Abstract
Genome-wide analysis of microbial pathogens and molecular pathogenesis processes has become an area of considerable activity in the last 5 years. These studies have been made possible by several advances, including completion of the human genome sequence, publication of genome sequences for many human pathogens, development of microarray technology and high-throughput proteomics, and maturation of bioinformatics. Despite these advances, relatively little effort has been expended in the bacterial pathogenesis arena to develop and use integrated research platforms in a systems biology approach to enhance our understanding of disease processes. This review discusses progress made in exploiting an integrated genome-wide research platform to gain new knowledge about how the human bacterial pathogen group A Streptococcus causes disease. Results of these studies have provided many new avenues for basic pathogenesis research and translational research focused on development of an efficacious human vaccine and novel therapeutics. One goal in summarizing this line of study is to bring exciting new findings to the attention of the investigative pathology community. In addition, we hope the review will stimulate investigators to consider using analogous approaches for analysis of the molecular pathogenesis of other microbes.
Introduction and Overview of Group A Streptococcus (GAS)
Diseases
GAS (also known as Streptococcus pyogenes) is a gram-positive bacterium that causes many types of human disease, including pharyngitis and/or tonsillitis, skin infections (impetigo, erysipelis, and other forms of pyoderma), acute rheumatic fever (ARF), poststreptococcal glomerulonephritis, scarlet fever, severe sepsis, puerperal sepsis (childbed fever), meningitis, necrotizing fasciitis, and toxic shock syndrome.1–3 For reasons that remain obscure, ARF and acute glomerulonephritis sometimes occur as late noninfectious sequelae in certain individuals. Globally, GAS causes immense human morbidity and mortality. For example, annual direct costs associated with pharyngitis in the US are thought to be in the range of 1 billion dollars. An estimated 10,000 to 15,000 invasive infections caused by GAS occur each year in the US, and a recent study highlighted the importance of this pathogen as a cause of childhood bloodstream infections in developing countries.4,5 Antibiotic treatment has essentially eliminated rheumatic fever and subsequent rheumatic heart disease in the US and Europe. However, these crippling forms of heart disease remain common in the developing world and in some populations in developed countries.6–8
The Infection Cycle
Because GAS is a human specialist bacterium, transmission occurs mainly person-to-person. Outbreaks of pharyngitis caused by ingestion of contaminated food have been reported, but these are rare. Study of the early molecular events resulting in GAS colonization of the host have focused predominantly on analysis of extracellular adhesin molecules made by the pathogen, showing that many distinct molecules participate.9 The relative contribution of each GAS adherence molecule seems to depend in part on the target host cell and the experimental system used. Some of the GAS outer surface molecules implicated in adherence include M protein, M protein-like molecules that bind immunoglobulins, lipoteichoic acid, fibronectin- and vitronectin-binding proteins, streptococcal pyrogenic exotoxin B, pullulanase, hyaluronic acid capsule, and C-carbohydrate.2,3,9 Host molecules implicated as receptors for GAS include fibronectin, vitronectin, fibrinogen, collagen, laminin, CD44, CD46, and galactose- and fucose-containing glycoproteins. However, this area of GAS pathobiology has been explored less than the bacterial products that contribute to adherence. A full discussion of these important topics is outside of the realm of this review.
Although adherence to host cells is a crucial early event in the infection cycle, the human oropharynx is bathed in saliva, which is the first substance with which GAS interacts. Saliva contains many components of the innate and acquired immune systems and plays a pivotal role in defending the oropharynx against invading microorganisms.10 Hence, infection and subsequent transmission to new hosts depends on the ability of GAS to survive and proliferate in saliva (see below).
After initial host interaction, GAS may either remain at the site of colonization in a relatively quiescent state or proliferate to cause local disease such as pharyngitis or pyoderma. In a few individuals, the pathogen avoids clearance or control by host immune mechanisms and initiates a more invasive infection characterized by extensive local tissue destruction and spread and/or intravascular entry and dissemination. The tissue destruction in many patients can be fulminant, resulting in complete destruction of the extracellular matrix and death. The exact molecular mechanism(s) responsible for tissue destruction are unknown, but production of substances that directly degrade host molecules or subvert normal host defenses is known to participate.11,12
Complex Regulatory Networks Participate in Virulence
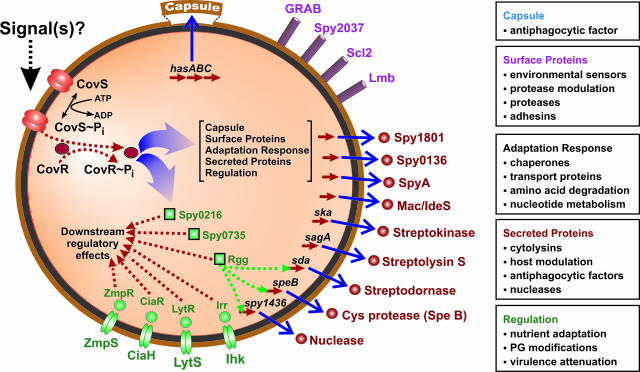
All bacterial pathogens, including GAS, have molecular mechanisms dedicated to coordinately regulating expression of virulence factors in response to environmental stimuli (Figure 1).13 This coordinate regulation provides a strategy for bacteria to adapt very rapidly to changes in their extracellular milieu and thereby enhance survival. A key feature of many of these networks is control by a two-protein interaction cascade involving a sensor and response protein, commonly referred to as a two-component gene regulatory system (TCS). These TCSs control expression of unlinked genes encoding toxins, adhesins, and other virulence-associated molecules that promote bacterial survival and pathogenesis.13 As an initial activation step, a transmembrane sensor protein, usually a histidine kinase, detects extracellular stimuli and undergoes autophosphorylation. The histidine kinase subsequently phosphorylates a response regulator, which in turn acts as a transcription factor for specific target genes. GAS has 12 conserved TCSs, but only 4 of these 12 (CovR-CovS, FasBCAX, Ihk-Irr, and Spy0874/0875, also known as SptR/SptS) have been well studied (see below).14 In addition, a GAS gene called mga (multiple gene activator) also serves as a crucial gene regulator.14 The mga gene encodes a trans-acting positive regulatory protein (Mga) that activates transcription of several genes encoding proven or putative GAS virulence factors, including M protein, M-like proteins, the streptococcal inhibitor of complement (C5a peptidase), and a lipoprotein lipase (opacity factor). Interestingly, although sequence homologies suggest that Mga is the transducer protein of a TCS, the putative sensor protein that responds to environment perturbations and subsequently activates mga transcription has not been identified. Several other important regulatory systems have been described in GAS, but space constraints do not permit us to review their contributions to host-pathogen interactions. We refer the interested reader to a recent review of this topic.14
Figure 1.
Gene-regulatory networks mediated by the CovR-CovS two-component system. Adapted from Graham and colleagues.52
Rapidly Emerging Discipline of Systems Biology Research
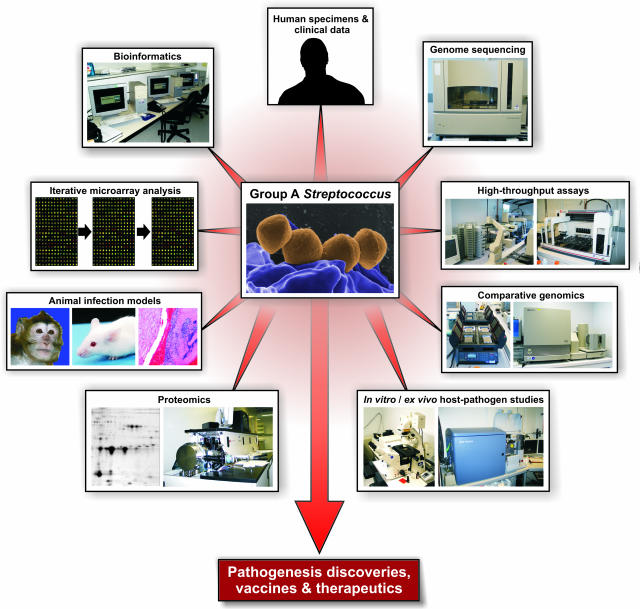
An investigative discipline now referred to as systems biology research has emerged rapidly in recent years.15,16 One publication15 defines systems biology as “… a scientific discipline that endeavors to quantify all of the molecular elements of a biological system to assess their interactions and to integrate that information into graphical network models that serve as predictive hypotheses to explain emergent behaviors.” Inherent in this definition is that genome-wide strategies will be used to generate very large multiparameter datasets, including but not limited to comparative genome sequences, transcriptome, proteome, metabolome, and so forth. The datasets are systematically integrated and interrogated to achieve the particular goal of interest. Much of the effort thus far has focused on cancer biology or analysis of model organisms such as yeast, with relatively little attention given to problems in infectious diseases research. Analysis of the molecular pathogenesis of infectious disease by a systems biology approach is especially complicated, in part because pathogens are highly diverse genetically, multiple phases of the infectious process can be prolonged and anatomically distinct (eg, skin, tissue, blood, cerebrospinal fluid, individual and multiple organs), and host immune processes are multiphasic. Thus, in the case of bacterial pathogenesis, there exists concern about parameters such as the genotype of the infecting strain and host, phase of infection, immune status, and treatment modality, to name a few variables. Clearly a systems biology analysis of this type of problem is quite complicated. However, it is reasonable to suspect that steps toward this goal will yield new insights about host-pathogen interaction at each juncture. Currently, we think of systems biology research of infectious agents with a holistic view of integrating molecular events (broadly defined) occurring in the pathogen with those occurring in the host. Initial steps toward this type of analysis in our laboratories involve a highly integrated, discovery-enabling infrastructure coupled with animal infection models and human patient studies (Figure 2).
Figure 2.
An integrated systems biology approach for investigating bacterial pathogenesis.
The systems biology approach toward understanding GAS-host interactions and pathogenesis has included sequencing the genome of multiple GAS strains. The wealth of resulting genome data were subsequently used to produce expression and DNA-DNA microarrays, from which many dozens of target genes of interest were identified. The role of specific GAS target molecules in pathogenesis was then examined in animal models and ex vivo/in vitro host-pathogen assays and was coupled in multiple instances with a repository of human clinical data (Figure 2). Proteomics also contributed significantly to the identification of GAS virulence molecules, although the topic is not discussed extensively in this review. This integrated methodology has led to a significant number of new discoveries in GAS pathogenesis, many of which are generally applicable to mechanisms of virulence in other human pathogens.
GAS Genome Sequences: an Embarrassment of Riches
Introduction and Background
We believe a systems biology approach to analysis of host-pathogen interaction must start with a reasonably complete description of the components of the system. From the pathogen side of the equation, an excellent starting point is the genome sequence of strains of interest. The first bacterial genome sequence was reported in 1995,17 and now more than 150 bacterial genome sequences have been published, including at least one strain of every major human pathogen (http:/www.tigr.org/tigr-scripts/CMR2/CMRHomePage.spl and http:/www.genomesonline.org/). Until relatively recently, it was common to have only one genome sequence available for a pathogenic bacterial species. However, it has long been known that the amount of genetic diversity present in most bacterial species far exceeds that for eukaryotic organisms. That knowledge, coupled with well-recognized differences in clinically relevant behavior between strains of individual bacterial species, relatively small genome size, decreasing DNA sequencing costs, therapeutics research, and concerns about illegitimate release of certain pathogens, fueled the effort to sequence the genome of multiple isolates of many important human pathogens. Thus, more than one genome sequence is now publicly available for many bacterial pathogens, and for several species, including GAS, more than three genomes have been sequenced. The availability of multiple genome sequences has been a tremendous asset to pathogenesis research.
For more than 50 years, GAS strains have been classified based on serological diversity in M protein, a major surface antigen and virulence factor.2 However, serological typing of strains has been replaced by sequencing of the hypervariable part of the emm gene encoding M protein. More than 125 emm types are recognized, and this allelic diversity has been useful for categorizing strains for epidemiological studies.4,18–21 Although no one M serotype or emm type is solely responsible for any single type of GAS infection, strains expressing certain M types have been repeatedly associated with specific diseases. For example, serotype M1 and M3 strains are the leading cause of invasive infections in many western countries, and M18 strains have been commonly associated with rheumatic fever.19–21 These epidemiological observations have led to the concept that certain serotypes are differentially capable of causing specific clinical diseases. A corollary of this idea is that genetic differences between GAS strains account for some of these nonrandom disease associations. Importantly, this line of thinking has stimulated much of the interest in sequencing the genome of GAS strains with distinct disease associations.
Complete genome sequences are available for eight GAS strains (serotypes M1, M3, M5, M6, M18, and M28) commonly causing pharyngitis and invasive disease (http://www.sanger.ac.uk/Projects/S_pyogenes/).22–28 The availability of genome sequences of M protein serotypes causing distinct diseases has yielded a tremendous amount of new information important for a systems biology approach to pathogenesis research.
General Concepts Revealed by Comparative Genomics
The eight genome sequences have provided extensive data regarding the breadth of strain variation within and between serotypes, molecular correlates of disease specificity, and many unexpected leads for pathogenesis research. All strains studied thus far have a genome size of ∼1.8 to 1.9 Mb, five or six highly conserved rRNA operons, and a core group of proven and putative virulence genes.29 Approximately 90% of gene content is shared among strains and constitutes the core GAS genome. Importantly, the genomes of all strains sequenced are polylysogenic, that is, they contain multiple prophages or prophage-like elements. (Prophages are bacterial viruses that are integrated into the chromosome.) Virtually all GAS prophages encode one or two proven or putative extracellular virulence factors such as pyrogenic toxin superantigens (PTSAgs), DNAses, novel phospholipase A2 (SlaA), efflux pump responsible for resistance to macrolide antibiotics, and a novel cell wall-anchored protein hypothesized to be an adhesin.23–29 Many of these prophage-associated virulence factor genes were unknown before their discovery by GAS genome sequencing projects. Additional specific discoveries revealed by the genome sequencing projects are described below.
Genome Sequence and Population Genomics of ARF-Associated GAS Serotype M18 Strains
ARF is the leading cause of global preventable pediatric heart disease. This disease is a devastating sequelae of GAS infection that usually occurs several weeks after an antecedent throat infection. The molecular pathogenesis of ARF and subsequent rheumatic heart disease has not been fully delineated, but autoimmune mechanisms likely play a prominent role.30 It is believed that only a relatively small number of the more than 125 M protein serotypes have rheumatogenic potential in humans who are predisposed, presumably for host genetic reasons. For example, serotype M18 GAS strains have been repeatedly implicated in ARF outbreaks in the US. As a first step toward gaining new insight into ARF pathogenesis, we sequenced the genome of a M18 strain cultured from a patient with ARF.24 In addition, we used DNA microarray analysis to compare the genomes of 36 M18 strains, including organisms cultured from patients with ARF and pharyngitis during two epidemics of ARF in Salt Lake City, UT.24,31
Many genes are unique to serotype M18 strains. Importantly, this serotype has genes encoding unique secreted proteins involved in human-GAS interactions, including two previously uncharacterized novel PTSAgs that are encoded by a prophage.32 Whole-genome DNA-DNA microarray analysis of 36 serotype M18 strains, collected from diverse localities throughout 6 decades, revealed that most regions of variation corresponded to prophages or prophage-like elements. Two epidemics of ARF occurring 12 years apart in Salt Lake City, UT, were caused by serotype M18 strains that were genetically identical, or nearly so. The analysis provided a critical foundation for accelerated research into ARF pathogenesis and a molecular framework to study the plasticity of GAS genomes.
Insight into the Emergence of Drug-Resistant GAS Strains: an Outbreak of Pharyngitis Caused by a Serotype M6 Macrolide-Resistant Clone
GAS infections are commonly treated with penicillin or a related β-lactam antibiotic. Strangely, despite decades of intense selective pressure, all GAS strains remain very susceptible to this class of antibiotics. Erythromycin and related macrolide antibiotics sometimes are used as an alternative treatment for GAS pharyngitis. In contrast to β-lactams, resistance of GAS to macrolide antibiotics was described as early as the late 1950s and has increased markedly worldwide in the last decade.33
Martin and colleagues34 reported an outbreak of pharyngitis caused by erythromycin-resistant serotype M6 GAS strains among schoolchildren in Pittsburgh, PA. This event was of particular concern because within a few months the frequency of macrolide-resistant GAS increased rapidly, and drug-resistant strains spread widely to surrounding communities. Molecular studies implicated serotype M6 strains of a single clone, and drug resistance was due to the presence of the mefA gene encoding a macrolide efflux pump.34
Inasmuch as serotype M6 strains are one of the more common causes of pharyngitis and invasive infections4,19–21,26,35 and there was considerable conflicting information in the literature about the molecular mechanism of acquisition and transfer of the mefA gene in GAS, we chose to sequence the genome of a genetically representative strain.26,36 Interestingly, we found that the mefA gene in the Pittsburgh clone was encoded by an unusual 58.8-kb foreign genetic element with characteristics of both a transposon and a prophage. It is likely that acquisition of the mefA element by lateral (horizontal) gene transfer and subsequent expansion of the drug-resistant clone were key contributors to the outbreak.
The chimeric element also encoded a novel extracellular protein that is expressed during GAS-human interaction.26,36 The protein is related to molecules that facilitate adhesion to host cells in other microbial pathogens, suggesting that it assists the organism in colonizing the posterior pharynx. Taken together, sequencing of this genome and the studies it facilitated shed new light on the important mechanism of increased antimicrobial resistance in GAS.
New Understanding about the Pathogenesis of Puerperal Sepsis Revealed by the Genome Sequence of a Serotype M28 GAS Strain
Puerperal sepsis, also known as childbed fever, killed up to one in every six mothers who delivered babies in some European hospitals in the 19th century.37 Clinical investigations conducted by Semmelweis38 in the 1840s determined that puerperal sepsis was transmitted to pregnant women in labor by attending doctors. Semmelweis’ studies eventually led to widespread implementation of infection control practices such as routine hand washing, thereby changing medical history. Puerperal sepsis is frequently caused by GAS, and serotype M28 strains are significantly overrepresented among puerperal sepsis and neonatal GAS infections.20,39,40
The molecular mechanisms responsible for the enrichment of serotype M28 strains in these infections are not known, indicative of a general lack of understanding of the pathogenic processes underlying intraspecies disease specificity. We hypothesized that serotype M28 strains possess genes encoding novel extracellular proteins that contribute to their enrichment in puerperal sepsis. To begin testing this hypothesis, we sequenced the genome of a genetically representative serotype M28 strain from an invasive infection.27 Genes were discovered that encoded a novel array of prophage virulence factors, cell-surface proteins, and other molecules likely to contribute to host-pathogen interactions. However, the biggest surprise was the finding that genes for seven secreted proteins are encoded by a 37.4-kb foreign DNA element that is shared with group B Streptococcus, the most abundant cause of neonatal sepsis. These proteins are produced in human infections, as assessed by analysis of sera obtained from patients with GAS disease.27 Our findings suggest that acquisition of genes encoding novel extracellular proteins has helped to create a disease specialist clone of GAS, thereby broadening the ecological niche of this pathogen and modifying infection character.
Molecular Mechanisms Underlying the Emergence of Severe GAS Infections: the Key Role of Lateral Gene Transfer (LGT)
The molecular events contributing to the evolution and emergence of unusually virulent pathogens are poorly understood.41 Bacteria can evolve slowly by accumulation of point mutations or rapidly in a quantum leap by acquisition of new genetic material through lateral (also known as horizontal) gene transfer (LGT).42 New combinations of genes or allelic variants can increase the fitness of a bacterium by conferring unique properties such as immune evasion, antimicrobial agent resistance, and the ability to colonize a previously unexploitable ecological niche. Three well-described processes known to promote LGT are transduction, conjugation, and transformation.
In the mid-1980s, the frequency and severity of invasive infections caused by serotype M1 GAS precipitously increased in the US and many other countries.1,43–46 Although serotype M1 GAS are usually the predominant serotype recovered from invasive disease episodes, the increase in disease frequency and severity (especially necrotizing fasciitis) caused by M1 strains was noteworthy. However, no information was available about the molecular processes responsible for this phenotypic shift at the time of the episodes.
To better understand the molecular events involved in the origin of new pathogenic bacteria, we studied the evolution and development of a virulent clone of serotype M1 GAS that emerged in the mid-1980s.28,46 This study was greatly facilitated by the availability of the genome sequence of one serotype M1 GAS organism (strain SF370).22 Inasmuch as strain SF370 is genetically distinct from the great majority of strains responsible for contemporary infections caused by M1 strains, and is less virulent, we hypothesized that acquisition of novel genetic elements had created an unusually successful clone of serotype M1.47,48 To test this hypothesis, we sequenced the genome of a strain that is genetically representative of organisms responsible for contemporary episodes of M1 infections. Genome sequencing, DNA-DNA microarray analysis, and high-throughput single nucleotide polymorphism analysis revealed that this clone evolved from an ancestral strain through a complex series of LGT episodes. These events included acquisition of prophages encoding SpeA and extracellular DNase virulence factors. Another LGT event involved reciprocal recombination of a 36-kb chromosomal segment encoding the extracellular toxins NAD+-glycohydrolase (NADase) and streptolysin O. Virtual identity in the 36-kb region present in contemporary M1 isolates and serotype M12 isolates suggested a model in which a serotype M12 strain served as the donor of this region. The most likely mechanism involved generalized transduction, a process in which a bacteriophage inadvertently packages a segment of GAS chromosomal DNA, rather than the phage genome, into its phage head. This LGT event was associated with significantly increased streptolysin O and NADase production. These findings have implications for understanding the molecular events underlying the emergence of other bacterial clones that are unusually virulent or have other characteristic disease phenotypes; they also stress the importance of using highly integrated genome-wide analyses to study clone emergence and abrupt changes in infection character.
Genome-Wide Dissection of the Molecular Events Underlying GAS Epidemics: the Serotype M3 Model
Molecular factors that directly contribute to bacterial epidemics remain elusive. GAS is an ideal pathogen to investigate this topic because the organism undergoes rapid changes in disease frequency and severity caused by strains expressing certain M protein serotypes.35 We conducted a molecular genetic dissection of two epidemic waves by using multipronged approach that involved sequencing the genome of a relevant strain, performing functional analysis of novel extracellular proteins encoded by the genome, and then using comparative genomic analysis of the strains.49
To initiate this line of investigation, the genome of a strain isolated from a patient with invasive disease was sequenced.23 As found for other GAS strains, prophage-like elements accounted for the great majority of variation in gene content relative to other GAS genomes. The strain had phage genes that encoded proven and putative virulence factors, including PTSAgs, and a novel phospholipase A2 virulence factor (designated SlaA) with similarity to snake venom toxins.23,50 Importantly, we discovered that serotype M3 strains with the prophage-encoded slaA gene increased dramatically in frequency late in the 20th century, commensurate with the rise in invasive infections caused by serotype M3 strains. The study showed that phage-mediated recombination involving virulence factor genes played a critical role in the emergence of a new, unusually virulent clone of GAS. Japanese investigators subsequently confirmed our findings.25
The availability of the genome sequence of a serotype M3 strain opened the door to a detailed analysis of molecular factors contributing to disease epidemics. We hypothesized that analysis of a comprehensive population-based strain sample recovered from patients with invasive infections together with genome-wide investigative methods would provide new insight into this fundamental infectious disease problem. To test the hypothesis, 255 contemporary M3 strains obtained from an 11-year population-based surveillance study of invasive disease in Ontario, Canada, were analyzed.49 Genetic diversity in these strains was investigated by several state-of-the-art methods, including DNA-DNA microarray and whole-genome PCR tiling. The core genome of all strains was identical in gene content. That is, variation in prophage content accounted for all differences in gene content between strains.
Sequence analysis of the emm gene and single nucleotide polymorphism analysis were used to extend analysis of genetic relationships among the 255 M3 strains. The strains were pauciclonal, with a limited number of subclone groups identified. Interestingly, statistically significant associations were discovered between prophage genotypes and GAS disease types. Hence, by integrating genome-wide molecular genetic analysis techniques with detailed epidemiological and clinical information, temporal changes in the gene content of M3 subclones were linked to disease phenotype. Collectively, acquisition or loss of prophages, allelic variation in chromosomal genes, expansion of subclone populations, and introduction of new subclone variants were shown to contribute to peaks of infection and different infection types.49
One finding from this study was particularly noteworthy and provided key insight into a potential mechanism accounting for clone emergence. Molecular genetic analysis, combined with immunological studies, implicated a 4-amino acid duplication in the extreme N-terminus of M protein as a factor contributing to rapid clone emergence. Linear epitope mapping with antisera raised against M protein-based synthetic peptides revealed differential reactivity to the peptides representing the extreme N-terminus of M protein. In addition, the difference in M protein sequence resulted in significantly decreased ability of human polymorphonuclear leukocytes (PMNs) to phagocytose the GAS strain that expressed the variant M3 protein. Taken together, the findings indicated that the 4-amino acid duplication conferred altered immune recognition to the M3 protein. Thus, it is possible that GAS strains with the variant M3 sequence rose to prominence as a consequence of host immunological selective pressure.
Expression Microarray Analysis of the GAS Transcriptome
Expression microarray (transcriptome) analysis has dramatically accelerated the pace of discovery in every area of biomedical research, and microbial pathogenesis is no exception. Several expression microarray studies have examined GAS, and extensive new information has been obtained about the nature and extent of changes in the pathogen’s transcriptome in response to various perturbations.28,51–56 This global approach to understanding pathogenesis was facilitated by the information obtained from GAS genome sequencing projects, as described above. Initial studies used conventional spotted microarrays and focused on in vitro analysis, but more recently a custom Affymetrix high-density oligonucleotide array has been used for ex vivo and in vivo studies.55,56 The result has been substantial new understanding about how GAS reacts to its environment and the mammalian host. Here we summarize key findings from several of these investigations.
Virulence Control and the covR-covS Gene Regulatory System
The CovR-CovS (Cov, control of virulence; also known as CsrR-CsrS) TCS has been investigated extensively.14,52,55,57,58 Genetic inactivation of this TCS produces a mutant strain that is hypervirulent in mice, has a mucoid colony phenotype associated with overexpression of hyaluronic acid capsule, and has increased resistance to in vitro killing by human PMNs. In addition, increased transcripts for proteins likely to promote survival and virulence, including streptokinase, streptolysin S, and a DNase, were identified in the isogenic mutant strain by conventional Northern blot analysis. However, expression microarray analysis afforded assessment of the full impact of the CovR-CovS TCS on GAS gene expression.52 In vitro, this TCS influences (directly or indirectly) transcription of 15% of all GAS genes, including many that encode surface and secreted proteins mediating host-pathogen interactions. CovR also plays a central role in gene regulatory networks by influencing expression of genes encoding transcriptional regulators, such as other TCSs. The key findings of the microarray analysis were confirmed in a mouse soft-tissue model of invasive infection and by ex vivo analysis of GAS growth in human blood (see below).52,55
Molecular Interactions of GAS with Human Saliva
The ability of microbes to adapt successfully to specific host environments by differential gene expression is essential for survival and causing human disease. Although many human bacteria persist for extended periods in distinct anatomical niches, relatively little is known about the molecular mechanisms that facilitate such persistence. The human oropharynx is the predominant site of GAS infections. Although saliva is a vital component of host defense in the human oropharynx, little is known about how GAS responds to interaction with this substance. To increase our understanding of this interaction, we investigated the growth of GAS in saliva obtained from healthy volunteers.59 We found that GAS responds in a complex manner to growth in human saliva. Importantly, two abundantly secreted proteins made by GAS (Sic, streptococcal inhibitor of complement, and SpeB, a potent cysteine protease) were discovered to contribute to prolonged survival in saliva.59
To gain enhanced understanding of the molecular mechanisms used by GAS to interact with saliva, we recently analyzed the transcriptome of a contemporary serotype M1 GAS strain (sequenced strains, see above) during growth in this fluid.60 An iterative expression microarray method was used. This powerful strategy involves expression array analysis of a wild-type organism to generate an initial dataset. The information that is revealed is then exploited to identify genes potentially critical to pathogen-host interaction under the condition of interest. Subsequently, a target gene of interest is genetically inactivated, and a second (iterative) expression microarray experiment is performed to compare the transcriptome of the wild-type and mutant organisms grown under identical conditions. Thus, the datasets obtained from these experiments rapidly yield information about complex gene regulatory networks operating in distinct settings. With respect to interaction of GAS with saliva, one example illustrates the power of this strategy. Expression microarray analysis of wild-type GAS revealed that a TCS (designated Spy0874/0875, also known as SptR/SptS) of unknown function had very high transcript levels in human saliva, suggesting a contribution to growth and persistence in this fluid. Consistent with this idea, an isogenic nonpolar mutant strain (ΔSptR) was dramatically less able to persist in human saliva compared to the parental strain. Transcriptome analysis revealed that compared to the parental strain, the isogenic mutant strain had decreased transcript levels of multiple genes encoding several well-known virulence factors and proteins involved in complex carbohydrate acquisition and utilization pathways. Thus, iterative microarray analysis identified a genetic program that significantly increased persistence in saliva of a major human pathogen, thereby providing new insight into how bacteria survive in the host. Taken together, the data suggest that the molecular processes that enhance human colonization and survival in the upper respiratory tract are underway well before the pathogen contacts the epithelial cell surface. From the standpoint of translational research, the data suggest that using novel inhibitors to target this particular two-component system would be fruitful.
Transcriptome Dynamics in the Upper Respiratory Tract
To what extent, if any, do molecular events that occur during in vitro growth reflect in vivo processes? This important question has long plagued investigators in all areas of biomedical research, including that of microbial pathogenesis. Identification of the genetic events that contribute to host-pathogen interactions is important not only for understanding the natural history of infectious diseases but also for developing new therapeutics. Central to this goal in recent years have been transcriptome studies conducted on pathogens, although most have focused on specific end points or disease phases rather than analysis of the entire time course of infection. To gain a more complete understanding of how bacterial gene expression in vivo changes throughout time, the transcriptome of our contemporary serotype M1 GAS strain was analyzed during an 86-day infection protocol in 20 cynomolgus macaques with experimental pharyngitis.56 A key discovery was that the temporal pattern of GAS gene expression in pharyngitis is intimately linked to three distinct phases of infection. For example, successful colonization and severe inflammation were significantly correlated with superantigen gene expression. Similarly, differential expression of TCS regulators covR and SptR was significantly associated with GAS colony-forming units, inflammation, and phases of disease. It is important to note that SptR was also identified as a key gene regulator by the ex vivo saliva transcriptome studies described above. This nonhuman primate study confirmed the in vivo relevance of many of the in vitro observations described recently.60 The study also provided extensive new knowledge of the gene expression patterns characterizing each clinical phase of pathogen-host interaction. The detailed understanding of GAS gene transcript patterns throughout time in an infected host provides new avenues for targeted investigation of proven and putative virulence factors and genes of unknown function and will assist therapeutics research. An analogous study of the changes in the monkey transcriptome occurring throughout the 86 days of this experiment is currently underway (K. Virtaneva and J. M. Musser, unpublished data).
GAS Transcriptome Dynamics during Growth in Human Blood
During the transition from a throat or superficial skin infection to an invasive infection, GAS must adapt to changing environments and host factors. Recently, we used transcript profiling and functional analyses to investigate the transcriptome of a serotype M1 GAS strain in human blood ex vivo.55 This was a particularly pertinent investigation because GAS sepsis is a devastating infection with high morbidity and mortality rates. Hence, insight into molecular events transpiring in blood is crucial. Using an Affymetrix custom microarray, we discovered that global changes in GAS gene expression occurred rapidly in response to human blood exposure. Increased transcription was identified for many genes that are likely to enhance bacterial survival, including those encoding PTSAgs and host-evasion proteins. For example, on blood exposure, expression increases for GAS genes encoding proteins that interact with host cell surfaces (adhesins, such as M protein; collagen-binding proteins; and capsule) and that contributes to the evasion of the host innate defenses (Sic, Mac, and SpeA). The microarray analysis confirmed and extended evidence that the CovR-CovS TCS enhances bacterial survival during disseminated host infections.57,58 This study provided crucial insights into strategies used by a bacterial pathogen to thwart host defenses and to survive in human blood and suggested new vaccine and therapeutic strategies.
Molecular Mechanisms Used by GAS to Circumvent Destruction by the Innate Immune System
Identification of a GAS Genetic Program that Protects against Killing by Human PMNs: the Ihk-Irr Two-Component System
Most microbes are destroyed by PMN-derived reactive oxygen species and/or microbicidal granule products, which are released into pathogen-containing phagosomes. However, GAS is relatively resistant to killing by these anti-bacterial components.53,54,61–64 Although progress has been made, the molecular basis for this resistance is incompletely understood. Two recent transcriptome-based studies shed new light on the gene regulation mechanisms used by GAS to avoid destruction by human PMNs.
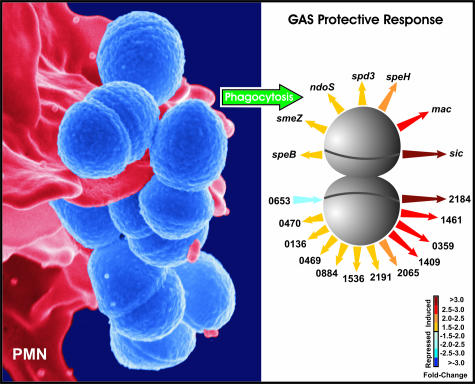
To begin analysis of the gene regulatory pathways used by GAS to circumvent the human innate immune system, we conducted a comprehensive analysis of changes in GAS gene expression during phagocytic interaction with human PMNs.53,54 Genes related to prophage, oxidative stress, cell-wall biosynthesis, virulence, and novel extracellular secreted proteins of unknown function were up-regulated during phagocytic interaction (Figure 3). Importantly, an essential role in immune evasion was discovered for the Ihk-Irr TCS using an isogenic mutant strain in which this gene regulatory system was inactivated. That is, the mutant strain was killed far more rapidly by human PMNs than the wild-type parental strain. Differences in survival were attributed to increased susceptibility of the irr-mutant strain to microbicidal components present within the PMN phagosome.53,54 Thus, the Ihk-Irr TCS controls expression of genes that facilitate evasion of PMN-mediated bacterial killing, thereby assisting GAS survival and pathogenesis (Figure 4). Consistent with these findings, the irr gene is highly expressed in human GAS pharyngitis.65
Figure 3.
GAS-PMN interaction. Left: Scanning electron micrograph of serotype M1 GAS (blue) during phagocytic interaction with a human PMN (red). Right: Induction of genes encoding GAS-secreted molecules during phagocytic interaction. The speB, smeZ, ndoS, spd3, speH, mac, and sic genes encode proven or putative GAS virulence factors. Numbers refer to GAS open reading frames encoding proteins of unknown or poorly understood function. Adapted from Voyich and colleagues53 and Kobayashi and colleagues.64
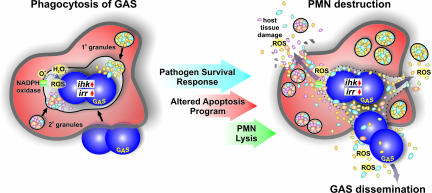
Figure 4.
GAS survival response and altered innate host defense. See text for details. Adapted from Voyich and colleagues.54 (Copyright 2004, The American Association of Immunologists, Inc.).
A subsequent study investigated the molecular basis for the inability of the irr-mutant strain to withstand destruction by neutrophils. Iterative expression microarray analysis revealed that Ihk-Irr significantly influenced 20% of all transcripts in the GAS genome. Importantly, genes implicated in cell wall metabolism, transcription, oxidative stress, and virulence were significantly down-regulated in the isogenic irr-mutant strain.54 Compared with the wild-type strain, the irr-mutant strain had significantly decreased resistance to killing by H2O2 and neutrophil primary granule components. Moreover, expression of ihk and irr were increased significantly in wild-type GAS strains exposed to H2O2 and PMN primary granules. Collectively, the data indicated that Ihk-Irr functions as a sensor for components of human innate host defense. Thus, these iterative expression microarray studies helped to identify a global pathogen-protective transcriptome that is engaged during interaction with the innate immune system.
GAS Modulates Human PMN Gene Expression and Function
Inasmuch as PMNs produce an array of cytotoxic molecules that can cause host tissue destruction, neutrophils typically undergo apoptosis after phagocytosis as a means to facilitate healthy turnover of effete or spent cells.64,66–68 This process is linked to the production of NADPH oxidase-derived reactive oxygen species.69–72 Using human oligonucleotide microarrays throughout the course of several independent studies, Kobayashi and colleagues64,67,71 discovered an apoptosis differentiation program in human PMNs that likely facilitates the resolution of infection. The apoptosis differentiation program is comprised of a core set of genes commonly induced or repressed after uptake of bacteria.71
During phagocytic interaction with human neutrophils, GAS modulates expression of the core set of PMN genes comprising the common apoptosis program.64 However, the pathogen also triggers changes in gene expression not observed with other bacteria tested, including repression of genes involved in specific cell survival pathways.64 Changes in the expression of these genes significantly parallels accelerated PMN apoptosis and host cell lysis (Figure 4). Based on those studies, we proposed that there are two fundamental outcomes of the interaction of neutrophils with bacteria.64,68 On one hand, neutrophils are activated, bacteria are killed, and there is induction of apoptosis leading to the resolution of infection. Alternatively, bacterial pathogens such as GAS circumvent killing by neutrophils and thus alter the apoptosis program to survive and cause disease. The ability of GAS to survive and cause eventual PMN lysis is in part regulated by the Ihk-Irr TCS as described above (Figure 4).
Genome-Wide Analysis and Therapeutics Research: Vaccines
In recent years, a paradigm shift in bacterial vaccine research has occurred due to the development of a strategy now referred to as “reverse vaccinology.”73–76 The basic concept behind the reverse vaccinology approach is to use the complete genome sequence of a pathogen to identify a large number of novel protective antigens, express each candidate in recombinant form, and test each protein in one or more assays for capacity to confer immunological protection. Potential candidates are initially identified by computer analysis of the bacterial genome sequence with algorithms that predict secreted or surface-exposed proteins. Testing of these antigens requires high-throughput cloning and expression of recombinant antigens as well as the use of in vitro and/or in vivo assays of protective capacity. Pioneering studies were performed using Neisseria meningitidis, an important cause of sepsis and meningitis worldwide.73,74 The complete genome sequence of a serogroup B strain of this pathogen was determined, 344 candidate proteins were overexpressed in Escherichia coli, and 28 novel proteins were ultimately identified as protective antigens.74 Subsequently, a similar strategy, with some improvements, was used to identify novel protective antigens against group B Streptococcus76 and GAS.77–79 The GAS efforts have involved an integrated systems biology approach that uses diverse techniques such as proteome, immunome, and in vitro and in vivo transcriptome analyses; comparative genomics and bioinformatics analysis; and experimental infection models in mice and monkeys.56,65,77–79 Importantly, this integrated strategy has enabled far faster identification of conserved candidate antigens than otherwise possible by conventional vaccine research methods.
Summary
The goal of this review was to summarize some of the key recent advances that have been made in GAS pathogenesis research by deploying a multipronged genome-wide systems biology strategy. The progress made thus far has provided many new avenues for basic pathogenesis research and translational research that is focused on developing novel therapeutics. Although many gaps in our knowledge of GAS-human interactions remain to be filled, we hope that our review stimulates other investigators to consider using analogous integrated strategies for studying the molecular basis of microbial pathogenesis.
Acknowledgments
We thank all members of our laboratories who provided thoughtful suggestions to improve the manuscript.
Footnotes
Address reprint requests to James M. Musser, M.D., Ph.D., Center for Molecular and Translational Human Infectious Diseases Research, The Methodist Hospital Research Institute, and Department of Pathology, The Methodist Hospital, 6565 Fannin St., F816, Houston, TX 77030. E-mail: jmmusser@tmh.tmc.edu.
Supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health (grant UO1 AI60595 and the Intramural Research Program).
References
- Musser JM, Krause RM. The revival of group A streptococcal diseases, with a commentary on staphylococcal toxic shock syndrome. Krause RM, editor. San Diego: Academic Press,; Emerging Infections. 1998:pp 185–218. [Google Scholar]
- Cunningham M. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisno AL, Brito MO, Collins CM. Molecular basis of group A streptococcal virulence. Lancet Infect Dis. 2003;3:191–200. doi: 10.1016/s1473-3099(03)00576-0. [DOI] [PubMed] [Google Scholar]
- O’Brien KL, Beall B, Barrett NL, Cieslak PR, Reingold A, Farley MM, Danila R, Zell ER, Facklam R, Schwartz B, Schuchat A. Epidemiology of invasive group A Streptococcus disease in the United States, 1995–1999. Clin Infect Dis. 2002;35:268–276. doi: 10.1086/341409. [DOI] [PubMed] [Google Scholar]
- Berkley JA, Lowe BS, Mwangi I, Williams T, Bauni E, Mwarumba S, Ngetsa C, Slack MP, Njenga S, Hart CA, Maitland K, English M, Marsh K, Scott JA. Bacteremia among children admitted to a rural hospital in Kenya. N Engl J Med. 2005;352:39–47. doi: 10.1056/NEJMoa040275. [DOI] [PubMed] [Google Scholar]
- Carapetis JR, Currie BJ, Kaplan EL. Epidemiology and prevention of group A streptococcal infections: acute respiratory tract infections, skin infections, and their sequelae at the close of the twentieth century. Clin Infect Dis. 1999;28:205–210. doi: 10.1086/515114. [DOI] [PubMed] [Google Scholar]
- McDonald M, Currie BJ, Carapetis JR. Acute rheumatic fever: a chink in the chain that links the heart to the throat? Lancet Infect Dis. 2004;4:240–245. doi: 10.1016/S1473-3099(04)00975-2. [DOI] [PubMed] [Google Scholar]
- Veasy LG, Wiedmeier SE, Orsmond GS, Ruttenberg HD, Boucek MM, Roth SJ, Tait VF, Thompson JA, Daly JA, Kaplan EL. Resurgence of acute rheumatic fever in the intermountain area of the United States. N Engl J Med. 1987;316:421–427. doi: 10.1056/NEJM198702193160801. [DOI] [PubMed] [Google Scholar]
- Courtney HS, Hasty DL, Dale JB. Molecular mechanisms of adhesion, colonization, and invasion of group A streptococci. Ann Med. 2002;34:77–87. doi: 10.1080/07853890252953464. [DOI] [PubMed] [Google Scholar]
- Amerongen AV, Veerman EC. Saliva—the defender of the oral cavity. Oral Dis. 2002;8:12–22. doi: 10.1034/j.1601-0825.2002.1o816.x. [DOI] [PubMed] [Google Scholar]
- von Pawel-Rammingen U, Bjorck L. IdeS and SpeB: immunoglobulin-degrading cysteine proteinases of Streptococcus pyogenes. Curr Opin Microbiol. 2003;6:50–55. doi: 10.1016/s1369-5274(03)00003-1. [DOI] [PubMed] [Google Scholar]
- Voyich JM, Musser JM, DeLeo FR. Streptococcus pyogenes and human neutrophils: a paradigm for evasion of innate host defense by bacterial pathogens. Microbes Infect. 2004;6:1117–1123. doi: 10.1016/j.micinf.2004.05.022. [DOI] [PubMed] [Google Scholar]
- Stock JB, Ninfa AJ, Stock AM. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreikemeyer B, McIver KS, Podbielski A. Virulence factor regulation and regulatory networks in Streptococcus pyogenes: impact on host-pathogen interactions. Trends Microbiol. 2003;11:224–232. doi: 10.1016/s0966-842x(03)00098-2. [DOI] [PubMed] [Google Scholar]
- Hood L, Heath JR, Phelps ME, Lin B. Systems biology and new technologies enable predictive and preventative medicine. Science. 2004;306:640–643. doi: 10.1126/science.1104635. [DOI] [PubMed] [Google Scholar]
- Khalil IG, Hill C. Systems biology for cancer. Curr Opin Oncol. 2005;17:44–48. doi: 10.1097/01.cco.0000150951.38222.16. [DOI] [PubMed] [Google Scholar]
- Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- Facklam RF, Marin DR, Lovgren M, Johnson DR, Efstratiou A, Thompson TA, Gowan S, Kriz P, Tyrrell GJ, Kaplan E, Beall B. Extension of the Lancefield classification for group A streptococci by addition of 22 new M protein gene sequences from the clinical isolates: emm103 to emm124. Clin Infect Dis. 2002;34:28–38. doi: 10.1086/324621. [DOI] [PubMed] [Google Scholar]
- Li Z, Sakota V, Jackson D, Franklin AR, Beall B, for the Active Bacterial Core Surveillance/Emerging Infections Program Network Array of M protein gene subtypes in 1064 recent invasive group A Streptococcus isolates recovered from the Active Bacterial Core Surveillance. J Infect Dis. 2003;188:1587–1592. doi: 10.1086/379050. [DOI] [PubMed] [Google Scholar]
- Tyrrell GJ, Lovgren M, Forwick B, Hoe HP, Musser JM, Talbot JA. 2002. M type of group A streptococcal isolates submitted to the National Centre for Streptococcus (Canada) from 1993–1999. J Clin Microbiol. 2002;40:4466–4471. doi: 10.1128/JCM.40.12.4466-4471.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shulman ST, Tanz RR, Kabat W, Kabat K, Cederlund E, Patel D, Li Z, Sakota V, Dale JB, Beall B. US Streptococcal Pharyngitis Surveillance Group. Group A streptococcal pharyngitis serotype surveillance in North America, 2000–2002. J Infect Dis. 2004;39:325–332. doi: 10.1086/421949. [DOI] [PubMed] [Google Scholar]
- Ferretti JJ, McShan WM, Ajdic D, Savic DJ, Savic G, Lyon K, Primeaux C, Sezate S, Suvorov AN, Kenton S, Lai HS, Lin SP, Qian Y, Jia HG, Najar FZ, Ren Q, Zhu H, Song L, White J, Yuan X, Clifton SW, Roe BA, McLaughlin R. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc Natl Acad Sci USA. 2001;98:4658–4663. doi: 10.1073/pnas.071559398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres SB, Sylva GL, Barbian KD, Lei B, Hoff JS, Mammarella ND, Liu MY, Smoot JC, Porcella SF, Parkins LD, Campbell DS, Smith TM, McCormick JK, Leung DY, Schlievert PM, Musser JM. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc Natl Acad Sci USA. 2002;99:10078–10083. doi: 10.1073/pnas.152298499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot JC, Barbian KD, Van Gompel JJ, Smoot LM, Chaussee MS, Sylva GL, Sturdevant DE, Ricklefs SM, Porcella SF, Parkins LD, Beres SB, Campbell DS, Smith TM, Zhang Q, Kapur V, Daly JA, Veasy LG, Musser JM. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc Natl Acad Sci USA. 2002;99:4668–4673. doi: 10.1073/pnas.062526099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa I, Kurokawa K, Yamashita A, Nakata M, Tomiyasu Y, Okahashi N, Kawabata S, Yamazaki K, Shiba T, Yasunaga T, Hayashi H, Hattori M, Hamada S. Genome sequence of an M3 strain of Streptococcus pyogenes reveals a large-scale genomic rearrangement in invasive strains and new insights into phage evolution. Genome Res. 2003;13:10442–10455. doi: 10.1101/gr.1096703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks DJ, Porcella SF, Barbian KD, Beres SB, Philips LE, Voyich JM, DeLeo FR, Martin JM, Somerville GA, Musser JM. Progress toward characterization of the group A Streptococcus metagenome: complete genome sequence of a macrolide-resistant serotype M6 strain. J Infect Dis. 2004;190:727–738. doi: 10.1086/422697. [DOI] [PubMed] [Google Scholar]
- Green NM, Zhang S, Porcella SF, Barbian KD, Beres SB, LeFebvre RB, Musser JM. Genome sequence of a serotype M28 strain of group A Streptococcus: new insights into puerperal sepsis and bacterial disease specificity. J Infect Dis. 2005;192:760–770. doi: 10.1086/430618. [DOI] [PubMed] [Google Scholar]
- Sumby P, Porcella SF, Madrigal AG, Kent KD, Ricklefs SM, Virtaneva K, Sturdevant DE, Graham MR, Vuopio-Varkila J, Hoe NP, Musser JM. Evolution and emergence of a highly successful clone of serotype M1 group A Streptococcus involved multiple horizontal gene transfer events. J Infect Dis. 2005;192:771–782. doi: 10.1086/432514. [DOI] [PubMed] [Google Scholar]
- Banks DJ, Beres SB, Musser JM. The contribution of phages to group A Streptococcus diversity and pathogenesis. Waldor M, Friedman D, Adhya S, editors. Washington DC: American Society for Microbiology Press,; Bacteriophages and Bacterial Pathogens. 2005:pp 319–334. [Google Scholar]
- Cunningham MW. Autoimmunity and molecular mimicry in the pathogenesis of post-streptococcal heart disease. Front Biosci. 2003;8:S533–S543. doi: 10.2741/1067. [DOI] [PubMed] [Google Scholar]
- Smoot JC, Korgenski EK, Daly DA, Veasy LG, Musser JM. Molecular analysis of group A Streptococcus type emm18 isolates temporally associated with acute rheumatic fever outbreaks in Salt Lake City, Utah. J Clin Microbiol. 2002;40:1805–1810. doi: 10.1128/JCM.40.5.1805-1810.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot LM, McCormick JK, Smoot JC, Hoe NP, Strickland I, Cole RL, Barbian KD, Earhart CA, Ohlendorf DH, Veasy LG, Hill HR, Leung DYM, Schlievert PM, Musser JM. Characterization of two novel pyrogenic toxin superantigens made by an acute rheumatic fever clone of Streptococcus pyogenes associated with multiple disease outbreaks. Infect Immun. 2002;70:7095–7104. doi: 10.1128/IAI.70.12.7095-7104.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdogan B, Appelbaum PC. Macrolide resistance in streptococci and Haemophilus influenzae. Clin Lab Med. 2004;24:455–475. doi: 10.1016/j.cll.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Martin JM, Green M, Barbadora KA, Wald ER. Erythromycin-resistant group A streptococci in children in Pittsburgh. N Engl J Med. 2002;346:1200–1206. doi: 10.1056/NEJMoa013169. [DOI] [PubMed] [Google Scholar]
- Colman G, Tanna A, Efstratiou A, Gaworzewska ET. The serotypes of Streptococcus pyogenes present in Britain during 1980–1990 and their association with disease. J Med Microbiol. 1993;39:165–178. doi: 10.1099/00222615-39-3-165. [DOI] [PubMed] [Google Scholar]
- Banks DJ, Porcella SF, Barbian KD, Martin JM, Musser JM. Structure and distribution of an unusual chimeric genetic element encoding macrolide resistance in phylogenetically diverse clones of group A Streptococcus. J Infect Dis. 2003;188:1898–1908. doi: 10.1086/379897. [DOI] [PubMed] [Google Scholar]
- Nuland SB. New York: W.W. Norton; The Doctors’ PlagueGerms, Childbed Fever, and the Strange Story of Ignac Semmelweis. 2003 [Google Scholar]
- Semmelweis I. The Etiology, Concept and Prophylaxis of Childbed Fever. Madison: The University of Wisconsin Press,; Translated by Carter KC. 1983 [Google Scholar]
- Chuang I, Van Beneden C, Beall B, Schuchat A, the Active Bacterial Core Surveillance/Emerging Infections Program Network Population-based surveillance for postpartum invasive group A Streptococcus infections, 1995–2000. Clin Infect Dis. 2002;35:665–670. doi: 10.1086/342062. [DOI] [PubMed] [Google Scholar]
- Eriksson BK, Norgren M, McGregor K, Spratt BG, Normark BH. Group A streptococcal infections in Sweden: a comparative study of invasive and noninvasive infections and analysis of dominant T28 emm28 isolates. Clin Infect Dis. 2003;37:1189–1193. doi: 10.1086/379013. [DOI] [PubMed] [Google Scholar]
- Morens DM, Folkers GK, Fauci AS. The challenge of emerging and re-emerging infectious diseases. Nature. 2004;430:242–249. doi: 10.1038/nature02759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- Stevens DL, Tanner MH, Winship J, Swarts R, Ries KM, Schlievert PM, Kaplan E. Severe group A streptococcal infections associated with a toxic shock-like syndrome and scarlet fever toxin A. N Engl J Med. 1989;321:1–7. doi: 10.1056/NEJM198907063210101. [DOI] [PubMed] [Google Scholar]
- Holm SE, Norrby A, Bergholm A-M, Norgren M. Aspects of pathogenesis of serious group A streptococcal infections in Sweden, 1988–1989. J Infect Dis. 1992;166:31–37. doi: 10.1093/infdis/166.1.31. [DOI] [PubMed] [Google Scholar]
- Muotiala A, Seppala H, Huovinen P, Vuopio-Varkila J. Molecular comparison of group A streptococci of T1M1 serotype from invasive and noninvasive infections in Finland. J Infect Dis. 1997;175:392–399. doi: 10.1093/infdis/175.2.392. [DOI] [PubMed] [Google Scholar]
- Musser JM, Kapur V, Szeto J, Pan X, Swanson DS, Martin DR. Genetic diversity and relationships among Streptococcus pyogenes strains expressing serotype M1 protein: recent intercontinental spread of a subclone causing episodes of invasive disease. Infect Immun. 1995;63:994–1003. doi: 10.1128/iai.63.3.994-1003.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoe N, Nakashima K, Grigsby D, Pan X, Dou SJ, Naidich S, Garcia M, Kahn E, Bergmire-Sweat D, Musser JM. Rapid molecular genetic subtyping of serotype M1 group A Streptococcus strains. Emerg Infect Dis. 1999;5:254–263. doi: 10.3201/eid0502.990210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukomski S, Nakashima K, Abdi I, Cipriano VJ, Ireland RM, Reid SD, Adams GG, Musser JM. Identification and characterization of the scl gene encoding a group A Streptococcus extracellular protein virulence factor with similarity to human collagen. Infect Immun. 2000;68:6542–6553. doi: 10.1128/iai.68.12.6542-6553.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres SB, Sylva GL, Sturdevant DE, Granville CN, Liu M, Ricklefs SM, Whitney AR, Parkins LD, Hoe NP, Adams GJ, Low DE, DeLeo FR, McGeer A, Musser JM. Genome-wide molecular dissection of serotype M3 group A Streptococcus strains causing two epidemics of invasive infection. Proc Natl Acad Sci USA. 2004;101:11833–11888. doi: 10.1073/pnas.0404163101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagiec MJ, Lei B, Parker SK, Vasil ML, Matsumoto M, Ireland RM, Beres SB, Hoe NP, Musser JM. Analysis of a novel prophage-encoded group A Streptococcus extracellular phospholipase A2. J Biol Chem. 2004;279:45909–45918. doi: 10.1074/jbc.M405434200. [DOI] [PubMed] [Google Scholar]
- Smoot LM, Smoot JC, Graham MR, Somerville GA, Sturdevant DE, Lux Migliaccio CA, Sylva GL, Musser JM. Global differential gene expression in response to growth temperature alteration in group A Streptococcus. Proc Natl Acad Sci USA. 2001;98:10416–10421. doi: 10.1073/pnas.191267598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham MR, Smoot LM, Lux Migliaccio CA, Sturdevant DE, Porcella SF, Federle MJ, Scott JR, Musser JM. Virulence control in group A Streptococcus by a two-component gene regulatory system: global expression profiling and in vivo infection modeling. Proc Natl Acad Sci USA. 2001;99:13855–13860. doi: 10.1073/pnas.202353699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voyich JM, Sturdevant DE, Braughton KR, Kobayashi SD, Lei B, Virtaneva K, Dorward DL, Musser JM, DeLeo FR. Genome-wide protective response used by group A Streptococcus to evade destruction by human polymorphonuclear leukocytes. Proc Natl Acad Sci USA. 2003;100:1996–2001. doi: 10.1073/pnas.0337370100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voyich JM, Braughton KR, Sturdevant DE, Vuong C, Kobayashi SD, Porcella SF, Otto M, Musser JM, DeLeo FR. Engagement of the pathogen survival response used by group A Streptococcus to avert destruction by innate host defense. J Immunol. 2004;173:1194–1201. doi: 10.4049/jimmunol.173.2.1194. [DOI] [PubMed] [Google Scholar]
- Graham MR, Virtaneva K, Porcella SF, Barry WT, Gowen BB, Johnson CR, Wright FA, Musser JM. Group A Streptococcus transcriptome dynamics during growth in human blood reveals bacterial adaptive and survival strategies. Am J Pathol. 2005;166:455–465. doi: 10.1016/S0002-9440(10)62268-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtaneva K, Porcella SF, Graham MR, Ireland RM, Johnson CA, Ricklefs SM, Babar I, Parkins L, Romero RA, Corn CJ, Gardner D, Bailey JR, Parnell MJ, Musser JM. Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc Natl Acad Sci USA. 2005;102:9014–9019. doi: 10.1073/pnas.0503671102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin JC, Wessels MR. Identification of csrR/csrS, a genetic locus that regulates hyaluronic acid capsule synthesis in group A Streptococcus. Mol Microbiol. 1998;30:209–219. doi: 10.1046/j.1365-2958.1998.01057.x. [DOI] [PubMed] [Google Scholar]
- Heath A, DiRita VJ, Barg NL, Engleberg NC. A two-component regulatory system, CsrR-CsrS, represses expression of three Streptococcus pyogenes virulence factors, hyaluronic acid capsule, streptolysin S, and pyrogenic exotoxin B. Infect Immun. 1999;67:5298–5305. doi: 10.1128/iai.67.10.5298-5305.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne SA, III, Granville C, Sitkiewicz I, Tokuyama M, Patel P, Musser JM. Growth characteristics and extracellular virulence factor production by group A Streptococcus during cultivation in human saliva. Infect Immun. 2005;73:4723–4731. doi: 10.1128/IAI.73.8.4723-4731.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelburne III SA, Sumby P, Granville C, Sitkiewicz I, DeLeo FR, Musser JM: Central role of a bacterial two-component gene regulatory system of previously unknown function in pathogen persistence in human saliva. Proc Natl Acad Sci USA 2005, (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staali L, Morgelin M, Bjorck L, Tapper H. Streptococcus pyogenes expressing M and M-like surface proteins are phagocytosed but survive inside human neutrophils. Cell Microbiol. 2003;5:253–265. doi: 10.1046/j.1462-5822.2003.00272.x. [DOI] [PubMed] [Google Scholar]
- Medina E, Rohde M, Chhatwal GS. Intracellular survival of Streptococcus pyogenes in polymorphonuclear cells results in increased bacterial virulence. Infect Immun. 2003;71:5376–5380. doi: 10.1128/IAI.71.9.5376-5380.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina E, Goldmann O, Toppel AW, Chhatwal GS. Survival of Streptococcus pyogenes within host phagocytic cells: a pathogenic mechanism for persistence and systemic invasion. J Infect Dis. 2003;187:597–603. doi: 10.1086/373998. [DOI] [PubMed] [Google Scholar]
- Kobayashi SD, Braughton KR, Whitney AR, Voyich JM, Schwan TG, Musser JM, DeLeo FR. Bacterial pathogens modulate an apoptosis differentiation program in human neutrophils. Proc Natl Acad Sci USA. 2003;100:10948–10953. doi: 10.1073/pnas.1833375100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtaneva K, Graham MR, Porcella SF, Hoe NP, Hua S, Graviss EA, Bailey JR, Parnell MJ, Musser JM. Group A Streptococcus gene expression in humans and cynomolgus macaques with acute pharyngitis. Infect Immun. 2003;71:2199–2207. doi: 10.1128/IAI.71.4.2199-2207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson RW, Redmond HP, Wang JH, Condron C, Bouchier-Hayes D. Neutrophils undergo apoptosis following ingestion of Escherichia coli. J Immunol. 1996;156:3986–3992. [PubMed] [Google Scholar]
- Kobayashi SD, Voyich JM, Buhl CL, Stahl RM, DeLeo FR. Global changes in gene expression by human polymorphonuclear leukocytes during receptor-mediated phagocytosis: cell fate is regulated at the level of gene expression. Proc Natl Acad Sci USA. 2002;99:6901–6906. doi: 10.1073/pnas.092148299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLeo FR. Modulation of phagocyte apoptosis by bacterial pathogens. Apoptosis. 2004;9:399–413. doi: 10.1023/B:APPT.0000031448.64969.fa. [DOI] [PubMed] [Google Scholar]
- Coxon A, Rieu P, Barkalow FJ, Askari S, Sharpe AH, von Andrian UH, Arnaout MA, Mayadas TN. A novel role for the beta 2 integrin CD11b/CD18 in neutrophil apoptosis: a homeostatic mechanism in inflammation. Immunity. 1996;5:653–666. doi: 10.1016/s1074-7613(00)80278-2. [DOI] [PubMed] [Google Scholar]
- Zhang B, Hirahashi J, Cullere X, Mayadas TN. Elucidation of molecular events leading to neutrophil apoptosis following phagocytosis: cross-talk between caspase 8, reactive oxygen species, and MAPK/ERK activation. J Biol Chem. 2003;278:28443–28454. doi: 10.1074/jbc.M210727200. [DOI] [PubMed] [Google Scholar]
- Kobayashi SD, DeLeo FR. An apoptosis differentiation programme in human polymorphonuclear leucocytes. Biochem Soc Trans. 2004;32:474–476. doi: 10.1042/BST0320474. [DOI] [PubMed] [Google Scholar]
- Kobayashi SD, Voyich JM, Braughton KR, Whitney AR, Nauseef WM, Malech HL, DeLeo FR. Gene expression profiling provides insight into the pathophysiology of chronic granulomatous disease. J Immunol. 2004;172:636–643. doi: 10.4049/jimmunol.172.1.636. [DOI] [PubMed] [Google Scholar]
- Tettelin H, Saunders NJ, Heidelberg J, Jeffries AC, Nelson KE, Eisen JA, Ketchum KA, Hood DW, Peden JF, Dodson RJ, Nelson WC, Gwinn ML, DeBoy R, Peterson JD, Hickey EK, Haft DH, Salzberg SL, White O, Fleischmann RD, Dougherty BA, Mason T, Ciecko A, Parksey DS, Blair E, Cittone H, Clark EB, Cotton MD, Utterback TR, Khouri H, Qin H, Vamathevan J, Gill J, Scarlato V, Masignani V, Pizza M, Grandi G, Sun L, Smith HO, Fraser CM, Moxon ER, Rappuoli R, Venter JC. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science. 2000;287:1816–1820. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- Pizza M, Scarlato V, Masignani V, Giuliani MM, Arico B, Comanducci M, Jennings GT, Baldi L, Bartolini E, Capecchi B, Galeotti CL, Luzzi E, Manetti R, Marchetti E, Mora M, Nuti S, Ratti G, Santini L, Savino S, Scarselli M, Storni E, Zuo P, Broeker M, Hundt E, Knapp B, Blair E, Mason T, Tettelin H, Hood DW, Jeffries AC, Saunders NJ, Granoff DM, Venter JC, Moxon ER, Grandi G, Rappuoli R. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science. 2000;287:1816–1820. doi: 10.1126/science.287.5459.1816. [DOI] [PubMed] [Google Scholar]
- Fraser CM, Rappuoli R. Application of microbial genome science to advanced therapeutics. Annu Rev Microbiol. 2005;56:459–474. doi: 10.1146/annurev.med.56.062904.144853. [DOI] [PubMed] [Google Scholar]
- Maione D, Ros IMY, Rinaudo D, Masignani V, Mora M, Scarselli M, Tettelin H, Brettoni C, Iacobini ET, Rosini R, D’Agostino N, Miorin L, Buccato S, Mariani M, Galli G, Nogarotto R, Dei VN, Vegni F, Fraser C, Mancuso G, Teti G, Madoff LC, Paoletti LC, Rappuoli R, Kasper DL, Telford JL, Grandi G. Identification of a universal group B Streptococcus vaccine by multiple genome screen. Science. 2005;309:148–150. doi: 10.1126/science.1109869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei B, Mackie S, Lukomski S, Musser JM. Identification and immunogenicity of group A Streptococcus culture supernatant proteins. Infect Immun. 2000;68:6807–6818. doi: 10.1128/iai.68.12.6807-6818.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei B, Liu M, Chesney GL, Musser JM. Identification of new candidate vaccine antigens made by Streptococcus pyogenes: purification and characterization of 16 putative extracellular lipoproteins. J Infect Dis. 2004;189:79–89. doi: 10.1086/380491. [DOI] [PubMed] [Google Scholar]
- Reid SD, Green NM, Sylva GL, Voyich JM, Stenseth ET, DeLeo FR, Palzkill T, Low DE, Hill HR, Musser JM. Postgenomic analysis of four novel antigens of group A Streptococcus: growth phase-dependent gene transcription and human serologic response. J Bacteriol. 2002;184:6316–6324. doi: 10.1128/JB.184.22.6316-6324.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]