of PCBs (25) and their degradation products to bacteria (10, 21) is scarce.
Complete sequencing of the LB400 genome (http://genome.ornl.gov/microbial/bfun) (11) led us to undertake a genomics-enabled investigation of the biology of PCB degradation. The ∼9.7-Mb genome of LB400 contains 8,958 predicted protein-coding genes (CDSs) in two chromosomes and one megaplasmid and is among the largest prokaryotic genomes closed to date. By use of genomic and proteomic approaches, an outline of genome-wide responses to a range of carbon and cell development conditions has been reported (13-15), establishing the groundwork for genomic analysis of PCB degradation.
As PCBs have been insinuated to impair microbial growth (25, 42), viability (10), and degradation (7, 23, 44, 47, 48), successful remediation should also consider the physiological and genetic responses limiting or overcoming these toxic effects. In this study, we investigate physiological and genome-wide cell responses and defenses against the toxicity posed by PCBs. To that end, we compare PCB tolerance levels of LB400 and other potential PCB-degrading bacteria and outline genome-wide gene expression patterns in response to different toxicity modes of PCBs. We further analyze LB400 differential gene expression patterns in response to PCBs with and without degradation. We find that LB400 is unusually tolerant to PCB degradation and suggest that some auxiliary pathways may remove toxic chlorinated intermediates.
MATERIALS AND METHODS
Bacteria, media, and growth conditions.
We determined the degrees of sensitivity of 18 strains previously isolated on biphenyl and naphthalene from Brazil, Puerto Rico, Japan, and the United States (9, 28) to 100-ppm (low) and 500-ppm (high) concentrations of the commercial PCB mixture Aroclor 1242 (Table 1). Sixteen strains previously characterized by Pellizari et al. (32) as potential PCB degraders and two well-studied PCB degraders, Burkholderia xenovorans LB400 (8) and Rhodococcus sp. strain RHA1 (28), were included in this study. To determine the effect of PCBs on growth, cultures of each strain were grown in triplicate on Luria-Bertani (LB) broth containing 0, 100, or 500 ppm Aroclor 1242 (Monsanto Co., St. Louis, Mo.) at 29 ± 1°C in 125-ml flasks agitated at 200 rpm. The percent growth, maximum optical density (OD), and estimated lag time were noted for PCB-containing cultures and compared with those for control medium that contained no PCBs. The maximum growth rate was calculated using OD values during logarithmic growth. Maximum biomass was measured as cultures reached stationary-phase growth.
TABLE 1.
Tolerance of potential PCB-degrading strains to 500 ppm Aroclor 1242 in LB mediuma
| Strainb | G+/G−c | μmax (h−1)d | Max OD600d | Rel growthe |
|---|---|---|---|---|
| Tolerant strains | ||||
| Comamonas testosteroni VP44 | G− | 1.07 ± 0.03 | 0.98 ± 0.01 | 1.05 |
| Burkholderia xenovorans LB400 | G− | 0.93 ± 0.04 | 0.99 ± 0.03 | 0.92 |
| Moderately tolerant strains | ||||
| Unidentified strain PR4 | G− | 0.81 ± 0.06 | 0.94 ± 0.01 | 0.76 |
| Staphylococcus warneri VP73 | G+ | 0.87 ± 0.22 | 0.83 ± 0.06 | 0.73 |
| Alcaligenes xylosoxidans VP98 | G− | 0.75 ± 0.06 | 0.95 ± 0.01 | 0.71 |
| Xanthomonas maltophilia VP48 | G− | 0.73 ± 0.02 | 0.94 ± 0.01 | 0.69 |
| Pseudomonas gladioli VP86 | G− | 0.68 ± 0.13 | 1.00 ± 0.01 | 0.68 |
| Unidentified strain PR2 | G− | 0.74 ± 0.05 | 0.87 ± 0.01 | 0.65 |
| Xanthomonas maltophilia VP90 | G− | 0.71 ± 0.07 | 0.89 ± 0.02 | 0.63 |
| Rhodococcus erythropolis NY05 | G+ | 0.57 ± 0.06 | 0.99 ± 0.02 | 0.57 |
| Moderately sensitive strains | ||||
| Xanthomonas maltophilia VP11 | G− | 0.68 ± 0.14 | 0.78 ± 0.04 | 0.53 |
| Unidentified strain VP69 | G− | 0.67 ± 0.03 | 0.68 ± 0.13 | 0.46 |
| Pseudomonas gladioli VP87 | G− | 0.58 ± 0.12 | 0.79 ± 0.03 | 0.46 |
| Pseudomonas gladioli VP49 | G− | 0.64 ± 0.08 | 0.65 ± 0.02 | 0.42 |
| Alcaligenes xylosoxidans PR5 | G− | 0.47 ± 0.16 | 0.81 ± 0.02 | 0.38 |
| Rhodococcus sp. strain RHA1 | G+ | 0.55 ± 0.08 | 0.56 ± 0.04 | 0.31 |
| Sensitive strains | ||||
| Pseudomonas gladioli VP71 | G− | 0.38 ± 0.05 | 0.26 ± 0.02 | 0.10 |
| Unidentified strain VP103 | G− | 0.29 ± 0.06 | 0.11 ± 0.01 | 0.03 |
Tolerance (relative growth) was determined by comparing the maximum growth rate (μmax) and maximum biomass (max OD600) in the presence and absence of PCBs.
Strains designated VP and PR are from the work of Pellizari et al. (32).
G+, gram positive; G−, gram negative.
Data are means ± standard errors. Standard error = standard deviation/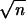 .
.
Relative (Rel) growth is calculated by multiplying μmax and maximum OD (relative to PCB-negative controls).
For studies in defined medium (46), batch cultures (25 ml) were prepared in 125-ml Wheaton flasks and sealed with Teflon-lined lids. Succinate at 1 g/liter (∼10 mM) and benzoate at 1 g/liter (∼5 mM) were added to the medium prior to autoclaving. Sterile biphenyl at 3 g/liter (∼20 mM) was added directly to the sterilized medium in each flask. The PCB-containing medium was allowed to equilibrate for at least 24 h prior to the addition of inoculum from a freshly grown, biphenyl-adapted culture. Triplicates were prepared for each growth condition. Microbial growth was monitored spectrophotometrically by measuring the absorbance at 600 nm. To account for any effect of the (in)solubility of biphenyl or PCBs on absorbance, CFU counts were conducted by plating serial dilutions on plates with R2A agar.
Distribution of PCBs.
The association of PCBs with bacteria was demonstrated by growing two strains incapable of PCB degradation (Burkholderia xenovorans strains LMG 27120 and LMG 16229 [20]) in 25 ml of K1 with succinate and 500 ppm PCBs. Following growth to stationary phase (OD at 600 nm [OD600], 1.2 and 1.0, respectively), the cultures were filtered using sterile glass wool and centrifuged at 4,000 rpm for 10 min. A control of medium with PCBs but no cells was also filtered using glass wool, centrifuged, and analyzed as a reference for the distribution of PCBs without cells. The supernatant and cell fraction were separated, and PCBs were extracted with octachloronaphthalene added as an internal standard.
PCB degradation by LB400 was measured in triplicate by measuring the disappearance of PCBs from cultures harvested at mid-logarithmic phase on each of the three carbon sources. Due to the difference in the growth rates on each carbon source, the harvest time for LB400 varied from 12 to 24 h following inoculation and was when the mid-logarithmic-growth phase was reached. Each PCB-containing culture was extracted three times using an equal volume of 1:1 hexane/acetone by shaking for 30 min and analyzed by gas chromatography as described previously (28). Percent degradation was calculated by determining the difference between the PCB concentrations in experimental cultures and in noninoculated controls.
Physiological adaptation to PCBs.
To determine membrane adaptation of LB400 and RHA1 to PCBs, cells were grown in the presence or absence of PCBs, and the fatty acid composition was analyzed as per the MIDI protocol (Microbial ID, Newark, DE). We also compared the morphologies of LB400 grown in the presence and absence of PCBs by analyzing scanning electron micrograph preparations (SEM). SEM fixation was performed as previously described (26). Multiple fields (at least five) with multiple cells (>10) from two biological replicates were subsequently analyzed using the CMEIAS image analysis software (27) (http://cme.msu.edu/cmeias). Due to the elongation of cells prior to cell division, we analyzed the smallest 25th percentile of the bacteria captured on electron micrographs for dimensional analysis. For transmission electron micrographs, cells were pelleted and the pellet was resuspended in fixative (2.5% glutaraldehyde and 2.0% paraformaldehyde in 0.1 M cacodylate buffer) overnight at 4°C. After fixation, cells were pelleted and the pellet was embedded in 2% agarose for postfixation treatment with osmium tetroxide and dehydrated in acetone series and embedded in Spurr's resin. Sections were stained with uranyl acetate and lead citrate for examination on a JEOL 100CX transmission electron microscope.
Gene expression analysis.
The genome-wide response to PCB stress was analyzed using a genomic microarray (Xeochip [13]) that contained 8,429 of the 8,989 total CDSs (93.8%) reported in the closed genome sequence of LB400 (http://genome.ornl.gov/microbial/bfun). LB400 cells were grown in succinate, benzoate, and biphenyl media in duplicate, each with parallel cultures containing 500 ppm Aroclor 1242. After three serial transfers in the same medium, bacterial cells were harvested by centrifugation from the exponential phase (OD600, 0.3 to 0.4) at 4,000 rpm for 10 min at 4°C. The culture was stored in RNAlater (Ambion) to protect against RNA degradation. Total RNA was extracted using an RNeasy mini kit (QIAGEN), and the contaminant DNA was degraded by using DNase I (Roche). The RNA was quantified spectrophotometrically, and the quality was verified by gel analysis. RNA was labeled by amino-allyl dUTP (Sigma) and hybridized as described before (13) by use of Cy5 and Cy3 fluorophores (Amersham). Two biological replicates were hybridized with at least 100 pmol of labeled material per Xeochip with dyes swapped. Hybridized chips were scanned with an Axon 4000B laser scanner and the data extracted using Genepix Pro 5.0 (Axon laboratories). Median expression data for each channel (Cy5 and Cy3) were imported into GeneSpring 7.2 (Silicon Genetics) and normalized using Lowess intensity-dependent normalization (13). Biological reproducibility was determined by calculating the coefficient of variation. Data were analyzed using GeneSpring software.
RESULTS
Tolerance to Aroclor 1242.
We analyzed the degrees of sensitivity or tolerance of potential PCB-degrading strains by determining the inhibition of growth due to the addition of Aroclor 1242 (Table 1) to LB broth. Of the 18 potential PCB-degrading strains tested, only Comamonas testosteroni VP44 and Burkholderia xenovorans LB400 demonstrated tolerance to high concentrations of Aroclor. In contrast, both growth rate and biomass production were decreased in Rhodococcus sp. strain RHA1 with exposure to PCBs. Several strains, including Alcaligenes xylosoxidans, exhibited extreme sensitivity to PCBs, with diminished biomasses, extremely low growth rates, and greater lag times. As expected, the ability to tolerate PCBs was better illustrated with 500 ppm than with 100 ppm in the medium (data not shown). The length of the lag period prior to growth in the presence of high concentrations of PCBs appears to be consistent with the bacterial tolerance to PCBs. Due to its PCB tolerance and the availability of genomics-enabled approaches, we selected LB400 for a detailed elucidation of PCB toxicity and tolerance responses.
We monitored the effect of PCBs on growth when the biphenyl pathway expression was low (minimal to no degradation of PCBs) in succinate- and benzoate-grown cells. Regardless of the presence of PCBs, RHA1 was unable to grow on succinate in K1 medium. Growth of RHA1 on benzoate was not inhibited in the PCB-containing treatments (0.48 ± 0.05 h−1) compared with controls (0.42 ± 0.03 h−1). Likewise, LB400 growth rates on both succinate (0.40 ± 0.02 h−1) and benzoate (0.27 ± 0.02 h−1) were not inhibited by PCBs (0.39 ± 0.02 h−1 and 0.28 ± 0.03 h−1, respectively). Plate counts confirmed this observation.
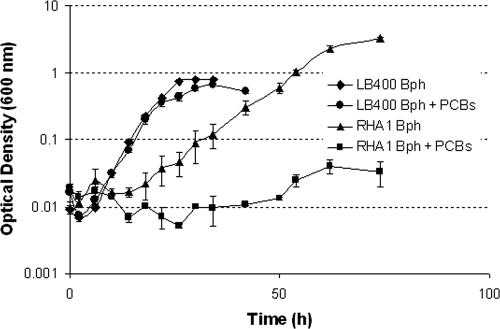
Upon expression of the biphenyl pathway during growth on biphenyl and subsequent PCB degradation, the growth of both RHA1 and LB400 was inhibited (Fig. 1). No growth of RHA1 was observed in batch cultures containing biphenyl and PCBs (0.10 ± 0.01 h−1 versus 0.01 ± 0.01 h−1). On the contrary, LB400 maintained growth rates similar to those of control cultures until mid-exponential phase before inhibition became apparent.
FIG. 1.
Growth curves of LB400 and RHA1 on biphenyl (Bph) (3 g/liter) as the carbon source with and without 500 ppm Aroclor 1242 (PCBs) in K1 medium.
Cell-associated PCBs.
Since PCBs have very low water solubility, we analyzed the amount of PCBs that partitioned to the biomass of two Burkholderia xenovorans strains incapable of PCB degradation after growth on succinate plus PCBs in K1 medium. Virtually all of the PCBs recovered were found in the biomass fraction (Table 2). Based on previous measurements of membrane content of cells (30), this amount of PCBs corresponds to roughly 9.5% of membrane weight if all cell-associated PCBs were localized in the membrane.
TABLE 2.
PCBs (ppm) extracted from two Burkholderia xenovorans strains incapable of PCB degradation after incubation with Aroclor 1242a
| Culture treatment | PCBs (ppm) extracted fromb:
|
|
|---|---|---|
| Supernatant | Cell pellet | |
| Uninoculated mediumc | 4.0 ± 0.1 | 5.1 ± 0.1 |
| LMG27120 | 2.9 ± 0.1 | 75 ± 5 |
| LMG16229 | 3.3 ± 0.3 | 57 ± 5 |
Cultures and control were filtered using sterile glass wool to remove insoluble PCBs prior to centrifugation.
Values are given as means ± standard deviations.
Control medium without bacterial inoculation.
PCB fate under degrading and nondegrading conditions.
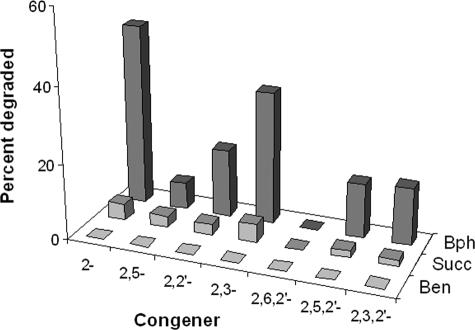
Insignificant disappearance of PCBs with LB400 growing on either benzoate or succinate during exponential growth indicated that PCBs were not degraded on these carbon sources (Fig. 2). However, during the transition to stationary phase, under carbon-limited conditions, some degradation did occur (not shown). Although RHA1 did not grow on succinate, it grew well on benzoate and similar to LB400 showed no degradation of PCBs. When the biphenyl pathway was induced in LB400, we found significant disappearance of ortho-chlorinated congeners such as 2-chlorobiphenyl and 2,3-dichlorobiphenyl, the more persistent congeners present in Aroclor 1242.
FIG. 2.
Percent degradation of ortho-chlorinated PCB congeners (500 ppm Aroclor 1242) by LB400 harvested during exponential-phase growth on benzoate (Ben), succinate (Succ), and biphenyl (Bph) with PCBs.
Morphological changes of LB400 due to PCBs.
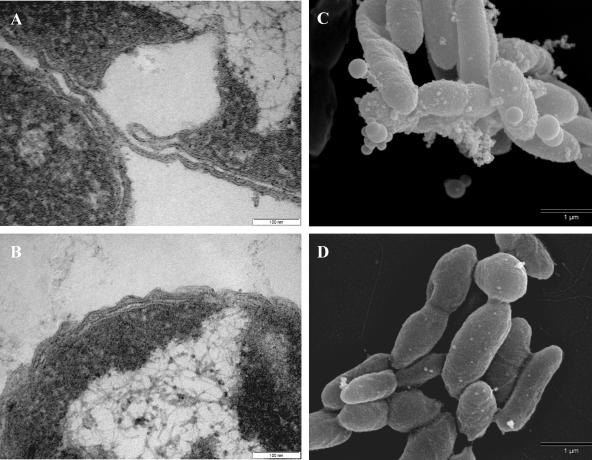
The morphological effects of PCBs on LB400 were analyzed by use of scanning electron micrographs of cells from exponential-phase growth of each of the treatments. Image analysis revealed a statistically significant reduction in the biosurface area caused by PCBs in cultures grown in succinate (14%) and benzoate (9%) (Table 3). When LB400 was grown with biphenyl as the growth substrate, no significant reduction was detected in the biosurface area due to PCBs; however, both biphenyl-grown treatments were significantly reduced (by 31%) compared to succinate- and benzoate-grown cells. Transmission electron micrographs from exponential-phase LB400 grown on succinate with PCBs revealed separation of the inner and outer membranes that was pervasive (Fig. 3A) compared to what was seen with succinate alone (Fig. 3B). Scanning electron micrographs of early-stationary-phase succinate-plus-PCB-grown cells show the formation of large membrane vesicles (Fig. 3C) that are absent in control samples (Fig. 3D). Nearly every micrograph collected from early-stationary-phase cells with PCBs contained evidence of vesicles at a ratio of approximately 3:4 (vesicle/cell).
TABLE 3.
Morphological measurements of the smallest quartile of LB400 cells grown on different carbon sources upon exposure to 500 ppm Aroclor 1242
| Growth condition | Mean measurement ± SDa
|
No. | Groupingb | ||
|---|---|---|---|---|---|
| Length (μm) | Width (μm) | Biosurface area (μm2) | |||
| Succinate | 1.86 ± 0.19 | 0.47 ± 0.04 | 3.2 ± 0.4 | 27 | A |
| Succinate + PCBs | 1.65 ± 0.16 | 0.44 ± 0.02 | 2.7 ± 0.2 | 23 | B |
| Benzoate | 1.75 ± 0.11 | 0.48 ± 0.03 | 3.2 ± 0.2 | 17 | A |
| Benzoate + PCBs | 1.61 ± 0.15 | 0.49 ± 0.04 | 2.9 ± 0.2 | 23 | B |
| Biphenyl | 1.41 ± 0.14 | 0.41 ± 0.06 | 2.1 ± 0.4 | 17 | C |
| Biphenyl + PCBs | 1.36 ± 0.16 | 0.42 ± 0.05 | 2.2 ± 0.3 | 19 | C |
As calculated by CMEIAS software.
Statistically significant groups are indicated by the different letters (determined by t test [P < 0.05]).
FIG. 3.
Electron micrographs of LB400 grown on succinate with and without PCBs: transmission electron microscopy images of exponential-phase cells with PCBs (A) and without PCBs (B) and SEM images of late-exponential-phase cells with PCBs (C) and without PCBs (D).
Fatty acid changes due to PCBs.
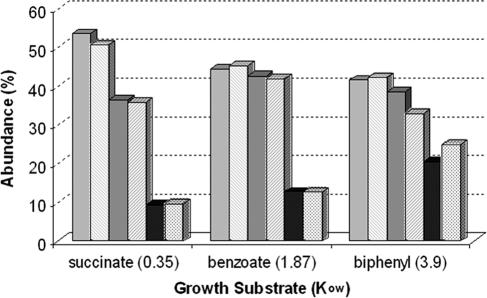
The change in fatty acid composition of LB400 varied slightly for each carbon source. As a general trend, unsaturated fatty acids, predominantly 18:1 and 16:1, decreased with an increasing octanol-water coefficient (Kow) of the C source (Fig. 4). Succinate has the lowest Kow (0.35), followed by benzoate (1.87), with biphenyl having the highest (3.90). Cyclic fatty acids (17:0 and 19:0) tended to increase with an increasing Kow of the C source. Upon degradation of PCBs (induction by biphenyl), there was a significant shift from saturated fatty acids to cyclic fatty acids.
FIG. 4.
Fatty acid analysis of LB400 during growth on carbon sources with different Kows in the presence (striped) and absence (solid) of PCBs. Fatty acids were grouped as unsaturated (light), saturated (gray), and cyclic (dark).
Gene expression in response to PCBs.
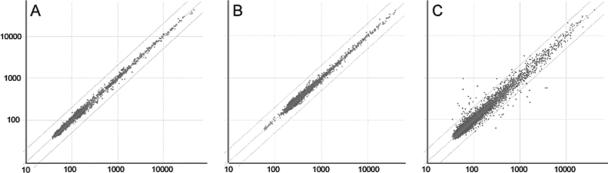
No significant differential gene expression was found between benzoate or succinate control cultures and PCB-containing cultures (Fig. 5A and B). The coefficients of variation for dye-swapped biological replicates were 9.5% for succinate plus PCBs with succinate as the reference and 5.4% for benzoate plus PCBs with benzoate as the reference.
FIG. 5.
Logarithmic scale plots of spot intensities (arbitrary units). Transcriptional profiles from mid-log growth on succinate versus succinate with PCBs (A), benzoate versus benzoate with PCBs (B), and biphenyl versus biphenyl with PCBs (C). Each represents a dye swap of biological replicates with Lowess normalization. Diagonal lines represent a twofold change in gene expression.
Gene expression patterns measured from PCB-degrading conditions versus biphenyl growth indicate that relatively few genes, only 47 of 8,429 CDSs, are differentially expressed more than twofold. Several of the structural genes in the upper biphenyl pathway were slightly more induced upon degradation of PCBs, as were several genes involved in (chloro)benzoate degradation via hydroxylation and coenzyme A (CoA) activation (Table 4). We also found induction of genes that may play an ancillary role, such as a cluster of genes involved in acetylacetone cleavage that contained a putative short-chain alcohol dehydrogenase as well as a group of genes that contained a putative chloroacetaldehyde dehydrogenase. Additionally, upon degradation of PCBs, gene expression patterns indicate an induction of genes potentially involved in methanol utilization, including those encoding a methanol dehydrogenase-like protein and formaldehyde-activating enzyme as well as genes responsible for cofactors involved in C1 metabolism. Down-regulated genes (Table 5) include those encoding several membrane proteins, that encoding an oxidoreductase that is similar to formate dehydrogenase (BxeC0084), that encoding a cyclohexanone monooxygenase (BxeA3588), and a cluster of genes possibly involved in lysine biosynthesis (BxeB1798, BxeB1802, and BxeB1806). The coefficient of variation for the biological replicates of PCB-degrading conditions (biphenyl plus PCBs versus biphenyl) was 15.3% (Fig. 5C).
TABLE 4.
Up-regulated genes of LB400
| Gene ID | Annotation | Expressionc |
|---|---|---|
| BxeA1129 | Chloromuconate cycloisomerase | 2.7 |
| BxeA1130 | Chlorocatechol-1,2-dioxygenase | 3.2 |
| BxeA1329 | Sugar ABC transporter, periplasmic binding protein | 2.7 |
| BxeA1427 | Amidase, hydantoinase/carbamoylase | 2.1 |
| BxeA2109 | Catechol 1,2-dioxygenase | 4.7 |
| BxeA2876 | Acetylacetone-cleaving enzyme | 2.9 |
| BxeA2877 | Hypothetical protein | 3.5 |
| BxeA2878 | Putative short-chain dehydrogenase/reductase | 2.1 |
| BxeA3675 | ABC sugar transporter, ATPase subunit | 2.5 |
| BxeA4207 | ABC nitrate/sulfonate/bicarbonate family transporter | 2.1 |
| BxeA4441 | Chloroacetaldehyde dehydrogenase | 4.9 |
| BxeA4442 | Hypothetical protein | 2.4 |
| BxeA4478 | Putative exported protein precursor | 3.1 |
| BxeB1279 | Glycogen synthase | 1.9 |
| BxeB1637 | Cytochrome o ubiquinol oxidase subunit II precursor | 1.9 |
| BxeB2286 | Unknown | 1.7 |
| BxeB2426 | Cytochrome c-555 precursorb | 5.4 |
| BxeB2427 | Methanol dehydrogenase large-subunit-like proteinb | 9.3 |
| BxeB2436 | Formaldehyde-activating enzymeb | 16.0 |
| BxeB2437 | Hypothetical proteinb | 5.5 |
| BxeB2469 | Coenzyme PQQ synthesis Db | 2.6 |
| BxeB2473 | TonB-dependent receptorb | 3.0 |
| BxeB2474 | Hypothetical proteinb | 3.2 |
| BxeC1082 | Lipopolysaccharide biosynthesis protein | 2.4 |
| BxeC1187 | 4-Hydroxy-2-oxovalerate aldolasea | 1.4 |
| BxeC1190 | Glutathione S-transferasea | 1.4 |
| BxeC1191 | 2,3-Dihydroxybiphenyl dioxygenasea | 1.5 |
| BxeC1192 | cis-2,3-Dihydrobiphenyl-2,3-diol dehydrogenasea | 1.5 |
| BxeC1196 | Biphenyl dioxygenase small subunita | 1.7 |
Corresponding CDS associated with the biphenyl pathway.
Corresponding CDS associated with C1 metabolism.
Expression is reported as the ratio between the growth on biphenyl with PCBs and the growth on biphenyl without PCBs (reference).
TABLE 5.
Down-regulated genes of LB400
| Gene ID | Annotationa | Expressionb |
|---|---|---|
| BxeA0676 | Putative membrane protein | 0.54 |
| BxeA1007 | 16S rRNA-processing protein | 0.54 |
| BxeA1178 | Hypothetical protein | 0.69 |
| BxeA1741 | RND efflux system, outer membrane lipoprotein | 0.73 |
| BxeA1939 | Putative permease from ABC transporter | 0.54 |
| BxeA2187 | Cytochrome c, class I precursor | 0.58 |
| BxeA3048 | Putative bacteriophage protein GP46 | 0.35 |
| BxeA3185 | Putative enoyl-CoA hydratase | 0.34 |
| BxeA3588 | Cyclohexanone monooxygenase | 0.52 |
| BxeA3722 | Acetyltransferase | 0.62 |
| BxeB0560 | Putative ADP-heptose-LPS heptosyltransferase | 0.40 |
| BxeB1798 | TRAP dicarboxylate transporter-DctP subunit | 0.49 |
| BxeB1802 | Dihydrodipicolinate synthetase | 0.18 |
| BxeB1806 | MFS transporter, phthalate permease family | 0.38 |
| BxeB2010 | Acriflavin resistance protein precursor | 0.69 |
| BxeB2146 | Putative membrane protein precursor | 0.15 |
| BxeB2160 | d-Lactate dehydrogenase | 0.38 |
| BxeB2536 | Putative LysR family transcriptional regulator | 0.30 |
| BxeC0084 | Formate dehydrogenase | 0.55 |
RND, resistance-nodulation-cell division; LPS, lipopolysaccharide; TRAP, tripartite ATP-independent periplasmic; MFS, major facilitator superfamily.
Expression is reported as the ratio between the growth on biphenyl with PCBs and the growth on biphenyl without PCBs (reference).
DISCUSSION
The physiological and genome-wide responses of B. xenovorans LB400 to PCBs enabled us to confine the source of toxicity to the by-products of PCB degradation. Although both LB400 and RHA1 are considered model organisms for the study of PCB degradation, our data indicate that LB400 is much more tolerant to PCB degradation-dependent toxicity (Fig. 1). In fact, LB400 is among the most tolerant PCB degraders identified in this study (Table 1). In neither case did PCBs per se affect the growth rate, viability, or fatty acid profile, and LB400 demonstrated no change in gene expression in the absence of degradation. Thus, the toxicity of PCBs to RHA1 and LB400 was a direct result of the production of deleterious metabolites during cometabolism. The difference in the growth rates of LB400 and RHA1 during PCB degradation may indicate mechanisms beyond the biphenyl pathway that contribute to tolerance. LB400 gene expression data under biphenyl-grown, PCB-degrading conditions indicate that the biphenyl, 3-chlorocatechol, and catechol pathways are further induced upon degradation of PCBs. The present data also indicate the induction of a putative chloroacetaldehyde dehydrogenase and components of the C1 metabolism pathway that may be involved in PCB product degradation.
Microorganisms responsible for the biodegradation of hydrophobic compounds, such as PCBs, are subject to membrane effects, such as fluidity disruption and interference with protein function (16, 42). Previous studies concerned with the adverse physiological responses to PCBs have attributed toxicity to PCBs themselves as well as to the products of their degradation (10, 21). While we confirmed that PCBs partition to the cell fraction of cultures (Table 2), this association did not correspond to a decrease in growth rate or viability in either the PCB-tolerant LB400 or the PCB-sensitive RHA1. Additionally, PCBs apparently do not interact with proteins that are responsible for cellular respiration, as seen for other chlorinated aromatic compounds (16).
Recent studies suggest that PCB degradation does not occur when LB400 is grown with simple carbon sources, such as glucose and succinate (6, 13). We found that PCBs were not degraded (Fig. 2) and mechanisms involved in PCB degradation were not induced by PCBs during exponential-phase growth on succinate or benzoate (Fig. 5A and B). Therefore, any physiological or genome-wide expression changes detected in succinate- or benzoate-grown LB400 cannot be attributed to degradation products of PCBs but rather are attributable to interaction with PCBs themselves, effectively allowing us to decouple and evaluate the potential sources of PCB toxicity.
One physiological response to membrane stress caused by lipophilic compounds involves altered fatty acid chain length or composition (36, 37, 42). Although the fatty acid composition of PCB-degrading Ralstonia eutropha HP850 shifts from saturated to unsaturated fatty acids during growth on biphenyl (25), the effect of PCBs themselves on membrane composition has not been documented. LB400 demonstrated no change in the fatty acid profile when grown on simple carbon sources (succinate or benzoate) with PCBs (Fig. 4). However, LB400 may be similar to pseudomonads that do not alter their saturated-to-unsaturated fatty acid profile upon exposure to lipophilic compounds (37). The only physiological changes we observed due to PCBs under nondegrading conditions appeared in electron micrographs of LB400 that indicate a reduction in the biosurface area of LB400 (Table 3). We also found frequent membrane aberrations caused by growth with PCBs (Fig. 3B and D). Similar membrane separation caused by PCBs has been noted previously (10), and such membrane aberrations are often indicative of stress conditions (5).
Although we found morphological evidence of PCB-membrane interactions in LB400, we found no differential gene expression (>2-fold) by microarray analysis in any of the 8,429 CDSs tested in either succinate or benzoate treatment with PCB treatment relative to no-PCB treatment (Fig. 5A and B). These analyses indicate that PCBs themselves do little to alter the composition of the membranes of LB400 and RHA1. Although PCBs do not affect growth rate or viability, the reduction in biosurface area and the separation of the inner and outer membranes do constitute a response to PCBs. However, dynamic morphological changes do not always elicit a change in gene expression (3). Either the responses reported here stem from gene expression changes below the sensitivity of microarray analysis or they are a direct physical effect of PCBs on membranes.
Past studies of the degradation of PCBs have focused exclusively on the biphenyl and chlorobenzoate pathways responsible for much of the degradation of simple congeners (4, 17-19, 22, 28, 38), and as expected, these pathways (biphenyl, catechol, and chlorocatechol) were induced upon degradation (Table 4). Although chlorinated aliphatic compounds derived from chlorinated pentadienoates may play an important role in PCB degradation, their fate and effect have been neglected. Potential metabolic derivatives of chlorinated pentadienoates have proven lethal to biodegrading microorganisms (2, 50, 51). Bacteria that are more capable of degrading or tolerating chlorinated aliphatic compounds would have a distinct advantage in environmental scenarios containing a mixture of PCB congeners. Among the putative genes induced upon degradation of PCBs was one encoding an acetaldehyde dehydrogenase that is 83% (amino acid) identical to the chloroacetaldehyde dehydrogenase found in Xanthobacter autotrophicus GJ10, a known dichloroethane (DCE) degrader (24). Chloroacetaldehyde dehydrogenase is responsible for the rapid conversion of the highly toxic chloroacetaldehyde intermediate, an essential step in DCE metabolism (24, 49). The alcohol dehydrogenase involved in DCE degradation is the same as the quinoprotein methanol dehydrogenase in methanol oxidation by gram-negative methylotrophs (24, 33). The chloroacetaldehyde dehydrogenase genes and methanol oxidation pathway (and cofactors) were the most differentially expressed genes induced by PCB degradation.
Down-regulated genes demonstrate equally significant characteristics associated with the degradation of PCBs (Table 5). In agreement with the change in fatty acid profile upon degradation of PCBs (Fig. 4), a putative enoyl-CoA hydratase involved in fatty acid metabolism (29) is repressed. Although lysine biosynthesis has been shown to have an indirect association with fatty acid beta-oxidation in oxidative stress (8), the specific role of lysine repression in this case is unknown. Finally, the down-regulation of oxygen-dependent proteins may be indicative of oxygen-limited conditions.
Although many of the mechanisms and components presented here may be involved in a general toxic response, such as oxygen limitation, some genes that are differentially expressed are almost certainly involved in minimizing the effects of toxic metabolites produced during degradation of PCBs. Unexpectedly, we found no growth inhibition or notable physiological response to PCBs under nondegrading conditions. Hence, factors beyond the upper biphenyl and benzoate degradation pathways contribute to successful PCB degradation. While RHA1 oxidizes a wide range of PCBs in resting cell assays (following growth on biphenyl) (27), it grew poorly on biphenyl in the presence of 500 ppm Aroclor 1242. Further comparison with other potential biodegrading bacteria, such as RHA1, shows that LB400 is extremely tolerant to the toxic metabolites that are created as a result of PCB degradation, possibly due to its superior ability to oxidize chlorinated aliphatic compounds. As LB400 is one of the bacteria most tolerant to PCB toxicity and has specificity to degrade a broad range of congeners, it becomes a model system for studying PCB degradation and its effects on bacteria, particularly for in situ biodegradation. Understanding the cell's responses to PCBs revealed in this study may provide information on the modes of toxicity and effective counterresponses, eventually leading to development of more-efficient PCB amelioration strategies.
Acknowledgments
This work was supported by Superfund Basic Research Program grant P42 ES 04911-12 from the U.S. National Institute of Environmental Health Sciences and by the Genomics:GTL Program of the U.S. Department of Energy.
We acknowledge Ewa Daniellewicz for assistance with scanning electron micrographs, Alicia Pastor-Lecha for transmission electron microscopy assistance, and Benli Chai for bioinformatic support.
Footnotes
Published ahead of print on 21 August 2006.
REFERENCES
- 1.Abramowicz, D. A. 1990. Aerobic and anaerobic biodegradation of PCBs: a review. Crit. Rev. Biotechnol. 10:241-248. [Google Scholar]
- 2.Alvarez-Cohen, L., and P. L. McCarty. 1991. Effects of toxicity, aeration, and reductant supply on trichloroethylene transformation by a mixed methanotrophic culture. Appl. Environ. Microbiol. 57:228-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arends, S. J. R., and D. S. Weiss. 2003. Inhibiting cell division in Escherichia coli has little if any effect on gene expression. J. Bacteriol. 186:880-884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bedard, D. L., and J. F. Quensen III. 1995. Microbial reductive dechlorination of polychlorinated biphenyls, p. 127-216. In L. Y. Young, and C. Cerniglia (ed.), Microbial transformation and degradation of toxic organic chemicals. Wiley-Liss Division, John Wiley & Sons, Inc., New York, N.Y.
- 5.Beveridge, T. J. 1999. Structures of gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 181:4725-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Billingsley, K. A., S. M. Backus, C. Juneson, and O. P. Ward. 1997. 1999. Comparison of the degradation patterns of polychlorinated biphenyl congeners in Aroclors by Pseudomonas strain LB400 after growth on various carbon sources. Can. J. Microbiol. 43:1172-1179. [DOI] [PubMed] [Google Scholar]
- 7.Blasco, R., M. Mallavarapu, R.-M. Wittich, K. N. Timmis, and D. H. Pieper. 1997. Evidence that formation of protoanemonin from metabolites of 4-chlorobiphenyl degradation negatively affects the survival of 4-chlorobiphenyl-cometabolizing microorganisms. Appl. Environ. Microbiol. 63:427-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bopp, L. H. 1986. Degradation of highly chlorinated PCBs by Pseudomonas strain LB400. J. Ind. Microbiol. 1:23-29. [Google Scholar]
- 9.Breitling, R., O. Sharif, M. L. Hartman, and S. K. Krisans. 2002. Loss of compartmentalization causes misregulation of lysine biosynthesis in peroxisome-deficient yeast cells. Eukaryot. Cell 6:978-986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Camara, B., C. Herrera, M. Gonzalez, E. Couve, B. Hofer, and M. Seeger. 2004. From PCBs to highly toxic metabolites by the biphenyl pathway. Environ. Microbiol. 6:842-850. [DOI] [PubMed] [Google Scholar]
- 11.Chain, P. S. G., V. J. Denef, K. Konstantinidis, L. M. Vergez, L. Agulló, V. L. Reyes, L. Hauser, M. Córdova, L. Gómez, M. González, M. Land, V. Lao, F. Larimer, J. J. LiPuma, E. Mahenthiralingam, S. A. Malfatti, C. J. Marx, J. J. Parnell, A. Ramette, P. Richardson, M. Seeger, D. Smith, T. Spilker, W.-J. Sul, T. V. Tsoi, L. E. Ulrich, I. B. Zhulin, and J. M. Tiedje. Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73 M bp genome shaped for versatility. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]
- 12.Demers, A., P. Ayotte, J. Brisson, S. Dodin, J. Robert, and E. Dewailly. 2002. Plasma concentrations of polychlorinated biphenyls and the risk of breast cancer: a congener-specific analysis. Am. J. Epidemiol. 155:629-635. [DOI] [PubMed] [Google Scholar]
- 13.Denef, V. J., J. Park, T. V. Tsoi, J.-M. Rouillard, H. Zhang, J. A. Wibbenmeyer, W. Verstraete, E. Gulari, S. A. Hashsham, and J. M. Tiedje. 2004. Biphenyl and benzoate metabolism in a genomic context: outlining genome-wide metabolic networks in Burkholderia xenovorans LB400. Appl. Environ. Microbiol. 70:4961-4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denef, V. J., M. A. Patrauchan, C. Florizone, J. Park, T. V. Tsoi, W. Verstraete, J. M. Tiedje, and L. D. Eltis. 2005. Growth substrate- and phase-specific expression of biphenyl, benzoate, and C1 metabolic pathways in Burkholderia xenovorans LB400. J. Bacteriol. 187:7996-8005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denef, V. J., J. A. Klappenbach, M. A. Patrauchan, C. Florizone, J. L. M. Rodrigues, T. V. Tsoi, W. Verstraete, L. D. Eltis, and J. M. Tiedje. 2006. Genetic and genomic insights into the role of benzoate-catabolic pathway redundancy in Burkholderia xenovorans LB400. Appl. Environ. Microbiol. 72:585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Donato, M. M., A. S. Jurado, M. C. Antunes-Madeira, and V. M. Madeira. 1997. Comparative study of the toxic actions of 2,2-bis(p-chlorophenyl)-1,1,1-trichloroethane and 2,2-bis(p-chlorophenyl)-1,1-dichloroethylene on the growth and respiratory activity of a microorganism used as a model. Appl. Environ. Microbiol. 63:4948-4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erickson, B. D., and F. J. Mondello. 1993. Enhanced biodegradation of polychlorinated biphenyls after site-directed mutagenesis of a biphenyl dioxygenase gene. Appl. Environ. Microbiol. 59:3858-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furukawa, K., J. R. Simon, and A. M. Chakrabarty. 1983. Common induction and regulation of biphenyl, xylene/toluene, and salicylate catabolism in Pseudomonas paucimobilis. J. Bacteriol. 154:1356-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibson, D. T., D. L. Cruden, J. D. Haddock, G. J. Zylstra, and J. M. Brand. 1993. Oxidation of polychlorinated biphenyls by Pseudomonas sp. strain LB400 and Pseudomonas pseudoalcaligenes KF707. J. Bacteriol. 175:4561-4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goris, J., P. De Vos, J. Caballero-Mellado, J. Park, E. Falsen, J. F. Quensen III, J. M. Tiedje, and P. Vandamme. 2004. Classification of the biphenyl- and polychlorinated biphenyl-degrading strain LB400 and relatives as Burkholderia xenovorans sp. nov. Int. J. Syst. Evol. Microbiol. 54:1677-1681. [DOI] [PubMed] [Google Scholar]
- 21.Hiraoka, Y., T.Yamada, K. Tone, Y. Futaesaku, and K. Kimbara. 2002. Flow cytometry analysis of changes in the DNA content of the polychlorinated biphenyl degrader Comamonas testosteroni TK102: effect of metabolites on cell-cell separation. Appl. Environ. Microbiol. 68:5104-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hrywna, Y.,T. V. Tsoi, O. V. Maltseva, J. F. Quensen III, and J. M. Tiedje. 1999. Construction and characterization of two recombinant bacteria that grow on ortho- and para-substituted chlorobiphenyls. Appl. Environ. Microbiol. 65:2163-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imbeault, N. Y. R., J. B. Powlowski, C. L. Colbert, J. T. Bolin, and L. D. Eltis. 2000. Steady-state kinetic characterization and crystallization of a PCB-transforming dioxygenase. J. Biol. Chem. 275:12430-12437. [DOI] [PubMed] [Google Scholar]
- 24.Janssen, D. B., A. Scheper, and B. Witholt. 1984. Biodegradation of 2-chloroethanol and 1,2-dichloroethane by pure bacterial cultures, p. 169-178. In E. H. Houwink and R. R. van der Meer (ed.), Innovations in biotechnology. Elsevier, Amsterdam, The Netherlands.
- 25.Kim, I. S., H. Lee, and J. T. Trevors. 2001. Effects of 2,2′,5,5′-tetrachlorobiphenyl and biphenyl on cell membranes of Ralstonia eutropha H850. FEMS Microbiol. Lett. 200:17-24. [DOI] [PubMed] [Google Scholar]
- 26.Klomparens, K., S. L. Flegler, and G. R. Hooper. 1986. Procedures for transmission and scanning electron microscopy for biological and medical science. Ladd Research Industries, Burlington, Vt.
- 27.Lui, J., F. B. Dazzo, O. Glagoleva, B. Yu, and A. K. Jain. 2001. CMEIAS: a computer aided system for the image analysis of bacterial morphotypes in microbial communities. Microb. Ecol. 41:173-194. [DOI] [PubMed] [Google Scholar]
- 28.Maltseva, O. V., T. V. Tsoi, J. F. Quensen III, M. Fukuda, and J. M. Tiedje. 1999. Degradation of anaerobic reductive dechlorination products of Aroclor 1242 by four aerobic bacteria. Biodegradation 10:363-371. [DOI] [PubMed] [Google Scholar]
- 29.Muller-Newen, G., and W. Stoffel. 1991. Mitochondrial 3-2 trans-enoyl-CoA isomerase. Purification, cloning, expression, and mitochondrial import of the key enzyme of unsaturated fatty acid beta-oxidation. Biol. Chem. Hoppe-Seyler 372:613-624. [DOI] [PubMed] [Google Scholar]
- 30.Neidhardt, F. C., J. L. Ingraham, and M. Schaechter. 1990. Physiology of the bacterial cell. Sinauer Associates, Inc., Sunderland, Mass.
- 31.Patandin, S., P. C. Dagnelie, P. G. H. Mulder, E. Op de Coul, J. E. van der Veen, N. Weisglas-Kuperus, and P. J. J. Sauer. 1999. Dietary exposure to polychlorinated biphenyls and dioxins from infancy until adulthood: a comparison between breast-feeding, toddler, and long-term exposure. Environ. Health Perspect. 107:45-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pellizari, V. H., S. Bezborodnikov, J. F. Quensen III, and J. M. Tiedje. 1996. Evaluation of strains isolated by growth on naphthalene and biphenyl for hybridization of genes to dioxygenase probes and polychlorinated biphenyl-degrading ability. Appl. Environ. Microbiol. 62:2053-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Poelarends, G. J. 2001. Genetic adaptation of bacteria to halogenated aliphatic compounds: the recruitment and distribution of dehalogenase genes. Dissertation. University of Groningen, Groningen, The Netherlands.
- 34.Quensen, J. F., III, J. M. Tiedje, and S. A. Boyd. 1988. Reductive dechlorination of polychlorinated biphenyls by anaerobic microorganisms from sediment. Science 242:752-754. [DOI] [PubMed] [Google Scholar]
- 35.Quensen, J. F., III, S. A. Boyd, and J. M. Tiedje. 1990. Dechlorination of four commercial polychlorinated biphenyl mixtures (Aroclors) by anaerobic microorganisms from sediments. Appl. Environ. Microbiol. 56:2360-2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramos, J. L., E. Duque, J.-J. Rodriguez-Herva, P. Godoy, A. Haidour, F. Reyes, and A. Fernandez-Barrero. 1997. Mechanisms for solvent tolerance in bacteria. J. Biol. Chem. 272:3887-3890. [DOI] [PubMed] [Google Scholar]
- 37.Ramos, J. L., E. Duque, M.-T. Gallegos, P. Godoy, M. I. Ramos-Gonzalez, A. Rojas, W. Teran, and A. Segura. 2002. Mechanisms of solvent tolerance in Gram negative bacteria. Annu. Rev. Microbiol. 56:743-768. [DOI] [PubMed] [Google Scholar]
- 38.Rodrigues, J. L. M., O. V. Maltseva, T. V. Tsoi, R. R. Helton, J. F. Quensen III, M. Fukuda, and J. M. Tiedje. 2001. Development of a Rhodococcus recombinant strain for degradation of products from anaerobic dechlorination of PCBs. Environ. Sci. Technol. 35:663-668. [DOI] [PubMed] [Google Scholar]
- 39.Rodrigues, J. L. M., C. A. Kachel, M. R. Aiello, J. F. Quensen, O. V. Maltseva, T. V. Tsoi, and J. M. Tiedje. 2006. Degradation of Aroclor 1242 dechlorination products in sediments by Burkholderia xenovorans LB400(ohb) and Rhodococcus sp. strain RHA1(fcb). Appl. Environ. Microbiol. 72:2476-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seegal R. F., and S. L. Schantz. 1994. Neurochemical and behavioral sequelae of exposure to dioxins and PCBs, p. 409-447. In A. Schecter (ed.), Dioxins and health. Plenum Press, New York, N.Y.
- 41.Seto, M., N. Okita, K. Sugiyama, E. Masai, and M. Fukuda. 1996. Growth inhibition of Rhodococcus sp. strain RHA1 in the course of PCB transformation. Biotechnol. Lett. 18:1193-1198. [Google Scholar]
- 42.Sikkema, J., J. A. M. de Bont, and B. Poolman. 1995. Mechanisms of membrane toxicity of hydrocarbons. Microbiol. Rev. 59:201-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smithwick, L. A., A. Smith, J. F. Quensen III, A. Stack, L. London, and P. J. Morris. 2003. Inhibition of LPS-induced splenocyte proliferation by ortho-substituted polychlorinated biphenyl congeners. Toxicology 188:319-333. [DOI] [PubMed] [Google Scholar]
- 44.Sondossi, M., M. Sylvestre, and D. Ahmad. 1992. 1992. Effects of chlorobenzoate transformation on the Pseudomonas testosteroni biphenyl and chlorobiphenyl degradation pathway. Appl. Environ. Microbiol. 58:485-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Swanson, G. M., H. E. Ratcliffe, and L. J. Fischer. 1995. Human exposure to polychlorinated biphenyls (PCBs): a critical assessment of the evidence for adverse health effects. Regul. Toxicol. Pharmacol. 21:136-150. [DOI] [PubMed] [Google Scholar]
- 46.Tsoi, T. V., E. G. Plotnikova, J. R. Cole, W. F. Guerin, M. Bagdasarian, and J. M. Tiedje. 1999. Cloning, expression, and nucleotide sequence of the Pseudomonas aeruginosa strain 142 ohb genes coding for oxygenolytic ortho dehalogenation of halobenzoates. Appl. Environ. Microbiol. 65:2151-2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vaillancourt, F. H., S. Han, P. D. Fortin, J. T. Bolin, and L. D. Eltis. 1998. Molecular basis for the stabilization and inhibition of 2,3-dihydroxybiphenyl 1,2-dioxygenase by t-butanol. J. Biol. Chem. 273:34887-34895. [DOI] [PubMed] [Google Scholar]
- 48.Vaillancourt, F. H., G. Labbe, N. M Drouin, P. D. Fortin, and L. D. Eltis. 2002. The mechanism-based inactivation of 2,3-dihydroxybiphenyl 1,2-dioxygenase by catecholic substrates. J. Biol. Chem. 277:2019-2027. [DOI] [PubMed] [Google Scholar]
- 49.Van der Ploeg, J., M. P. Smidt, A. S. Landa, and D. B. Janssen. 1994. Identification of chloroacetaldehyde dehydrogenase involved in 1,2-dichloroethane degradation. Appl. Environ. Microbiol. 60:1599-1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Hylckama Vlieg, J. E. T., W. de Koning, and D. B. Janssen. 1997. Effect of chlorinated ethene conversion on viability and activity of Methylosinus trichosporium OB3b. Appl. Environ. Microbiol. 63:4961-4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yeager, C. M., P. J. Bottomley, and D. J. Arp. 2001. Cytotoxicity associated with trichloroethylene oxidation in Burkholderia cepacia G4. Appl. Environ. Microbiol. 67:2107-2115. [DOI] [PMC free article] [PubMed] [Google Scholar]