Abstract
Human adenocarcinoma (AC) is the most frequently diagnosed human lung cancer, and its absolute incidence is increasing dramatically. Compared to human lung AC, the A/J mouse-urethane model exhibits similar histological appearance and molecular changes. We examined the gene expression profiles of human and murine lung tissues (normal or AC) and compared the two species’ datasets after aligning ∼7500 orthologous genes. A list of 409 gene classifiers (P value <0.0001), common to both species (joint classifiers), showed significant, positive correlation in expression levels between the two species. A number of previously reported expression changes were recapitulated in both species, such as changes in glycolytic enzymes and cell-cycle proteins. Unexpectedly, joint classifiers in angiogenesis were uniformly down-regulated in tumor tissues. The eicosanoid pathway enzymes prostacyclin synthase (PGIS) and inducible prostaglandin E2 synthase (PGES) were joint classifiers that showed opposite effects in lung AC (PGIS down-regulated; PGES up-regulated). Finally, tissue microarrays identified the same protein expression pattern for PGIS and PGES in 108 different non-small cell lung cancer biopsies, and the detection of PGIS had statistically significant prognostic value in patient survival. Thus, the A/J mouse-urethane model reflects significant molecular details of human lung AC, and comparison of changes in orthologous gene expression may provide novel insights into lung carcinogenesis.
In North America and developed countries, annual lung cancer deaths account for more deaths than the combined mortality due to prostate, breast, and colorectal cancers.1 Approximately 170,000 new cases of lung cancer will be diagnosed this year with a 5-year survival rate <15%. Women are showing a faster increase in occurrence than men, presumably due to their increased tobacco usage after World War II.2 Better survival prognosis is correlated with earlier detection of the disease, with stage IA patients showing ∼60% 5-year survival while later stage detection (II to IV) 5-year survival declines to ∼5%.3 The poor prognosis for lung cancer patients is generally attributed to the limited success of early detection screening methods, combined with an inability to treat the resultant late stage, metastatic disease. Lung cancer is divided into small cell and non-small cell histological types, with the most common form being non-small cell lung adenocarcinoma (AC) whose incidence is becoming more predominant.4
The earliest published description of a primary lung tumor in mice was of a spontaneously appearing tumor in a wild mouse in 1896.5 During the first three-quarters of the 20th century, experimental study of murine lung tumors focused on whether their induction by chemicals constituted an effective means of evaluating putative carcinogens. These early studies described primary lung tumor development, tested the effectiveness of using tumor antigenicity to protect recipient mice inoculated with tumor-specific antibodies, and determined patterns of inheritance among inbred strains of spontaneously appearing and chemically induced tumors. An incisive review of this system6 stimulated molecular approaches used throughout the past 25 years to investigate progression-dependent biochemical changes that guide neoplastic development,7 the molecular basis of genetic susceptibility,8 detection of chemoprevention agents,9 and application of genetically altered mice to study each of these aspects.10
Human lung AC is often detected late in disease progression; sequential changes in the human lung leading to the development of lung AC are infrequently observed. The current model for the development of human lung AC begins with atypical adenomatous hyperplasia with low-grade histological features.11 For the A/J mouse-urethane model, lung tumors proceed through hyperplastic and adenoma stages, ultimately developing into AC.12 Urethane-induced murine AC appear to arise from alveolar type II epithelial cells, and a high percentage of human ACs have characteristics suggesting some of these tumors are also derived from alveolar type II cells. In addition, murine and human ACs have a high frequency of activating Kras mutations. Significant effort has been put toward creating transgenic murine models of human cancers with more than a dozen different transgenic mouse models of pulmonary cancer recently reviewed by the Mouse Models of Human Cancer Consortium13 and others.14 The majority of the transgenic models produced lung adenomas and AC histological types through various genetic combinations, whereas one model produced neuroendocrine tumors with small cell lung cancer characteristics. Although transgenic mouse studies are elegant in their approach, epidemiological analyses repeatedly demonstrate the overwhelming contribution of environmental effects, such as tobacco usage and occupational exposures, which together account for ∼90% of the lung cancer cases in the human population. A study of the Swedish Family-Cancer Database estimated the genetic component as 14% of the lung cancer burden in this population.15 In addition, analysis of nonsmoking probands suggest lung cancer incidence is best modeled through environmental exposure rather than on a genetic basis except for some genetic contribution in early onset disease.16
Because carcinogen exposure is responsible for the vast majority of human lung ACs, we decided to compare lung tissue from the A/J mouse-urethane model and human AC using microarray analysis17 to assess global gene expression changes. This approach allowed us to quantify the degree of molecular similarity between AC in humans and the A/J mouse-urethane model. We hypothesized that common molecular events leading to AC in either species would result in conserved gene expression changes between adjacent normal and tumor tissues. A number of previously published microarray studies of human non-small cell lung cancer (NSCLC)18–21 have clearly shown that different histological subtypes are distinguishable and prognostic information can be obtained by this approach. Murine lung tissue microarray studies have been reported that emphasized discerning strain-specific gene expression differences in normal lung.22,23 Two recent reports examined human and mouse lung tumors in relation to murine lung development,24,25 although these studies were complicated by the difficulties of microarray cross-platform data analysis.26 In addition, identification of orthologous genes with similar expression changes may elucidate the most conserved pathways underlying development of this lung cancer type. Toward this end, a gene expression analysis of the transgenic murine KrasLA model27 identified a murine activated Kras expression signature.28 The murine activated Kras expression signature was able to correctly classify human lung AC tumor samples. Significant molecular similarities between human disease and the A/J mouse-urethane model would strongly support using this model to identify early markers for disease and to test a wide range of chemoprevention and therapeutic agents.
Materials and Methods
Mouse Lung Carcinogenesis
Male A/J mice, obtained from Jackson Laboratories (Bar Harbor, ME), at 5 to 7 weeks of age, were allowed to acclimate for 10 to 14 days before their use in experiments. The mice were given access to Teklad-8640 standard laboratory chow (Harlan Teklad, Madison, WI) and water ad libitum and maintained on a hardwood bedding under a 12-hour light/dark cycle. Mice were injected once intraperitoneally with 1 mg/g body weight urethane dissolved in 0.9% NaCl (saline) or saline control as described.29 Mice were sacrificed 24 to 26 weeks or 42 weeks later by lethal injection of 100 μl of 90 mg/ml sodium pentobarbital containing 1000 U/ml heparin (Sigma, St. Louis, MO).
Lung Tissue Preparation
Mouse
At each time point, male mice were sacrificed for either histological examination or RNA preparation. For histology, the lungs were perfused with saline through the right pulmonary artery, gently inflated through a cannulated trachea with 600 to 900 μl of formalin (10% neutral buffered formaldehyde solution), and incubated for 1 hour at room temperature. Lungs were then removed from the chest cavity and individual lobes dissected and submerged in formalin overnight. Fixed lungs were transferred to 95% ethanol, embedded in paraffin, and cut in 4-μm sections for histological analysis. For RNA preparation, the lung tissue was examined using a dissection microscope immediately after the mice were sacrificed. Tumor or normal-appearing lung parenchyma tissues from tumor-bearing mice (adjacent tissue) were dissected and placed in 300 to 750 μl of RNAlater at 4°C (Ambion, Austin, TX) and subsequently stored at −20°C. At the early time point, nine independent mice generated nine adjacent and nine tumor tissue samples with an additional three tumor samples from other mice that did not yield sufficient quality RNA from their adjacent tissues (total of 9 adjacent and 12 tumor samples). At 42 weeks, eight independent mice generated 8 adjacent and 19 tumor samples from different regions of the lung.
Human
All patients participating in this study were enrolled in a local Colorado Multiple Institutional Review Board (COMIRB) approved protocol for use of remnant tissue with anonymization and analysis of specimens and clinical data. All but one of the patients had a history of smoking. Patients range in age from 45 to 73 years of age. Tumors from five males and five females were used in the study. All specimens for microarray analysis were obtained at surgery with nine patients undergoing lobectomy and one wedge resection. Specimens were examined immediately after removal from the patient and grossly visible solid tumor tissue was snap-frozen for RNA extraction. The tumors were all invasive ACs, but five specimens exhibited evidence of bronchoalveolar differentiation at the edge of tumor nests. Most tumors were low to intermediate grade and low stage, although two stage III tumors were included in the analysis. The degree of contamination of tumor cells by stromal cells was variable and ranged from 10 to 90% but did not affect the ability of the microarray analysis to correctly distinguish adjacent from tumor tissue samples. Tumor and adjacent normal tissue samples from the 10 different AC patients were stored in liquid nitrogen until total RNA was extracted. Approximately 100 to 300 mg of tissue samples were cut from the frozen tissue pieces for RNA purification.
Sample and array experiment naming convention used the following number/letter combinations to indicate species (Hs = human; AJ or LZ = A/J mouse), and sample type [N or A = adjacent (normal), T = tumor]. The designation adjacent is used for normal appearing lung tissue sample from either a human or a mouse with AC. This designation recognizes that adjacent tissue from a lung with AC may not be identical to normal tissue from an untreated or disease-free lung. For the A/J mouse experiments, the late time points at 42 weeks after urethane injection include the number 42 in their names. The two A/J array experiments that were consistently misclassified are labeled with a q (questionable).
Microarray Data Collection
To minimize the difficulties of cross-platform data analysis,26 our study was designed around the Affymetrix GeneChip platform for samples from either species performed within one core laboratory. A/J mouse samples included adjacent (normal histology) and tumor samples from urethane-treated male animals. Human lung samples were from resectioned AC patients and included both their tumor and adjacent tissue taken ∼1 cm from the tumor site. Total RNA was purified from all lung tissue samples using RNeasy kits (Qiagen, Valencia, CA). Total RNA quality was assessed by UV spectral characterization (A260/A280 > 1.9) and Agilent Bioanalyzer separation (undegraded 18S and 28S rRNA). Total RNA (2 to 5 μg) was used as starting material following the Affymetrix labeling protocol. After conversion to cRNA and fragmentation, the probe was hybridized to the corresponding species’ Affymetrix GeneChip microarray (human HG-U95Av2 or mouse MG-U74Av2). Image data from each microarray was scaled and normalized using Microarray Suite 5.0 (MAS 5, Affymetrix) with the target intensity set at 500 and normalized to 1.0. Thirty-nine human arrays were completed from adjacent and tumor tissues derived from 10 different patients run in duplicate. The human arrays had an average correlation coefficient r = +0.83 (SD = 0.009) between duplicates with an average scaling factor of 4.51 (SD = 0.76). A total of 45 mouse arrays were completed, with 44 included in this data set with an average scaling factor of 2.52 (SD = 1.06); one array was excluded due to a significantly higher scaling factor (8.36). All of the gene expression datasets have been deposited at Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under the following accession numbers: human adjacent (GSM47958 to GSM47976); human tumor (GSM36757 to GSM36776); murine adjacent (early GSM47977 to GSM47984, late GSM47997 to GSM48003); murine tumor (early GSM47985 to GSM47996, late GSM48004 to GSM48020).
Microarray Data Analysis
The MAS 5 pivot table was imported into Biometric Research Branch (BRB)-ArrayTools suite [http://linus.nci.nih.gov/BRB-ArrayTools.html; version 3.0.1a (6/03)] for analysis. All output data were handled in Excel 2000 and converted into Filemaker Pro 6 files. Each individual file was linked via Affymetrix’s probe ID using Netaffx (http://www.affymetrix.com/analysis/index.affx) orthologous gene listing as the central database.30 Statistical analysis used BRB-ArrayTools and JMPIN version 4 (SAS Institute, Cary, NC). Gene ontology (GO) annotation31 was also derived from the Netaffx website using the human probe IDs.30 Because of redundancies present on each species’ Affymetrix GeneChip and in the NetAffx ortholog alignment, our analytical approach is limited to only ∼60% (∼7500/∼12,400 probe IDs per GeneChip) of the complete dataset collected from each microarray experiment. The BRB-ArrayTools suite makes extensive use of parametric and permutation analyses, as well as estimation of significance by false discovery rate.32 Classifiers are cross-validated by a leave one out strategy by several different methods.
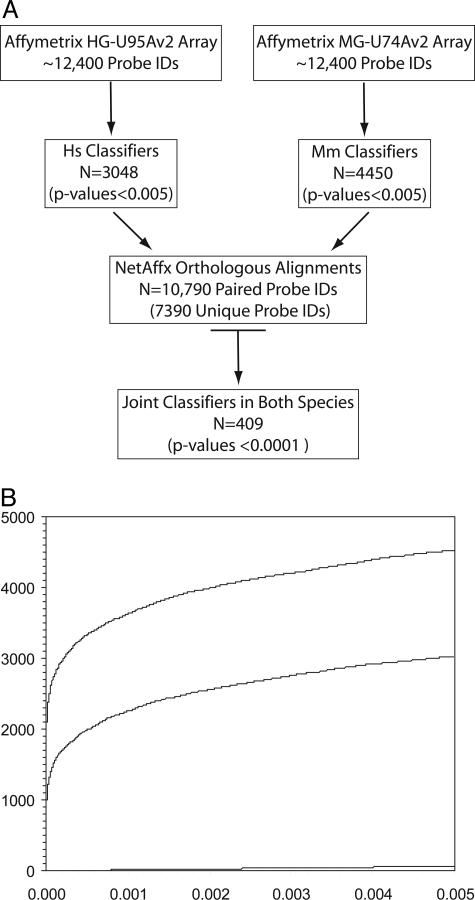
To define a classifier, the gene expression data for a given probe ID (data point) across all of a species’ microarrays are hypothesis tested as to whether it can reliably distinguish between an adjacent or tumor tissues at a preset level of significance (parametric P value). The null hypothesis is that the data points within a given probe ID do not distinguish between adjacent and tumor samples. One difficulty with microarray studies is the asymmetry in the richness of the data obtained relative to the number of samples. In these studies, ∼10,800 data points are considered per sample after orthologous alignment. The Bonferroni threshold is often used in this situation as a conservative approximation to provide a P value correction at the desired level of significance. To calculate the Bonferroni threshold, the desired level of significance (P value) is divided by the number of data points in the experiment. For this dataset, a desired P value of 0.05 would need to be corrected to a required P value of 0.000005 (0.05/10,800). Even at this conservative measure, the analysis would include ∼950 human and ∼2000 murine probe IDs as classifiers, respectively, with P value ≤0.000005. However, an unbiased measure for determining a statistical cutoff value is using an overabundance plot to indicate where the cutoff can be reliably placed.33 Based on this approach, the threshold was set at a P value ∼0.005 to include no more than an false discovery rate of 100 false discoveries, increasing the probe IDs in this study to 3048 human and 4450 murine classifiers (P values <0.005). One hundred false discoveries within the datasets would result in a potential of 3.3% (human) and 2.2% (murine) false-positives of the probe IDs identified. The null hypothesis would predict only 62 probe IDs, based on all microarray probe IDs (0.005 × 12,400 probe IDs on each microarray), meeting these statistical criteria by random chance. The two species’ individual classifier lists were then aligned using Netaffx as an annotated source of orthologous genes. From this alignment, 409 unique genes were identified as joint classifiers that had P values <0.0001 for additional study. As a test of validity of this approach, the least statistically significant joint classifiers (P values >0.001; n = 47) were tested for their discriminating ability. The mouse data set had two microarray experiments, one early and one late tumor sample (LZ30 Tq and LZ73 Tq42), that were consistently misclassified, while the human data set had two tumor samples occasionally misclassified (Hs28 7T1 and Hs31 8T2).
Expression heat maps were generated using Cluster and TreeView.34 The log2 expression data from the human and mouse microarrays was imported for the probe IDs corresponding to the 409 unique genes. To keep the output diagrams in a similar gene order, the GORDER option was used to list the human and mouse data in the same order. The raw data were median centered (by genes and arrays) and clustered using the Spearman correlation to use the high correlation by ranking.
Tissue Microarray Construction
Paraffin blocks of tumor tissue from 110 patients diagnosed with NSCLC (stages I to III) between 1993 through 1999 were obtained from the archives of the University of Colorado Cancer Center (Denver, CO) and Johns Hopkins Medical Institutions (Baltimore, MD) according to institutional review board-approved protocols. Patients were followed by the tumor registries for survival time and outcome with median follow-up of 51 months (ranging from 18 to 100 months). The tumors were staged according to the tumor-node-metastasis classification and histologically classified according to the World Health Organization guidelines. The NSCLC tissue samples were classified as 51 squamous, 45 AC, 7 large cell, and 7 bronchioloalveolar carcinoma histological subtypes. A detailed listing of histological subtype, stage, and grade is included in the on-line supplemental material at http://ajp.amjpathol.org (Supplemental Table 1).
The tissue microarrays were assembled using a tissue-arraying instrument (Beecher Instruments, Silver Spring, MD), consisting of thin-walled stainless steel biopsy needles and stylets used to empty and transfer the needle content. The assembly is held in an X-Y position guide that is adjusted manually. A large diameter stylet (1.5 mm) was used for sampling, and nonnecrotic areas of the blocks were routinely oversampled with three replicate core samples of tumor (different areas) regions from each donor block to account for tumor heterogeneity. Normal lung and 15 other control tissues were included in each tissue array block. Four-μm sections of the resulting microarray blocks were cut with a Leitz microtome. Sections were transferred to adhesive-coated slides using the adhesive-coated tape-sectioning system (Instrumedics Inc., Hackensack, NJ). Subsequently, UV light treatment of the slides for 60 seconds polymerized the adhesive coating into a plastic layer and sealed the sections to the slides. Thereafter, the tape could be removed in a solvent (Instrumedics Inc.).
Tissue Microarray Immunohistochemistry and Analysis
The tissue sections were deparaffinized with standard xylene and hydrated through graded alcohols into water. Antigen retrieval was performed by heating slides in citrate buffer for 20 minutes at 105°C in a Biocare Medical decloaking chamber (Walnut Creek, CA). Peroxide blocking was performed with 3% hydrogen peroxide in water for 10 minutes. Avidin and biotin blocks were done for 10 minutes each using the DAKO A/B blocking kit (DAKO, Carpinteria, CA). The sections were incubated with a goat polyclonal anti-cyclooxygenase 2 (COX-2) antibody (Santa Cruz Biotechnology, Santa Cruz, CA) at a 1:400 dilution in 0.05 mol/L Tris-buffered saline with 10% bovine serum albumin and 1% sodium azide for 1 hour at room temperature. The secondary biotinylated rabbit anti-goat antibody (DAKO) was applied at a 1:400 dilution with 40% normal human serum for 30 minutes at room temperature. The DAKO LSAB Plus horseradish peroxidase detection reagent was applied for 30 minutes at room temperature followed by application of diaminobenzidine chromogen for 5 minutes. The slides were then counterstained in hematoxylin and coverslipped. For prostaglandin E2 synthase (PGES) staining, the peroxide block was followed by a 10-minute universal block using Power Block (BioGenex, San Ramon, CA). The sections were incubated with a rabbit polyclonal anti-PGES antibody (Cayman Chemical, Ann Arbor, MI) at a 1:500 dilution overnight at 4°C. The DAKO Envision Plus horseradish peroxidase detection reagent was applied for 30 minutes at room temperature followed by the application of diaminobenzidine chromogen for 5 minutes. For cytosolic phospholipase A2 (cPLA2) and prostacyclin synthase (PGIS) staining, a peroxidase anti-peroxidase system was used and antigen retrieval was increased to 30 minutes in citrate buffer. For cPLA2, the peroxidase block was followed by incubation with a goat polyclonal anti-cPLA2 antibody (Santa Cruz Biotechnology) at 1:10 dilution overnight at 4°C. Next, a rabbit anti-goat bridging antibody (Zymed, San Francisco, CA) was applied at a 1:200 dilution for 30 minutes at room temperature. A goat peroxidase anti-peroxidase complex (Jackson Immunoresearch, West Grove, PA) was applied at a 1:400 dilution for 30 minutes at room temperature followed by incubation with the diaminobenzidine chromogen. For PGIS, a 10-minute universal block with Power Block followed the peroxide block. A primary polyclonal rabbit anti-PGIS antibody (gift from Dr. David DeWitt, Michigan State University) was applied at a 1:25 dilution overnight at 4°C. Slides were incubated with a goat anti-rabbit bridging antibody (Zymed) at 1:200 dilution plus 40% normal human serum for 30 minutes at room temperature followed by application of a rabbit peroxidase anti-peroxidase complex (Zymed) at 1:250 dilution for 30 minutes before visualization with diaminobenzidine. Either goat or rabbit IgG (Sigma) was applied at the same concentration as the primary antibodies for negative controls. Sections were dehydrated through a series of alcohols and xylene and covered with a glass slip.
Each core on the tissue microarray was examined by conventional white light microscopy and the observed staining pattern for each core graded independently by two pathologists without knowledge of the patients’ histories. A semiquantitative grading score was obtained by multiplying the intensity of staining (0 = negative, 1 = trace, 2 = weak, 3 = intermediate, 4 = strong) by the percentage of tumor cells stained (0 to 100%) for scores ranging from 0 to 400. The final score is an average among the three core samples for each patient and the scores for both pathologists. SAS/STAT statistical package (Cary, NC) was used for the analysis of the immunohistochemical data. Univariate analysis was performed using a Cox proportional hazards model to examine the association between each of the four enzymes and survival adjusting for age, gender, and stage.35 No significant association was found between expression level and survival for PGES, COX-2, or cPLA2.
Results
Murine Lung Tumor Histology
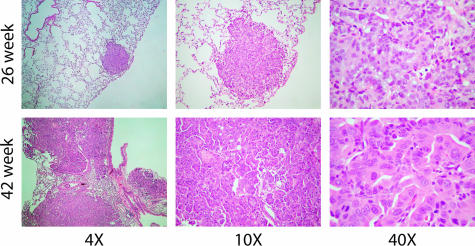
As previously described,7 the A/J mouse-urethane model provides a reproducible time course for tumor initiation, progression, and metastasis. Typically, ∼30 independent benign tumors per mouse are produced after a single urethane injection. At the early time in these experiments (24 to 26 weeks after urethane injection), mouse tumors were small, self-contained nodules (adenomas) within the lung. At the late time (42 weeks), tumors were significantly larger, covering most of the lung volume, and showed a strong invasive phenotype (ACs). The typical histology observed for the A/J mouse lung AC is shown in Figure 1. Comparisons of early and late mouse tumor samples showed increased disorganization of tumor cells and invasion into neighboring stroma. At higher magnification, late mouse AC samples had more cells with enlarged nuclei, a higher mitotic index, and frequency of visible nucleoli. These histological findings closely parallel that observed in human AC development.
Figure 1.
A/J mouse lung tissues stained with H&E at early (26 weeks) and late (42 weeks) time points after urethane administration shown at three different magnifications. The early lesions are well circumscribed whereas the late lesions show evidence of invasion and more dysplasia at higher magnification.
Gene Expression Data Analysis
BRB-ArrayTools has several classification/prediction modes we used for identifying probe IDs of high statistical significance common to both species.32 Each species’ data were analyzed to determine which probe IDs reproducibly distinguish adjacent from tumor samples (Figure 2A). No bias was imposed for whether the genes were up- or down-regulated in tumors relative to adjacent tissue in either species, just that they met the statistical criteria described in the Materials and Methods section. One way to visualize the significance of these probe IDs is through an overabundance plot (Figure 2B).33 The two individual species’ classifier lists contain 3048 human probe IDs and 4450 murine probe IDs. The null hypothesis predicts only ∼62 probe IDs would be found at the statistical levels used in our analysis. Sixty-two false-positives represent only ∼2.0% of the probe IDs in our classifier lists. The murine dataset consistently yielded a larger number of classifiers than the human dataset at any P value (Figure 2B). The increased number of murine classifiers presumably reflects the genetic in-bred nature of the A/J mouse as compared to humans, resulting in smaller standard deviations in the expression levels.
Figure 2.
A: Schematic representation of gene expression data analyses highlighting the approach used to define the overlapping set of joint classifiers between human and murine AC. In each case, the number of probe IDs underlying each subset is indicated. Human and murine data are indicated as Hs and Mm, respectively. B: Overabundance plot demonstrating the vast excess of highly significant genes identified by this analysis at any P value. The murine genes (top line), human genes (middle line), and null hypothesis (bottom line) are shown as a cumulative sum versus P value. The sharp rise for human and murine genes shown in the plot at a P value of 0.000 included probe IDs with P values <0.0001.
NetAffx30 supplies a sequence-based orthologous alignment of human and mouse entries cross-referenced through the Affymetrix probe IDs. The most recent version of NetAffx had a total of 10,790 entries without accounting for redundancies within the individual species or the alignments between them (Figure 2A). After accounting for these multiple layers of redundancies, the orthologous alignment of human and murine Affymetrix probe IDs allows the direct comparison of 7390 unique gene alignments between the two species (Figure 2A). Our approach was to first identify each species’ classifiers that can distinguish between adjacent and tumor samples. Of the probe IDs that could be compared (through the NetAffx orthologous alignment), an overlapping set of joint classifiers, common to both human and mouse, were identified. A list of 409 unique genes was generated from the overlap of human and mouse classifiers (complete listing available as an on-line spreadsheet supplement at http://ajp.amjpathol.org).
Identification of Orthologous Classifiers
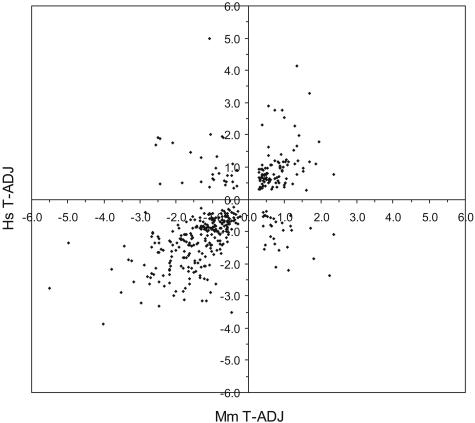
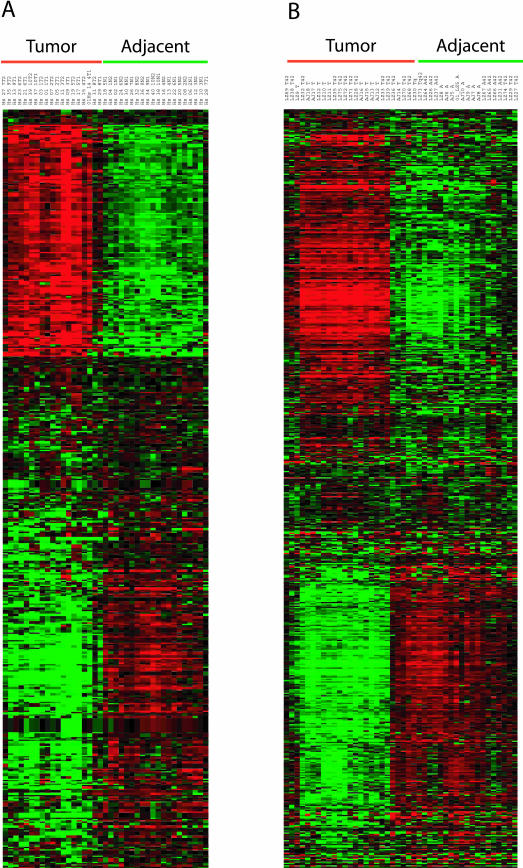
The log2 intensity difference between tumor and adjacent samples for each of the 409 unique genes is plotted for the human and mouse data (Figure 3). The two species’ log2 intensity differences were positively correlated (Pearson coefficient r = +0.61; P value <0.000136). Of the 409 unique genes, 256 joint classifiers were down-regulated (63%), 95 were up-regulated (23%), and 58 (14%) were discordant in expression (the human and mouse log2 intensity differences were of opposite sign). An alternative way to visualize the strong correlation between the two species is an expression heat map (Figure 4).34 The log2 intensity data from each species’ probe IDs displayed striking similarity in the pattern of up- and down-regulated genes. Finally, a comparison of our results to those from the transgenic murine KrasLA model37 indicated remarkable similarities in the unique genes identified (42% in common) and their relative gene expression levels between AC tumors and tissue from adjacent or age-matched normal littermates (Pearson coefficient r = +0.82, P value <0.0001; see Supplemental Figure S1 at http://ajp.amjpathol.org).
Figure 3.
Plot of human versus murine log2 difference (tumor minus adjacent) intensities for the overlapping set of 409 unique genes. Different probe IDs representing the same gene were averaged (in either species). Genes along the diagonal represent concordant expression changes (Pearson correlation r = +0.61, P value <0.0001). The genes in the top left and bottom right quadrants had discordant expression changes between human and murine AC.
Figure 4.
Gene expression data for the 409 gene in the highly statistically significant subset is displayed using the Cluster and Treeview programs for the human (A) and murine (B) datasets. The data were log-transformed, median centered by genes and arrays, followed by hierarchical clustering by Spearman rank correlation centering. The GORDER option maintained a similar vertical ordering of the genes in both species. Green underlining indicates lung tissue adjacent to AC, whereas red underlining indicates AC tumor samples.
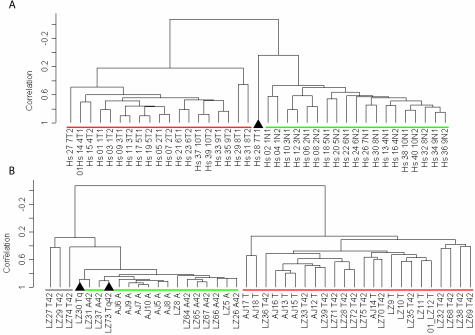
The 409 unique genes were used for supervised clustering34 and representation by multidimensional scaling (Figure 5; multidimensional scaling shown in Supplemental Figure S2 at http://ajp.amjpathol.org) of the human and murine datasets. The majority (∼95%) of adjacent and tumor samples from either species were correctly distinguished by both analytical approaches. The cluster tree shows that all except one pair of human tumor duplicate samples are nearest neighbors with high correlation values; human adjacent duplicate samples are nearest neighbors most of the time, probably reflecting differing degrees of stromal and/or tumor cell contamination. In the murine data set, most nearest neighbors are from the same time points and have extremely high correlation values. Multidimensional scaling analysis was used as a separate measure of correlation between the human and murine data. Multidimensional scaling was able to capture a large amount of the variation within either species as the first three principal components calculated by this approach covered 65% and 68% of the total variation in human and mouse, respectively, and were highly statistically significant (P value ∼0.0 for either species; Supplemental Figure S2 at http://ajp.amjpathol.org). Two mouse tumor samples were consistently misclassified as normal, while one human tumor sample (Hs28 7T1) was placed midway between the two groupings (multidimensional scaling). The 409 unique genes included in this group were each required to have a P value <0.0001 in testing the hypothesis that individually they could distinguish between adjacent and tumor tissues. The probability of finding >400 unique genes together that each individually meet this level of statistical significance is vanishingly small (P value ∼0.0), supporting the hypothesis that there is a high degree of molecular similarity between human and murine lung AC.
Figure 5.
Supervised clustering of human (A) and murine (B) gene expression data for the 409 genes in the highly statistically significant subset is displayed as a dendrogram. Black triangles indicate tissue samples not correctly identified (human 7T1 and murine LZ30Tq and LZ73 Tq42). The three murine samples underlined in gray were not clearly associated with either group. Green underlining indicates lung tissue adjacent to AC, whereas red underlining indicates AC tumor samples.
Analysis of Extended Set of Joint Classifiers
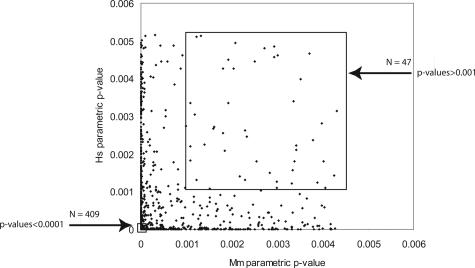
Besides the 409 joint classifiers of high statistical confidence described above, several sets of lower statistically significant joint classifiers were identified to test the potential value of all joint classifiers (Figure 6, Supplemental Figure S3 at http://ajp.amjpathol.org). Plots similar to Figure 3 are shown in Supplemental Figure S4 at http://ajp.amjpathol.org, for decreasing levels of statistical significance within the joint classifier lists. As the statistical significance is relaxed from a P value <0.0001 to include all joint classifiers (1354 probe IDs with P values <0.005), the Pearson correlation decreased to r = +0.51, indicating a strong relationship remained between the gene expression patterns seen in both species even after including lower ranking probe IDs. The least significant set of 47 probe IDs having P values >0.001 (indicated in Figure 6 within the upper box) are shown in comparison to the 409 joint classifiers with P value <0.0001(contained within the lower box). Using the 47 probe IDs in supervised clustering of the data resulted in the dendrogram shown in Supplemental Figure S5 at http://ajp.amjpathol.org. The majority (>90%) of the samples were correctly placed on their respective branch of the dendrograms. Further, BRB-Array Tools includes five different algorithms for class prediction that can also be used in a supervised manner.32 When the datasets are tested using only the 47 probe IDs, both human and murine tissues were correctly identified >85% of the time using any of the different algorithms contained in the BRB-Microarray Tools software (data not shown). A second approach is to use only half of the microarray datasets for training and then test if the 47 probe IDs are able to correctly predict the other half. In fact, >90% of the unused half of the microarray datasets was correctly identified. Therefore, the least statistically significant joint classifiers contain sufficient information for the correct identification of tissues in the majority of cases. The conclusion from this analysis is that there is significant amount of gene expression parallelism between human and murine lung ACs, and the genes identified through this study should lead to testable hypotheses as to important targets for chemoprevention and therapeutic development in preclinical murine models.
Figure 6.
Joint classifier subsets used in subsequent analysis are shown as the highly statistically significant subset of 409 genes (P values <0.0001; bottom box at left) and the least statistically significant subset of 47 genes (P values >0.001; top box at right).
Orthologous Similarities in the Hallmarks of Cancer
Having established a strong correlation in gene expression changes in human and murine AC, we queried the identified 409 genes for biological commonalities in well-documented hallmarks of cancer.38 We have specifically listed our findings in Table 1 for three of these hallmarks: glycolysis, cell-cycle control, and angiogenesis. One of the earliest recognized hallmark of cancer is a change in glucose metabolism giving rise to the increased aerobic production of lactic acid, a finding generally recognized as the Warburg effect.39 The glycolytic pathway has become a reinvigorated cancer research area through the combination of proteomics and metabolomics.40,41 Table 1A lists the glycolytic pathway enzymes that were joint predictors from our list of 409 genes. In all but one case, the gene expression changes were concordant and of the expected direction early glycolytic steps were up-regulated including lactate dehydrogenase A, while the terminal alcohol dehydrogenases were down-regulated. Only phosphofructokinase (platelet) (PFKP) exhibited discordant gene expression changes between human and murine AC. Interestingly, the other phosphofructokinase isotype (liver; PFKL), which is more highly expressed in lung (http://symatlas.gnf.org), was up-regulated in both species. The human classifier had a P value = 0.00004 while the murine classifier had a P value = 0.0014, just slightly higher than our P value cutoff, and thus not included in Table 1A. Deoxyribonucleotide synthetic enzymes and many cell-cycle genes were up-regulated in tumor compared to adjacent tissues and the changes were concordant between the two species (Table 1B). In particular, the late stage cyclins controlling G2/M transition (cyclins A2, B1, and B2) were up-regulated. The negative regulator of G1/S transition, the cyclin-dependent kinase inhibitor KIP2 (CDKN1C), was down-regulated in both species and has been previously shown in murine lung cancer.42 Finally, angiogenesis is an important feature in cancer because it involves many cell types and biological changes. However our studies indicate that in general, the archetypical cancer angiogenic markers are in fact down-regulated in lung AC, specifically vascular endothelial growth factor (VEGF and VEGFC), its receptor (KDR), and the hypoxia-inducible transcription factor HIF-2α (Table 1C). Markers of neovascularization (angiopoietin 1, thrombomodulin, and von Willebrand factor) were also down-regulated in both species.
Table 1.
Gene Expression Changes in Three Hallmark Pathways of Cancer
| Name | Gene symbol | Human | Murine | Comments |
|---|---|---|---|---|
| A: Glycolytic enzymes | ||||
| Glucose phosphate isomerase | GPI | ↑ | ↑ | |
| Phosphofructokinase (platelet) | PFKP | ↑ | ↓ | Discordant* |
| Aldolase A | ALDOA | ↑ | ↑ | |
| Aldolase C | ALDOC | ↑ | ↑ | |
| Triose phosphate Isomerase | TPI | ↑ | ↑ | |
| Enolase | ENO1 | ↑ | ↑ | |
| Lactate dehydrogenase A | LDHA | ↑ | ↑ | Lactate production |
| Alcohol dehydrogenase 1B | ALDH1B | ↓ | ↓ | |
| Alcohol dehydrogenase 1C | ALDH1C | ↓ | ↓ | |
| B: DNA biosynthesis, repair, and cell cycle | ||||
| Thymidylate synthetase | TYMS | ↑ | ↑ | DNA synthesis |
| Thymidine kinase 1 | TK1 | ↑ | ↑ | |
| IMP dehydrogenase 2 | IMPDH2 | ↑ | ↑ | |
| Spermidine synthase | SRM | ↑ | ↑ | |
| Growth arrest/DNA-damage inducible | GADD45B | ↓ | ↓ | DNA repair |
| Growth arrest-specific 1 | GAS1 | ↓ | ↓ | |
| Xeroderma pigmentosus group C | XPC | ↓ | ↓ | |
| Cell division cycle 2 (CDK1) | CDC2 | ↑ | ↑ | Multiple steps |
| CDC28 protein kinase regulatory subunit 1B | CKS1B | ↑ | ↑ | |
| CDC28 protein kinase regulatory subunit 2 | CKS2 | ↑ | ↑ | |
| Cell division cycle 6 | CDC6 | ↑ | ↑ | G1→S |
| Cyclin D3 | CCND3 | ↓ | ↓ | |
| Cyclin-dependent kinase inhibitor KIP2 | CDKN1C | ↓ | ↓ | |
| Replication factor C 4 | RFC4 | ↑ | ↑ | S |
| Cell division cycle 20 | CDC20 | ↑ | ↑ | G2→M |
| Cyclin B1 | CCNB1 | ↑ | ↑ | |
| Cyclin B2 | CCNB2 | ↑ | ↑ | |
| Cyclin A2 | CCNA2 | ↑ | ↑ | |
| Chromosome condensation 1 | CHC1 | ↑ | ↑ | |
| C: Angiogenesis-related genes | ||||
| Angiopoietin 1 | ANGPT1 | ↓ | ↓ | |
| Endothelial PAS Domain protein 1 (HIF-2α) | EPAS1 | ↓ | ↓ | |
| Kinase insert domain receptor (FLK1, VEGFR2) | KDR | ↓ | ↓ | |
| Thrombomodulin | THBD | ↓ | ↓ | |
| Vascular endothelial growth factor | VEGF | ↓ | ↓ | |
| Vascular endothelial growth factor C | VEGFC | ↓ | ↓ | |
| von Willebrand factor | VWF | ↓ | ↓ |
Genes identified as joint classifiers in glycolysis, cell cycle, or angiogenesis are listed with their HUGO name, and arrow indicating up-regulation (↑) or down-regulation (↓) in tumor tissues relative to adjacent normal. The single asterisk indicates PFKP showed discordant gene expression changes between human (↑) and murine (↓) data, although the ubiquitously expression isoform PFKL was up-regulated in both species (see text). (The on-line web supplement at http://ajp.amjpathol.org, contains the human and mouse Affymetrix Probe IDs and log2 intensity data, as well as the human gene symbol, chromosome location, title, and GO information for the complete set of 409 joint classifiers.)
Identification of Lung AC Biomarkers
One stated goal of using gene expression profiles is to help identify potentially useful biomarkers for the diagnosis and prognosis of disease.18,19 We have listed five genes in Table 2 that are often referenced in the recent cancer biomarker literature as potential candidates. As one would expect, potential biomarkers would typically be induced in the new cell type (cancer) being diagnosed. Four of the potential biomarkers were up-regulated in both species as well, while caveolin 1 was down-regulated in tumor tissues. Interestingly, melanoma antigen family members have been of high interest in lung and other cancers,43 but to date no one has examined MAGED1 as a candidate biomarker in lung AC. Claudin 3 has become a therapeutic target as well as a potential biomarker in ovarian cancer,44 and our results suggest investigating both of these possibilities in lung AC.
Table 2.
Genes Recently Identified as Potential Biomarkers in Human Cancers
| Name | Gene symbol | Human | Murine | Reference |
|---|---|---|---|---|
| Caveolin 1 | CAV1 | ↓ | ↓ | 55 |
| Claudin 3 | CLDN3 | ↑ | ↑ | 44** |
| Matrix metalloproteinase 12 | MMP12 | ↑ | ↑ | 56 |
| Melanoma antigen, family D1* | MAGED1 | ↑ | ↑ | 43 |
| Osteopontin (secreted phosphoprotein 1) | SPP1 | ↑ | ↑ | 57 |
Gene expression changes are shown for five previously reported potential biomarkers, as designated in Table 1. The single asterisk refers to MAGED1, which has not been reported as a biomarker but is a member of MAGE gene family, which has been reported as a biomarker for numerous cancer types. CLDN3 has not been reported as a biomarker for lung cancer (indicated by the double asterisks) but has been reported as a biomarker and potential therapeutic target in ovarian cancer.
Eicosanoid Pathway Gene Expression and Patient Survival
Our laboratories have a long standing interest in the arachidonic acid pathway, how their metabolites may play an important role in human lung cancer, and new avenues of therapeutic approaches.45 We are currently testing Iloprost, a synthetic, stable PGI2 analogue, in randomized phase II chemoprevention trials to determine whether it can reverse the histological changes in the bronchial epithelium of patients at high risk to develop lung cancer.
We and others have shown that lung cancer tissues have increased expression of PGES and decreased expression of PGIS in comparison to the adjacent normal tissue.46 These results were mirrored in the microarray datasets for both species, with both PGES (up-regulated) and PGIS (down-regulated) identified as 2 of the 409 joint classifiers. Within the context of lung cancer development, these results, along with many others,47,48 suggest that PGES and its product PGE2 have protumorigenic effects while PGIS and its product, PGI2, are anti-tumorigenic.
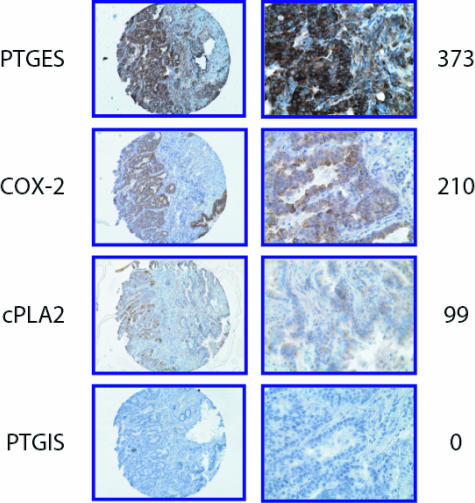
For these reasons, we investigated the expression of PGES, COX-2, cPLA2, and PGIS by immunohistochemistry in 110 human NSCLC tissue samples using tissue microarrays. Figure 7 shows staining of each of the four factors in representative AC samples and the distribution of scores for each of the four enzymes is shown in Supplemental Figure S6 at http://ajp.amjpathol.org. PGES expression was observed in 100% of the tumors with 92% showing strong staining (score = 301 to 400). COX-2 was expressed in 98% of tumors with 36% showing low staining (score = 0 to 200), 29% showing moderate staining (score = 201 to 300), and 33% showing strong staining. cPLA2 was expressed in all of the tumors with a majority showing low (66%) to moderate (28%) staining. COX-2 and PGES expression was also seen in macrophages and occasionally in normal bronchial epithelial cells. In many cases there was a low level of cPLA2 expressed in normal stroma and macrophages. Expression of PGIS was primarily absent in tumor cells, occurring at very low levels (score ≤17) in only 13% of the tumors.
Figure 7.
Immunohistochemical staining of human lung AC for eicosanoid pathway enzymes using a tissue microarray. Four enzymes [inducible prostaglandin E synthase (PGES), cyclooxygenase-2 (COX-2), cytosolic phospholipase A2 (cPLA2), and prostacyclin synthase (PGIS)] were stained and scored for intensity. Example of the relative staining intensity scoring system is show at the right.
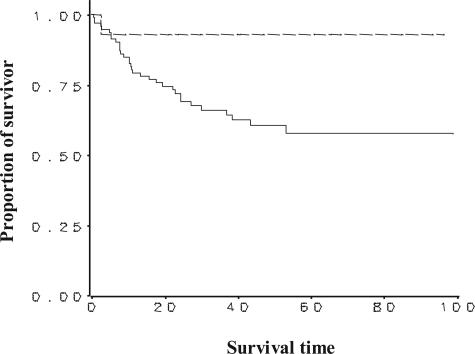
Kaplan-Meier survival curves were generated to test the survival benefit of positive PGIS staining versus negative PGIS staining for all NSCLC samples on the tissue microarray (Figure 8). All of the NSCLC samples were included in this analysis because of the limited number of patients (14 of 108) who had positive PGIS immunostaining. There was a statistically significant correlation between positive PGIS staining and increased survival (log-rank test P values = 0.047). The hazard ratio equaled 0.201, indicating an 80% reduction of mortality in patients with positive PGIS staining. The hazard ratio was not statistically significant (95% CI = 0.027 to 1.51; P value = 0.119), presumably due to the small number of deaths observed in the positive PGIS staining group throughout the available follow-up time. Survival analysis was also done categorically for PGES, COX-2, and cPLA2 by grouping expression as low (0 to 200), intermediate (201 to 300), or high (301 to 400), and no significant association was found between expression and survival for any of these three proteins.
Figure 8.
Kaplan-Meier survival curves are shown relating PGIS immunohistochemical staining to patient survival time (months) from the tissue microarray. Analysis for all NSCLC regardless of tumor type (total number = 110 samples), was completed (PGIS-negative = 94; PGIS-positive = 14; 2 not scoreable). Cox regression model was used to examine the association between positive PGIS staining and survival after adjusting for age, gender, and tumor stage. Survival was significantly correlated to positive PGIS staining using log-rank test (χ2 = 3.942, P value = 0.047) and gave a hazard ratio = 0.20 (95% CI = 0.027 to 1.51, P value = 0.119). The lack of significance in the hazard ratio is probably a function of the small number of deaths in the positive PGIS staining group relative to the amount of follow-up and variability in time to death.
Discussion
Our results represent one of the first studies completed using a single microarray technology within one laboratory comparing a murine model system to its human disease counterpart. We have shown, by several different statistical measures, that the gene expression changes between tumor and adjacent normal lung tissue in human AC are recapitulated in the A/J mouse-urethane lung AC model with striking detail. Two hallmarks of cancer cell biology (Warburg effect on glycolysis and cell-cycle alterations) were examined within the microarray datasets and were found to be consistent with the current thinking in what makes cancer cells different from normal cells. The expression pattern for the cell-cycle genes would predict that therapeutic agents active at the G2/M phase of the cycle may offer better efficacy than at other points in the cycle. New agents are currently being investigated that affect this point in cell growth.49,50 A third hallmark of cancer investigated was angiogenesis, which is associated with increased hypoxia-inducible factors’ (HIFs) transcription targets being up-regulated. The mechanism for the stabilization of HIFs is through the loss of von Hippel-Lindau (VHL) protein, a classic tumor suppressor gene, best exemplified in renal clear cell carcinoma. Surprisingly, lung AC did not show the expected pattern of general up-regulation of the angiogenesis genes but showed uniformly down-regulation, particularly for the transcription factor HIF-2α. Lung tissue is uniquely situated as the internal interface to atmospheric oxygen (21%), which may preclude the actions of the HIF pathway in lung AC. These results would predict that therapeutic agents targeting angiogenesis may not prove to be effective in treating lung AC. Clinical trials for lung cancer of various anti-angiogenic agents are underway,51 although the interim results have not been encouraging.52
In addition, we examined the arachidonic acid pathway enzymes PGIS and PGES, which are considered anti- and protumorigenic, respectively. The microarray results showed that PGIS and PGES were joint classifiers but showed opposite transcriptional effects with PGIS down-regulated and PGES up-regulated in lung AC tumor tissues compared to adjacent normal tissues. This confirmed others46 and our results on the divergent levels of these two enzymes seen at the protein level. We then showed that any detectable PGIS expression in human NSCLC lung cancer tissue microarray samples was correlated with increased survival of those patients. The analysis of just these two genes (from microarray expression level to patient survival) highlights the impact the general approach of comparing human diseases with their murine models will have in the future.
We asked whether the orthologous gene expression similarities could result from an explanation other than revealing the common pathobiology in AC disease between humans and mice. Both human and murine ACs are derived from alveolar epithelial type II cells, so one possibility was the common cellular origin may be reflected in the microarray data. Surfactant protein C (SP-C) is a highly characterized alveolar epithelial type II-specific protein.53 Examination of the human and mouse microarray datasets indicated similar SP-C gene expression in tumors and adjacent normal tissues, suggesting the tumors do not have a transcriptional bias due to its type II cellular origin. In addition, Clara cell-specific protein CC-10 (uteroglobulin) was underexpressed in tumors. This suggests the strong correlation in gene expression changes between species is not a result of common cellular origin. A second possibility is that orthologous gene expression similarities may be derived from a general proliferation phenotype. This is a much more difficult question to resolve as, in general, proliferation is part of the neoplastic phenotype.38 Although cell-cycle and proliferation genes are highlighted in Table 1, general proliferation markers, such as proliferating cell nuclear antigen and Ki-67, were up-regulated in tumors compared to normal tissue but did not meet the statistical tests that defined our joint classifiers.
In conclusion, orthologous gene expression data demonstrated that many changes in human lung AC disease are accurately replicated in the A/J-urethane murine AC model. More than 85% of these genes had concordant expression levels between adjacent and tumor samples in both species. Importantly, 256 (63%) genes were down-regulated while 95 (23%) were up-regulated in tumors. The chromosomal positions for many of the human genes match known regions of frequent loss in human lung cancer,54 suggesting a significant role for genomic instability in lung cancer etiology in both species. Examination of the full set of classifier genes should help identify common affected pathways and extend the correspondence between down-regulation and genomic loss. We speculate that genomic instability and the relative scarcity of up-regulated targets may explain the limited success thus far in developing therapeutic agents for lung AC. Orthologous gene expression analysis of AC may lead to better therapeutic target identification. This study represents one of the first orthologous comparisons by gene expression analyses of a murine model and its corresponding human disease. Our findings help validate a well-defined, preclinical murine carcinogenesis model as a platform for investigating chemoprevention and therapeutic interventions for human lung AC. In addition, the gene expression studies led to the investigation of PGIS protein expression in a NSCLC tissue microarray. We found that detection of PGIS immunostaining in tissue samples had strong prognostic value in predicting patient survival. The use of orthologous gene expression analysis in other disease systems should prove useful in defining biomarkers and novel molecular targets.
Supplementary Material
Acknowledgments
We thank Dr. Chris Coldren for critically reading this manuscript, Todd Woessner for his expert technical assistance in the University of Colorado Health Sciences Center Microarray Core Facility, and Dr. David Dewitt for the PGIS antibody.
Footnotes
Address reprint requests to Robert S. Stearman, Box C272, Room BB 3B10, 4200 East Ninth Ave., Denver, CO 80262. E-mail: robert.stearman@uchsc.edu.
Supported by the National Institutes of Health (grants R01 CA 03618 and R01 CA 108610 to R.A.N.; R01 HL 72340 to M.W.G.; R01 CA 96133 to A.M.M. and M.W.G.; R01 CA33497 to A.M.M.; P30 CA 46934 to P.A.B.; and P50 CA 58187 to P.A.B., W.A.F., and M.W.G.).
Supplemental material for this article can be found on http://www.amjpathol.org.
References
- Greenlee RT, Hill-Harmon MB, Murray T, Thun M. Cancer statistics, 2001. CA Cancer J Clin. 2001;51:15–36. doi: 10.3322/canjclin.51.1.15. [DOI] [PubMed] [Google Scholar]
- Patel JD, Bach PB, Kris MG. Lung cancer in US women: a contemporary epidemic. JAMA. 2004;291:1763–1768. doi: 10.1001/jama.291.14.1763. [DOI] [PubMed] [Google Scholar]
- Mountain CF. Staging classification of lung cancer. A critical evaluation. Clin Chest Med. 2002;23:103–121. doi: 10.1016/s0272-5231(03)00063-7. [DOI] [PubMed] [Google Scholar]
- Janssen-Heijnen ML, Coebergh JW. The changing epidemiology of lung cancer in Europe. Lung Cancer. 2003;41:245–258. doi: 10.1016/s0169-5002(03)00230-7. [DOI] [PubMed] [Google Scholar]
- Livingood LE. Bull Johns Hopkins Hosp. 1896;7:177–178. [Google Scholar]
- Shimkin MB, Stoner GD. Lung tumors in mice: application to carcinogenesis bioassay. Adv Cancer Res. 1975;21:1–58. doi: 10.1016/s0065-230x(08)60970-7. [DOI] [PubMed] [Google Scholar]
- Malkinson AM. Primary lung tumors in mice: an experimentally manipulable model of human adenocarcinoma. Cancer Res. 1992;52:2670s–2676s. [PubMed] [Google Scholar]
- Devereux TR, Kaplan NL. Use of quantitative trait loci to map murine lung tumor susceptibility genes. Exp Lung Res. 1998;24:407–417. doi: 10.3109/01902149809087377. [DOI] [PubMed] [Google Scholar]
- Malkinson AM. Primary lung tumors in mice as an aid for understanding, preventing, and treating human adenocarcinoma of the lung. Lung Cancer. 2001;32:265–279. doi: 10.1016/s0169-5002(00)00232-4. [DOI] [PubMed] [Google Scholar]
- Tuveson DA, Jacks T. Technologically advanced cancer modeling in mice. Curr Opin Genet Dev. 2002;12:105–110. doi: 10.1016/s0959-437x(01)00272-6. [DOI] [PubMed] [Google Scholar]
- Kayser K, Nwoye JO, Kosjerina Z, Goldmann T, Vollmer E, Kaltner H, Andre S, Gabius HJ. Atypical adenomatous hyperplasia of lung: its incidence and analysis of clinical, glycohistochemical and structural features including newly defined growth regulators and vascularization. Lung Cancer. 2003;42:171–182. doi: 10.1016/s0169-5002(03)00289-7. [DOI] [PubMed] [Google Scholar]
- Malkinson AM. Molecular comparison of human and mouse pulmonary adenocarcinomas. Exp Lung Res. 1998;24:541–555. doi: 10.3109/01902149809087385. [DOI] [PubMed] [Google Scholar]
- Nikitin AY, Alcaraz A, Anver MR, Bronson RT, Cardiff RD, Dixon D, Fraire AE, Gabrielson EW, Gunning WT, Haines DC, Kaufman MH, Linnoila RI, Maronpot RR, Rabson AS, Reddick RL, Rehm S, Rozengurt N, Schuller HM, Shmidt EN, Travis WD, Ward JM, Jacks T. Classification of proliferative pulmonary lesions of the mouse: recommendations of the mouse models of human cancers consortium. Cancer Res. 2004;64:2307–2316. doi: 10.1158/0008-5472.can-03-3376. [DOI] [PubMed] [Google Scholar]
- Meuwissen R, Berns A. Mouse models for human lung cancer. Genes Dev. 2005;19:643–664. doi: 10.1101/gad.1284505. [DOI] [PubMed] [Google Scholar]
- Hemminki K, Lonnstedt I, Vaittinen P, Lichtenstein P. Estimation of genetic and environmental components in colorectal and lung cancer and melanoma. Genet Epidemiol. 2001;20:107–116. doi: 10.1002/1098-2272(200101)20:1<107::AID-GEPI9>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Yang P, Schwartz AG, McAllister AE, Swanson GM, Aston CE. Lung cancer risk in families of nonsmoking probands: heterogeneity by age at diagnosis. Genet Epidemiol. 1999;17:253–273. doi: 10.1002/(SICI)1098-2272(199911)17:4<253::AID-GEPI2>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Mohr S, Leikauf GD, Keith G, Rihn BH. Microarrays as cancer keys: an array of possibilities. J Clin Oncol. 2002;20:3165–3175. doi: 10.1200/JCO.2002.12.073. [DOI] [PubMed] [Google Scholar]
- Meyerson M, Franklin WA, Kelley MJ. Molecular classification and molecular genetics of human lung cancers. Semin Oncol. 2004;31:4–19. doi: 10.1053/j.seminoncol.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, Lizyness ML, Kuick R, Hayasaka S, Taylor JM, Iannettoni MD, Orringer MB, Hanash S. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nat Med. 2002;8:816–824. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, Loda M, Weber G, Mark EJ, Lander ES, Wong W, Johnson BE, Golub TR, Sugarbaker DJ, Meyerson M. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proc Natl Acad Sci USA. 2001;98:13790–13795. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heighway J, Knapp T, Boyce L, Brennand S, Field JK, Betticher DC, Ratschiller D, Gugger M, Donovan M, Lasek A, Rickert P. Expression profiling of primary non-small cell lung cancer for target identification. Oncogene. 2002;21:7749–7763. doi: 10.1038/sj.onc.1205979. [DOI] [PubMed] [Google Scholar]
- Gariboldi M, Spinola M, Milani S, Pignatiello C, Kadota K, Bono H, Hayashizaki Y, Dragani TA, Okazaki Y. Gene expression profile of normal lungs predicts genetic predisposition to lung cancer in mice. Carcinogenesis. 2003;24:1819–1826. doi: 10.1093/carcin/bgg133. [DOI] [PubMed] [Google Scholar]
- Lemon WJ, Bernert H, Sun H, Wang Y, You M. Identification of candidate lung cancer susceptibility genes in mouse using oligonucleotide arrays. J Med Genet. 2002;39:644–655. doi: 10.1136/jmg.39.9.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner AE, Lemon WJ, Devereux TR, Lubet RA, You M. Molecular profiling of mouse lung tumors: association with tumor progression, lung development, and human lung adenocarcinomas. Oncogene. 2004;23:1166–1176. doi: 10.1038/sj.onc.1207234. [DOI] [PubMed] [Google Scholar]
- Borczuk AC, Gorenstein L, Walter KL, Assaad AA, Wang L, Powell CA. Non-small-cell lung cancer molecular signatures recapitulate lung developmental pathways. Am J Pathol. 2003;163:1949–1960. doi: 10.1016/S0002-9440(10)63553-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yauk CL, Berndt ML, Williams A, Douglas GR. Comprehensive comparison of six microarray technologies. Nucleic Acids Res. 2004;32:e124. doi: 10.1093/nar/gnh123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, Jacks T. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- Sweet-Cordero A, Mukherjee S, Subramanian A, You H, Roix JJ, Ladd-Acosta C, Mesirov J, Golub TR, Jacks T. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat Genet. 2005;37:48–55. doi: 10.1038/ng1490. [DOI] [PubMed] [Google Scholar]
- Malkinson AM, Beer DS. Major effect on susceptibility to urethane-induced pulmonary adenoma by a single gene in BALB/cBy mice. J Natl Cancer Inst. 1983;70:931–936. [PubMed] [Google Scholar]
- Liu G, Loraine AE, Shigeta R, Cline M, Cheng J, Valmeekam V, Sun S, Kulp D, Siani-Rose MA. NetAffx: Affymetrix probesets and annotations. Nucleic Acids Res. 2003;31:82–86. doi: 10.1093/nar/gkg121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McShane LM, Radmacher MD, Freidlin B, Yu R, Li MC, Simon R. Methods for assessing reproducibility of clustering patterns observed in analyses of microarray data. Bioinformatics. 2002;18:1462–1469. doi: 10.1093/bioinformatics/18.11.1462. [DOI] [PubMed] [Google Scholar]
- Barash Y, Dehan E, Krupsky M, Franklin W, Geraci M, Friedman N, Kaminski N. Comparative analysis of algorithms for signal quantitation from oligonucleotide microarrays. Bioinformatics. 2004;20:839–846. doi: 10.1093/bioinformatics/btg487. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DR. Regression models and life tables (with discussion). J Royal Stat Soc Series B. 1972;34:187–220. [Google Scholar]
- Zou KH, Tuncali K, Silverman SG. Correlation and simple linear regression. Radiology. 2003;227:617–622. doi: 10.1148/radiol.2273011499. [DOI] [PubMed] [Google Scholar]
- Sweet-Cordero A, Mukherjee S, Subramanian A, You H, Roix JJ, Ladd-Acosta C, Mesirov J, Golub TR, Jacks T. An oncogenic KRAS2 expression signature identified by cross-species gene-expression analysis. Nat Genet. 2005;37:48–55. doi: 10.1038/ng1490. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Altenberg B, Greulich KO. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics. 2004;84:1014–1020. doi: 10.1016/j.ygeno.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Unwin RD, Craven RA, Harnden P, Hanrahan S, Totty N, Knowles M, Eardley I, Selby PJ, Banks RE. Proteomic changes in renal cancer and co-ordinate demonstration of both the glycolytic and mitochondrial aspects of the Warburg effect. Proteomics. 2003;3:1620–1632. doi: 10.1002/pmic.200300464. [DOI] [PubMed] [Google Scholar]
- Kang Y, Ozbun LL, Angdisen J, Moody TW, Prentice M, Diwan BA, Jakowlew SB. Altered expression of G1/S regulatory genes occurs early and frequently in lung carcinogenesis in transforming growth factor-beta1 heterozygous mice. Carcinogenesis. 2002;23:1217–1227. doi: 10.1093/carcin/23.7.1217. [DOI] [PubMed] [Google Scholar]
- Sugita M, Geraci M, Gao B, Powell RL, Hirsch FR, Johnson G, Lapadat R, Gabrielson E, Bremnes R, Bunn PA, Franklin WA. Combined use of oligonucleotide and tissue microarrays identifies cancer/testis antigens as biomarkers in lung carcinoma. Cancer Res. 2002;62:3971–3979. [PubMed] [Google Scholar]
- Santin AD, Zhan F, Bellone S, Palmieri M, Cane S, Bignotti E, Anfossi S, Gokden M, Dunn D, Roman JJ, O’Brien TJ, Tian E, Cannon MJ, Shaughnessy J, Jr, Pecorelli S. Gene expression profiles in primary ovarian serous papillary tumors and normal ovarian epithelium: identification of candidate molecular markers for ovarian cancer diagnosis and therapy. Int J Cancer. 2004;112:14–25. doi: 10.1002/ijc.20408. [DOI] [PubMed] [Google Scholar]
- Bunn PA, Jr, Keith RL. The future of cyclooxygenase-2 inhibitors and other inhibitors of the eicosanoid signal pathway in the prevention and therapy of lung cancer. Clin Lung Cancer. 2002;3:271–277. doi: 10.3816/clc.2002.n.012. [DOI] [PubMed] [Google Scholar]
- Ermert L, Dierkes C, Ermert M. Immunohistochemical expression of cyclooxygenase isoenzymes and downstream enzymes in human lung tumors. Clin Cancer Res. 2003;9:1604–1610. [PubMed] [Google Scholar]
- Keith RL, Miller YE, Hoshikawa Y, Moore MD, Gesell TL, Gao B, Malkinson AM, Golpon HA, Nemenoff RA, Geraci MW. Manipulation of pulmonary prostacyclin synthase expression prevents murine lung cancer. Cancer Res. 2002;62:734–740. [PubMed] [Google Scholar]
- Meyer AM, Dwyer-Nield LD, Hurteau GJ, Keith RL, O’Leary E, You M, Bonventre JV, Nemenoff RA, Malkinson AM. Decreased lung tumorigenesis in mice genetically deficient in cytosolic phospholipase A2. Carcinogenesis. 2004;25:1517–1524. doi: 10.1093/carcin/bgh150. [DOI] [PubMed] [Google Scholar]
- Chung FL, Jiao D, Conaway CC, Smith TJ, Yang CS, Yu MC. Chemopreventive potential of thiol conjugates of isothiocyanates for lung cancer and a urinary biomarker of dietary isothiocyanates. J Cell Biochem Suppl. 1997;27:76–85. [PubMed] [Google Scholar]
- Singh SV, Herman-Antosiewicz A, Singh AV, Lew KL, Srivastava SK, Kamath R, Brown KD, Zhang L, Baskaran R. Sulforaphane-induced G2/M phase cell cycle arrest involves checkpoint kinase 2-mediated phosphorylation of cell division cycle 25C. J Biol Chem. 2004;279:25813–25822. doi: 10.1074/jbc.M313538200. [DOI] [PubMed] [Google Scholar]
- D’Amico TA. Angiogenesis in non-small cell lung cancer. Semin Thorac Cardiovasc Surg. 2004;16:13–18. doi: 10.1053/j.semtcvs.2004.01.003. [DOI] [PubMed] [Google Scholar]
- Blackhall FH, Shepherd FA. Angiogenesis inhibitors in the treatment of small cell and non-small cell lung cancer. Hematol Oncol Clin North Am. 2004;18:1121–1141. doi: 10.1016/j.hoc.2004.06.005. [DOI] [PubMed] [Google Scholar]
- Boggaram V. Regulation of lung surfactant protein gene expression. Front Biosci. 2003;8:751–764. doi: 10.2741/1062. [DOI] [PubMed] [Google Scholar]
- Balsara BR, Testa JR. Chromosomal imbalances in human lung cancer. Oncogene. 2002;21:6877–6883. doi: 10.1038/sj.onc.1205836. [DOI] [PubMed] [Google Scholar]
- Wikman H, Seppanen JK, Sarhadi VK, Kettunen E, Salmenkivi K, Kuosma E, Vainio-Siukola K, Nagy B, Karjalainen A, Sioris T, Salo J, Hollmen J, Knuutila S, Anttila S. Caveolins as tumour markers in lung cancer detected by combined use of cDNA and tissue microarrays. J Pathol. 2004;203:584–593. doi: 10.1002/path.1552. [DOI] [PubMed] [Google Scholar]
- Cho NH, Hong KP, Hong SH, Kang S, Chung KY, Cho SH. MMP expression profiling in recurred stage IB lung cancer. Oncogene. 2004;23:845–851. doi: 10.1038/sj.onc.1207140. [DOI] [PubMed] [Google Scholar]
- Zhang J, Takahashi K, Takahashi F, Shimizu K, Ohshita F, Kameda Y, Maeda K, Nishio K, Fukuchi Y. Differential osteopontin expression in lung cancer. Cancer Lett. 2001;171:215–222. doi: 10.1016/s0304-3835(01)00607-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.