Abstract
In cathepsin D-deficient (CD−/−) and cathepsins B and L double-deficient (CB−/−CL−/−) mice, abnormal vacuolar structures accumulate in neurons of the brains. Many of these structures resemble autophagosomes in which part of the cytoplasm is retained but their precise nature and biogenesis remain unknown. We show here how autophagy contributes to the accumulation of these vacuolar structures in neurons deficient in cathepsin D or both cathepsins B and L by demonstrating an increased conversion of the molecular form of MAP1-LC3 for autophagosome formation from the cytosolic form (LC3-I) to the membrane-bound form (LC3-II). In both CD−/− and CB−/−CL−/− mouse brains, the membrane-bound LC3-II form predominated whereas MAP1-LC3 signals accumulated in granular structures located in neuronal perikarya and axons of these mutant brains and were localized to the membranes of autophagosomes, evidenced by immunofluorescence microscopy and freeze-fracture-replica immunoelectron microscopy. Moreover, as in CD−/− neurons, autofluorescence and subunit c of mitochondrial ATP synthase accumulated in CB−/−CL−/− neurons. This suggests that not only CD−/− but also CB−/−CL−/− mice could be useful animal models for neuronal ceroid-lipofuscinosis/Batten disease. These data strongly argue for a major involvement of autophagy in the pathogenesis of Batten disease/lysosomal storage disorders.
Autophagy is a highly regulated process involving the bulk degradation of cytoplasmic macromolecules and organelles in eukaryotic cells via the lysosomal/vacuolar system.1 Although it is induced under starvation, differentiation, and normal growth control,2 the participation of autophagy has also been demonstrated in various neurodegenerative disorders.3 Moreover, it has been shown that autophagy can trigger a form of cell death distinct from apoptosis in neurons.4–6 Thus autophagy appears to be involved in neurodegenerative disorders.
The most common inherited neurodegenerative disease in childhood is neuronal ceroid-lipofuscinosis (NCL, or Batten disease), which is categorized as a lysosomal storage disorder and pathologically characterized by the accumulation of proteolipids, such as subunit c of mitochondrial ATP synthase and sphingolipid activator proteins in the lysosomes of neurons.7–10 We have previously demonstrated that the central nervous system (CNS) neurons in cathepsin D-deficient (CD−/−) mice show a new form of lysosomal accumulation disease with a phenotype resembling NCLs and subunit c of mitochondrial ATP synthase accumulates in the lysosomes of the affected neurons.11,12 Double membrane-bound vacuoles containing part of the cytoplasm are frequently detected in CNS neurons of CD−/− mouse brains near the terminal stage.
It has also been demonstrated that the phenotypes of mice deficient in both cathepsins B and L (CB−/−CL−/− mice) resemble NCLs, but the accumulated substances show no autofluorescence and are immunonegative for subunit c and saposins.13 However, in neurons of these CD−/− and CB−/−CL−/− mouse brains, the exact nature and biogenesis of the accumulated lysosomal structures remain unknown.
It has recently been found that LC3, light chain 3 of neuronal microtubule-associated protein 1A/B, is a mammalian homolog of yeast Atg8p that is required for autophagosome formation.14 Immediately after the synthesis of LC3, the protein is converted to the cytoplasmic form, LC3-I, by cleavage at the C-terminal region,14 which is further converted into a membrane-bound form, LC3-II, by Atg7p and Atg3p, respectively, when autophagy is induced.15–17 Moreover, LC3-II is localized mainly in the membranes of autophagosomes and, to a lesser extent, within autophagolysosomes.14 We therefore investigated whether autophagy participates in the accumulation of lysosomal structures in the brain and peripheral tissues of mice deficient in cathepsin D and those doubly deficient in cathepsins B and L by characterizing molecular forms of LC3. Our data showing that the conversion of LC3-I to LC3-II in CD−/− and CB−/−CL−/− mouse brains coincides with the accumulation of lysosomal structures or LC3-positive granules in the neurons, suggest that autophagy is involved in the abnormal storage accumulation in CNS neurons of NCL/lysosomal storage disorders.
Materials and Methods
Animals
The experiments described here were performed in compliance with the regulations of Osaka University Medical School Guideline for the Care and Use of Laboratory Animals. For cathepsin D-, B-, or L-deficient mice,18–20 heterozygous (+/−) mice were transferred to the Institute of Experimental Animal Sciences, Osaka University Graduate School of Medicine, and kept in conventional or specific pathogen-free facilities on a 12-hour light/dark cycle. To obtain homozygous mice (CD−/−, CB−/−, and CL−/−) mice, heterozygous mice (CD+/−, CB+/−, and CL+/−, respectively) were intercrossed. Selection of homozygous mice from littermates obtained by heterozygous coupling was performed by genomic polymerase chain reaction (PCR) as described previously.21,22 Briefly, for the selection of CD−/− mice, the template genomic DNA isolated from tail biopsies was examined by CD-exon 4-specific PCR with primers having the following sequences: MCD14 (5′-AGACTAACAGGCCTGT-TCCC-3′) and MCD15 (5′-TCAGCTGTAGTTGCTCACATG-3′). The selection of CB−/− mice from littermates obtained by heterozygous coupling was performed by examining the template genomic DNA isolated from tail biopsies, using CB-exon 4-specific PCR with primers of MCB11 (5′-GGTTGCGTTCGGTGAGG-3′) and MCBGT (5′-AACAAGAGCCGCAGGAGC-3′). The selection of CL−/− mice from littermates obtained by heterozygous coupling was performed by the triple PCR method; the template genomic DNA isolated from tail biopsies was examined using primers of MCL5 (5′-GGAGGAGAGCGATATGGG-3′), MCL9 (5′-AGCCATTCACCACCTGC-C-3′), and neo756-777 (5′-CGCAGAACCTGCGTGCAATCC-3′).
For the generation of CB and CL double-knockout (CB−/−CL−/−) mice, CB−/− mice were bred to CL−/− mice to create progeny that were heterozygous for both cathepsins B and L deletions (CB+/−CL+/− mice). These double heterozygotes were intercrossed to produce mice doubly deficient in both cathepsins B and L (CB−/−CL−/− mice). Most died at approximately P14, as previously reported.13 The heterozygous mice (CD+/− and CB+/−CL+/− mice) were used as control animals in the present study and showed no pathological phenotypes when examined by histological, immunocytochemical, and biochemical methods.
For the generation of CD−/− mice expressing GFP-LC3 (GFP-LC3/CD−/− mice), six GFP-LC3 transgenic mice (line no. 53)23 were obtained from Riken BioResource Center (Tsukaba, Japan), crossed with C57BL/6 mice, and maintained as heterozygotes for the GFP-LC3 transgene in the Institute of Experimental Animal Sciences. As mentioned above, the incorporation of the transgene was confirmed by PCR using primers GFP1 (5′-TCCTGCTGGAGTTCGTGACCG-3′) and LC3* (5′-TTGCGAATTCTCAGCCGTCTTCATCTCTCTCGC-3′). Another primer set mLC3ex3GT (5′-TGAGCGAGCTCATCAAGATAATCAGGT-3′) and mLC3ex4AG (5′-GTTA-GCATTGAGCTGCAAGCGCCGTCT-3′) amplifying the third intron of the LC3 genome as an internal control was used. The positive mice were also crossed with CD+/− mice and CD+/− mice heterozygous for the GFP-LC3 transgene (GFP-LC3/CD+/− mice) were generated. GFP-LC3/CD+/− mice were further crossed with CD+/− mice and CD−/− mice heterozygous for the GFP-LC3 transgene (GFP-LC3/CD−/− mice) were obtained.
Antisera
Rabbit antibodies against rat LC3 and rat subunit c of mitochondrial F1F0ATPase were produced and purified by affinity chromatography, as reported previously.8,24 Monoclonal antibodies against mouse lamp1 (the Developmental Studies Hybridoma Bank, Iowa City, IA) and mouse microglial cells (F4/80) (Serotec, Oxford, UK), respectively, and rabbit polyclonal antibodies against GFP (Abcam, Cambridge, UK) were commercially obtained.
Sampling
CD−/− and CD+/− littermates obtained at postnatal day 8 (P8) to P24 (n = 3 for each stage and each genotype), CB−/−CL−/− and their littermates at P13 (n = 3 each), and GFP-LC3/CD+/+ and GFP-LC3/CD−/− mice at P20 (n = 3 each) were deeply anesthetized with pentobarbital (25 mg/kg i.p.) and fixed by cardiac perfusion with 4% paraformaldehyde buffered with 0.1 mol/L phosphate buffer (pH 7.2), containing 4% sucrose for immunohistochemistry, detection of autofluorescence, and periodic acid-Schiff staining, with 1% paraformaldehyde buffered with 0.1 mol/L phosphate buffer (pH 7.2), containing 4% sucrose for freeze-fracture replica immunogold labeling,25 and with 2% paraformaldehyde-2% glutaraldehyde buffered with 0.1 mol/L phosphate buffer for ordinary electron microscopy.11 Immediately after perfusion fixation, brains and peripheral tissues were excised from the mice and processed for immunohistochemistry/cytochemistry and ordinary electron microscopy.11,26
For light and electron microscopy, brain tissues were quickly removed from the mice and further immersed in the same fixatives for 2 hours. Samples processed for paraffin embedding were cut at 5 μm with a microtome, and those for cryosections were cut at 10 μm with a cryostat (CM3050; Leika, Nussloch, Germany). These sections were placed on silane-coated glass slides and stored at −80°C until used. Samples for electron microscopy were postfixed with 2% OsO4, dehydrated with a graded series of alcohol, and embedded in Epon 812. Ultrathin sections were cut with an ultramicrotome (Ultracut N; Reichert-Nissei, Toyko, Japan), stained with uranyl acetate and lead citrate, and observed with a Hitachi H7100 electron microscope (Hitachi, Tokyo, Japan). For freeze-fracture replica immunogold labeling, the brain tissue was quickly excised from the mice, while cerebral cortical and cerebellar regions were separated from each brain and cut at 100 μm with a vibratome (VT1000S; Leica) at 4°C. Tissue slices were infiltrated with 30% glycerol buffered with 0.1 mol/L phosphate buffer (pH 7.2) until used.
Morphometry
Morphometric analyses were performed according to the method of Uchiyama and Watanabe.27 In samples obtained from CD−/− and CD+/− littermates (n = 4 each) at P8, P15, and P23 and CB−/−CL−/− and CB+/−CL+/− littermates (n = 4 each) at P13, electron micrographs (40 per sample) of the cerebral cortex (layer 5) were taken with systematic random sampling at each corner of 100-mesh grids with a primary magnification of ×7000. Three to five Epon blocks from each mouse were used. After printing at 2.6× the original magnification on projection papers, we estimated the cytoplasmic (perikaryal) volume fraction of lysosomal structures including various types of autophagic vacuoles (AVs), dense bodies, granular osmiophilic deposit (GROD)-like inclusions, and fingerprint profiles by point counting, using a double-lattice test system of 1.5-cm spacing. AVs were classified as early AVs (AVi), which contain the morphologically intact cytoplasm (see Figure 2A), and late AVs (AVd), which contain partially degraded but identifiable cytoplasmic materials (see Figure 2B).28–30 In CD−/− and CB−/−CL−/− brains, inclusions that, by electron microscopy, were dense granular and amorphous in neurons were quite similar to the so-called GRODs, a typical hallmark in infantile NCL;31 these granules were defined as GROD-like inclusions. In CD−/− neurons, distinct points falling on GROD-like inclusions or those surrounded by double-layered membranes resembling AVi were counted (see Figure 2D). Moreover, the multilayered lamellar structures that were counted as AVi always contained morphologically intact cytoplasm, and were different from fingerprint profiles that did not contain part of the cytoplasm and possessed tightly and concentrically arrayed membranes as in the structures observed in juvenile NCL.31 The volume density (Vv) of each lysosomal structure was expressed as the percent volume: Vv = (Pi/Pt) × 100 (%), where Pi is the number of points falling on each lysosomal structure and Pt is the number of points falling on the perikarya of large pyramidal neurons located in the layer V of the cerebral cortex.
Figure 2.
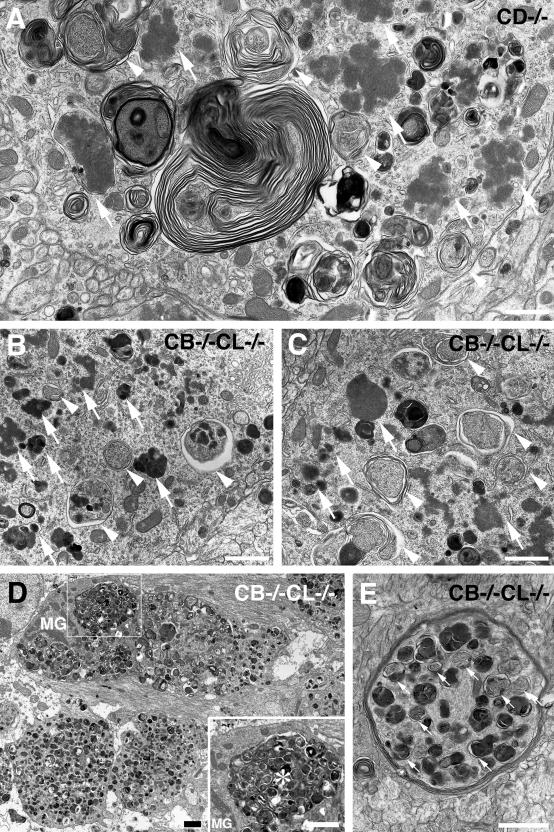
Electron micrographs of neuronal cells from brains of a CD−/− mouse at P23 (A) and a CB−/−CL−/− mouse at P12 (B–E). A–C: In the perikarya of cerebellar Purkinje cells (A, C) and a cerebral cortical neuron (B), numerous dense bodies, which resemble granular osmiophilic deposit-like inclusions (GROD-l) (arrows), can be seen, and vacuolar/autophagosome-like structures (arrowheads) encircled by ER-like membrane saccules and containing cytoplasmic organelles together with part of the cytoplasm are clearly visible. These autophagosome-like structures and GROD-like inclusions are often encircled by ER-like membrane saccules or multilamellated structures. D and E: In axons located in the corpus callosum of the same mouse as in B and C, accumulated autophagosome-like structures can be seen (D), while these structures accumulated in a myelinated axon are distinctly encircled by ER-like membrane saccules (E, small arrows). Moreover, a microglia-like cell (MG) appears in this field and possesses electron-dense heterophagosomes (asterisk) containing degenerating axons with numerous autophagosome-like structures (inset in D). The cytoplasm around the heterophagosome in the cell appears intact. Scale bars, 1 μm.
Immunohistochemical Analyses for Light Microscopy
For the detection of LC3 and subunit c, deparaffinized or frozen sections were used and immunostained according to the method described previously.32 The samples were incubated at 4°C with anti-LC3 (20 μg/ml), anti-subunit c (5 μg/ml), or anti-GFP (4 μg/ml) for 1 to 3 days and further processed for visualization. For double-immunofluorescent staining, deparaffinized or frozen sections of brain tissues from CD−/− and CB−/−CL−/− mice and those from GFP-LC3/CD+/+ and GFP-LC3/CD−/− mice were incubated with anti-LC3 and monoclonal anti-lamp1 or F4/80 for 24 hours at 4°C and further with goat anti-rabbit IgG coupled with Alexa 594 (Molecular Probes, Eugene, OR) and goat anti-rat IgG coupled with fluorescein isothiocyanate (Cappel, Durham, NC), or coupled with Alexa 594 for 1 hour at room temperature. The stained tissues were viewed with a confocal laser-scanning microscope (LSM510; Carl Zeiss, Jena, Germany).
Freeze-Fracture Replica Immunogold Labeling
To determine the subcellular localization of LC3, the sodium dodecyl sulfate (SDS)-digested freeze-fracture replica labeling (FRL) technique33 was applied to chemically fixed brain tissues.25 Samples were mounted between two gold specimen disks in a specimen-mounting medium, polyvinyl alcohol. The disks were quickly frozen in Freon 22 cooled to its freezing point, mounted in a Balzers complementary freeze-fracture apparatus (BAF 400D; Balzers, Furstentum, Lichtenstein) and immediately shadowed unidirectionally by evaporating platinum-carbon at 45° to the vertical carbon support. The thickness of platinum (2.5 nm) was controlled with a quartz crystal monitor. To release the replicas from the specimen carrier, the carrier was immersed gently in phosphate-buffered saline (PBS). After floating off, the pieces of the replicas were transferred to a detergent solution (2.5% SDS, 10 mmol/L Tris-HCl, pH 9). SDS digestion was performed for 24 hours at room temperature with vigorous constant stirring. The replicas were moved to the same detergent solution, washed for at least one additional 1 hour, rinsed four times in PBS (5 minutes each), and placed on drops of 1 to 10% bovine serum albumin (BSA) in PBS (BSA-PBS) for 30 minutes at room temperature. For immunostaining, rabbit polyclonal anti-LC3 antibody and 10-nm colloidal gold conjugated goat anti-rabbit IgG were used. The replicas, picked up on grids, were observed with a Hitachi H7100 electron microscope.
Immunoblot Analysis
Anesthetized mice were killed by decapitation and each excised tissue was independently homogenized in 2 ml of 0.05 mol/L Tris-buffered 0.15 mol/L saline containing 1% Triton X-100 and a protease inhibitor cocktail (Boehringer Mannheim, Mannheim, Germany) by a Polytron homogenizer (Kinematica, Littau, Switzerland) at 80% maximal speed. After centrifuging twice at 10,500 × g for 10 minutes at 4°C, the protein concentration in the supernatants were determined using the BCA protein assay system (Pierce, Rockford, IL). The samples were analyzed by immunoblotting according to the method of Towbin and colleagues.34 After immunostaining with anti-LC3 (1 μm/ml) or polyclonal anti-GFP (0.4 μm/ml), immunodetection was performed with a chemiluminescent ECL kit (Amersham, Arlington Heights, IL). The density of each protein band in the blotting was measured using a densitometer (Molecular Dynamics, Sunnyvale, CA) and the ratio of LC3-I to LC3-II molecules was calculated for brain tissues. In the case of peripheral tissues, the density of LC3-II was measured and presented as a mean ± SD from three independent experiments.
Reverse Transcriptase (RT)-PCR Analysis
RT-PCR analysis of LC3 was performed according to the method described previously.35 Total RNAs were extracted from neuronal tissues of CD +/− and −/− mice at P8, P15, and P22 and CB+/−CL+/− and CB−/−CL−/− mice at P13, using a Polytron homogenizer for 1 minute in Isogen (Nippon Gene, Tokyo, Japan). To avoid contamination with genomic DNA, 40 to 50 μg of total RNAs were treated with 0.2 U/ml DNase I (Nippon Gene) for 20 minutes at 37°C. Then, 2.0 μg/ml of the total RNAs were used for cDNA synthesis using AMV reverse transcriptase (Takara, Tokyo, Japan) and amplification of the LC-3 and β-actin cDNAs were performed by PCR using EX taq HS (Takara). The PCR amplification was performed at 95°C for 5 minutes, followed by 30 cycles of 95°C for 30 seconds, 55°C (for LC-3) or 60°C (for β-actin) for 30 minutes, and 72°C for 30 minutes using the primer sets for LC-3 (GenBank accession number NM_026160) (forward: 5′-GTGGGCCGCTCTAGGCACCAA-3′, reverse: 5′-CTCTTTGATGTCACGCACGATTTC-3′; 540 bp) and β-actin (GenBank accession number M12481) (forward: 5′-GTGGGCCGCTCTAGGCACCAA-3′, reverse: 5′-CTC-TTTGATGTCACGCACGATTTC-3′; 540 bp).
Results
Accumulation of Autophagosome-Like Inclusion Bodies in Both CD−/− and CB−/−CL−/− Mouse Neuronal Perikarya
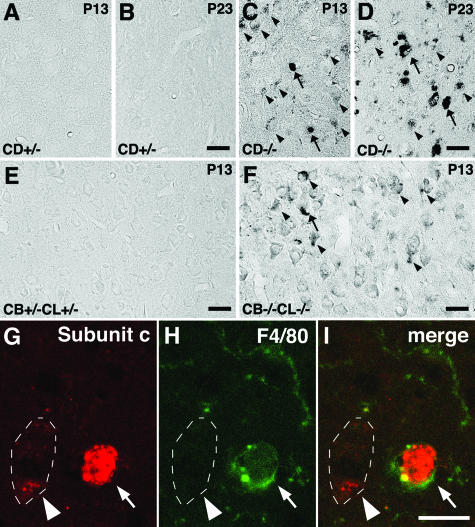
As described previously,11 massive autofluorescent granules, which are immunopositive for subunit c of mitochondrial ATP synthase and lysosomal cathepsin B, had accumulated in the neuronal cell perikarya of CD−/− mouse brains near the terminal stage (data not shown). Indeed, immunoreactivity for subunit c was intensely detected in neurons and microglial cells of CD−/− mouse brains at P13 and P23, but not in those in the control littermate mice (Figure 1, A–D).
Figure 1.
Immunostaining for subunit c of mitochondrial ATP synthase in the cerebral cortex obtained from CD+/− (A, C) and CD−/− (B, D) mice at P13 (A, B) and P23 (C, D) and from CB+/−CL+/− (E) and CB−/−CL−/− (F) mice at P13. A–D: Positive staining for subunit c is intense in neurons (arrowheads) and microglial cells (arrows) of the CD−/− mouse brain (B, D), but not in those of the control littermate brain (A, C). The immunoreactivity is weaker in neurons of CD−/− at P13 (C, arrowheads). E and F: Similar localization patterns of subunit c to those in A to D are observed in the cerebral cortex of CB+/−CL+/− (E) and CB−/−CL−/− (F) mice, although the immunoreactivity is weak in neurons (arrowheads). G–I: Double immunostaining of subunit c and F4/80 in the cerebral cortex of a CB−/−CL−/− mouse at P13. Positive signals for subunit c (red) are large and intense and co-localized in a F4/80-positive microglial cell (arrows), whereas they are fine granular and localized in a F4/80-negative neuron that is marked by arrowheads. Scale bars, 20 μm.
It has been shown that the phenotype of CB−/−CL−/− is highly reminiscent of NCLs, both of which demonstrate the early and severe brain atrophy; however, the former does not show immunoreactivity for subunit c in brain tissue and ultrastructural characteristics of accumulated lysosomal compartments in CB−/−CL−/− neurons are different from those in NCLs.13 The present study also generated CB−/−CL−/− mice by crossing CB+/−CL+/− mice and examined morphological and biochemical characteristics of these mutant mice. In CB−/−CL−/− mice at P13, localization patterns of subunit c in brain tissue were very similar to those in CD−/− mice at the same postnatal age, although the immunoreactivity for subunit c in these two models at P13 was much weaker than in CD−/− brains at P23. The immunoreactivity was more intense in microglial cells than in neurons in CD−/− and CB−/−CL−/− brains at any stages examined (Figure 1, C, D, and F). The positive signal for subunit c was not detected in the control littermate brains (Figure 1, A, B, and E). To confirm the presence of the subunit c immunoreactivity in neurons and microglial cells, double immunostaining of subunit c and F4/80 was performed in the cerebral cortex of CB−/−CL−/− mice at P13; intense staining for subunit c was clearly visible in F4/80-immunopositive microglial cells, while granular fluorescence for subunit c was also detected in F4/80-negative neurons (Figure 1, G–I). Moreover, autofluorescence was discerned in neurons and microglial cells in CB−/−CL−/− mouse brains at P13, while these mutant neurons were intensely positive for periodic acid-Schiff staining (Supplemental Figure 1 at http://ajp.amjpathol.org).
Electron microscopic observations in neurons of CD−/− mouse brain tissues showed that these lysosomal structures that have been shown to possess subunit c11 coincided with granular inclusions and autophagosome/autophagolysosome-like structures (Figure 2A; Figure 3, A, B, D, and E), the former of which have strong similarity to GRODs, a typical hallmark of NCLs.31 Autophagosome-like structures containing morphologically intact cytoplasm are often surrounded with multilayered lamellar structures (Figure 2A; Figure 3, A and D). Moreover, these multilayered lamellar structures were different from fingerprint-like structures that did not contain part of the cytoplasm and had tightly and concentrically arrayed membranes and strongly resembled fingerprint profiles, a typical hallmark of NCLs.31 In CB−/−CL−/− mouse brains, multilayered lamellar structures appeared only occasionally, while fingerprint profiles were totally absent (Table 1).
Figure 3.
Morphometric analysis of various lysosomal compartments in the perikarya of CD−/− and CD+/− mouse neurons. A–E: Representative examples of early (AVi; A, D) and late (AVd; B) AVs, granular osmiophilic deposit-like inclusions (GROD-l; E), and a fingerprint profile (FP; F) from CD−/− neurons obtained at P23, and a normal dense body (C) from a CD+/− neuron obtained at P23 are shown. In AVi, a mitochondrion (m) together with part of the cytoplasm is clearly visible (A), while in AVd, a mitochondrion with partially degraded cytoplasmic materials is also identifiable. D: AVi that contains GROD-l is detectable. Small arrows show double membranes surrounding GROD-l (asterisks). To show the fingerprint pattern clearly, a boxed area in F is enlarged in an inset. G: The volume densities (%) of AVi, AVd, dense body, GROD-l, and fingerprint profiles (FPs) in the perikarya of neurons in the layer V of the cerebral cortex obtained from CD+/− and CD−/− mice at P8, P15, and P23 and CB−/−CL−/− and CB+/−CL+/− littermates at P13 are shown as a stack bar chart. Scale bars: 0.1 μm (A–F); 0.2 μm (inset in F).
Table 1.
Pathological Findings Observed in CD−/− and CB−/−CL−/− Mice
| CD−/− mice | CB−/−CL−/− mice | |
|---|---|---|
| Duration of life | Around P26 | Around P14 |
| Tremor | Positive | Positive |
| Onset of the tremor | Around P20 | Around P10 |
| Autofluorescence in neurons | Fine granular at P13 and massive at P23 | Fine granular at P13 |
| Autofluorescence in microglial cells | Massive | Massive |
| PAS | Positive | Positive |
| Accumulation of subunit c in neurons | Fine granular at P13 and massive at P23 | Fine granular at P13 |
| Accumulation of subunit c in microglial cells | Massive | Massive |
| AVs in the perikarya and dendrites of neurons | Positive | Positive |
| Multilayered AVs | Frequent | Occasional |
| GROD-like inclusions | Positive | Positive |
| GROD-like inclusions within AVs | Positive | Positive |
| Fingerprint profiles | Positive | Negative |
| Curvilinear profiles | Negative | Negative |
PAS, periodic acid-Schiff staining; AV, autophagic vacuole; GROD, granular osmiophilic profiles.
In semithin sections of CB−/−CL−/− mouse brains, numerous granules that were stained by toluidine blue accumulated in neuronal cell perikarya, dendrites, and axons (data not shown), as has been shown in CD−/− mouse brains.11 By electron microscopy, most of the lysosome-like structures in the neurons of CB−/−CL−/− mouse brains resembled GRODs (GROD-like inclusions) and autophagosomal (autophagosome-like) structures (Figure 2, B and C). In the corpus callosum of the mouse brains, autophagosome-like structures abundantly accumulated in unmyelinated and myelinated nerve fibers (Figure 2, D and E), while microglia-like cells were often observed to possess heterophagosomes containing numerous autophagosome-like structures (Figure 2D, inset). Most vacuolar structures in the axons were encircled by double-membrane saccules (Figure 2E). In dendrites of cerebral cortical neurons and Purkinje neurons of CD−/− and CB−/−CL−/− mice, granular structures also accumulated, but these accumulated structures were similar to those observed in the neuronal perikarya (Supplemental Figure 2A at http://ajp.amjpathol.org). Like axons in the corpus callosum, autophagosome-like structures that were encircled by double-membrane saccules accumulated in axons of the cerebellum in CB−/−CL−/− mice (Supplemental Figure 2B at http://ajp.amjpathol.org).
It has been shown that autophagosomes undergo stepwise maturation by fusing with endosomes and/or lysosomes,36 while two types of AVs in this maturation process are distinguishable by electron microscopy;28,30 nascent or immature AVs encircled by endoplasmic reticulum (ER)-like membrane saccules contain part of the morphologically intact cytoplasm (AVi) (Figure 3, A and D) and mature AVs encircled by a single membrane possess degraded but morphologically identifiable cytoplasmic materials and structures (AVd) (Figure 3B). It is interesting that lysosome-like structures such as dense bodies, GROD-like inclusions, and autophagosome-like structures themselves were often surrounded, along with part of the cytoplasm, by membrane saccules (Figure 2, A–C; Figure 3, A–D), indicating that autophagosome formation is abundant. In addition to GROD-like inclusions and AVs, fingerprint profiles were seen in CD−/− neurons but not in CB−/−CL−/− neurons (Figure 3F). In CD+/− or CB+/−CL+/− mouse neurons, typical dense bodies were detected (Figure 3C), while autophagosome-like structures were only occasionally seen. In these control mouse neurons, however, GROD-like inclusions were totally absent (Supplemental Figure 3 at http://ajp.amjpathol.org). These results suggest that the common neuropathological findings of these two mutant mice are the accumulation of GROD-like inclusions and autophagosome-like structures in neuronal perikarya. Neuropathological findings of CD−/− and CB−/−CL−/− mouse brains are summarized in Table 1.
The Volume Density of Autophagosome-Like Inclusion Bodies Increases with Age in CD−/− and CB−/−CL−/− Mouse Neuronal Perikarya
To understand characteristics of inclusions in CD−/− and CB−/−CL−/− mouse neurons, the volume density (percent volume) of various lysosomal structures in the neuronal perikarya was analyzed using the point counting method. According to the morphological criteria as shown in Figure 3G, the volume densities of AVi, AVd, GROD-like inclusions, fingerprint profiles, and dense bodies were determined. In CD+/− neurons, the volume density of dense bodies gradually increased and became 2.5 ± 0.48% (mean ± SE) at P23 (Figure 3G). Although autophagosomes were also detected in these control neurons, the volume densities of AVi and AVd were very low and 0.39 ± 0.19% and 0.37 ± 0.16% at P23, respectively (Figure 3G). In CD−/− neurons, dense bodies were hardly seen from P8 to P23, while, as stated above, GROD-like inclusions was abundant and its density changed from 4.9 ± 0.62% at P8 to 20.5 ± 1.63% at P23 (Figure 3G). As for AVi and AVd, their volume densities increased rapidly from P15 to P23; in particular, the volume density of AVd at P23 was 5.94 times larger than that at P15 and was 4.48 ± 0.7%, while that of AVi at P23 was 3.6 times higher than that at P15 and was 10.1 ± 1.31% (Figure 3G). As shown in Figure 2D, GROD-like inclusions was often enwrapped by double-membrane saccules together with part of the cytoplasm and half of AVi contained GROD-like inclusions at P23, while 20% of GROD-like inclusions was found in AVi. In addition to GROD-like inclusions, fingerprint profiles that were also characteristic ultrastructures in NCLs, showed their volume density as 6.7 ± 0.97% at P23 (Figure 3, F and G). Moreover, the volume density of total lysosomal structures at P23 was 3.3% in control mouse neurons, whereas it reached nearly 42% in CD−/− neurons. These data suggest that abundant autophagosome formation in CD−/− mouse neurons from P15 to P23 contributes to the accumulation of lysosomal structures.
In CB+/−CL+/− neurons, the volume density of dense bodies was 1.8 ± 0.18% (mean ± SE) at P13, while that of AVd was very low (0.32 ± 0.18%) (Figure 3G). In CB−/−CL−/− neurons, dense bodies were hardly seen at P13, while the volume densities of AVi and AVd were 4.91 ± 3.61% and 1.14 ± 1.28%, respectively (Figure 3G). As stated above, GROD-like inclusions were abundant and its density reached 24.1 ± 0.53% at P13; this value corresponded well with that in CD−/− neurons at P23 (Figure 3G). Like CD−/− neurons, GROD-like inclusions in CB−/−CL−/− neurons were often enwrapped by double-membrane saccules together with part of the cytoplasm and ∼75% of AVi contained GROD-like inclusions at P13, while 15% of GROD-like inclusions were found in AVi. The volume density of total lysosomal structures at P13 was 2.1% in control mouse neurons, whereas it reached nearly 30% in CB−/−CL−/− neurons. These data suggest that like CD−/− neurons, the contribution of abundant autophagosome formation to the accumulation of lysosomal structures is also evident in CB−/−CL−/− mouse neurons.
The Level of Autophagosome-Associated LC3-II Increases in CD−/− and CB−/−CL−/− Mouse Brains
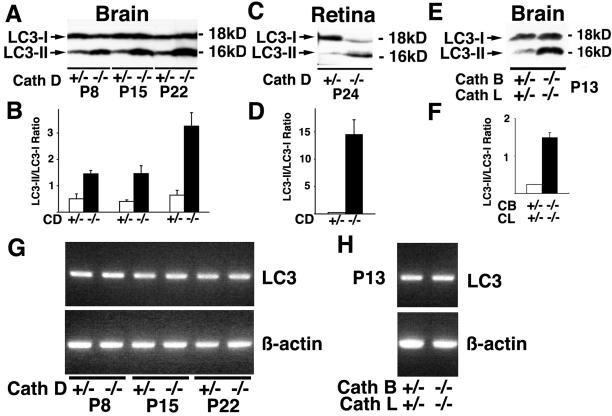
LC3-I, a cytosolic form, is known to be converted to LC3-II, a membrane-bound form when autophagy is induced.14 Because numerous autophagosome-like structures were detected in the perikarya of mutant mouse brains, we examined the molecular forms of LC3 in the tissues by Western blotting. As shown in Figure 4, A and B, LC3-I predominated in control brains obtained at any of the stages examined, whereas the LC3-II form gradually increased in CD−/− mouse brains in an age-dependent manner. In fact, the ratios of the amounts of LC3-II to LC3-I were significantly higher in the CD−/− mouse brains than in the control brains after P8 (Figure 4B). An increase in the amount of the LC3-II form was also found in CD−/− retina at P24 when massive retinal degeneration occurred (Figure 4, C and D).21 We also examined changes in the molecular forms of LC3 in CB+/−CL+/− and CB−/−CL−/− mouse brains at P13. Like CD−/− mouse brains, the LC3-II form predominated only in brain extracts obtained from CB−/−CL−/− mice (Figure 4, E and F).
Figure 4.
Molecular forms of LC3 in nervous tissues of CD−/− and CB−/−CL−/− mice and their littermate controls. A and B: Immunoblotting of LC3 in brains of CD−/− mice and their littermate controls (CD+/−) obtained at P8, P15, and P22 (A), and ratios of densities of LC3-II to LC3-I protein bands (B). C and D: Immunoblotting of LC3 in retinae of CD−/− mice and their littermate controls (CD+/−) obtained at P24 (C), and ratios of densities of LC3-II to LC3-I protein bands (D). E and F: Immunoblotting of LC3 in brains from CB−/−CL−/− and CB+/−CL+/− mice at P13 (E), and the ratios of densities of LC3-II to LC3-I protein bands (F). G and H: Expression of LC-3 mRNA in brain tissues of CD−/− and CB−/−CL−/− mice and their littermate controls. A RT-PCR study. Expression levels of LC-3 mRNA do not differ between CD−/− and CD+/− mice at all stages examined (G, top), and between CB−/−CL−/− and CB+/−CL+/− mice (H, top). Bottom panels show β-actin mRNA, used as internal controls.
Because molecular forms of LC3 in CD−/− mouse brains changed drastically from P8 to P23 and differed from those in CD+/− mouse brains at each stage, expression of LC3 mRNA in these mouse brains were also examined by RT-PCR. As shown in Figure 4G, no clear-cut changes in expression levels of LC3 mRNA were detected between CD−/− and CD+/− mouse brains at each stage and in CD−/− mouse brains between stages. Expression levels of LC3 mRNA were also analyzed between CB−/−CL−/− and CB+/−CL+/− mouse brains at P13, but no difference was discerned between them (Figure 4H). These results indicate that the cytosolic form of LC3 is predominant in intact brains at any of the stages examined, while the membrane-bound form of LC3 became a major form in both CD−/− and CB−/−CL−/− brains at their terminal stages, respectively.
Immunolocalization of LC3 in CD−/− and CB−/−CL−/− Mouse Brains
Because autophagosome-like structures were abundant in mutant neurons and the membrane-bound form of LC3 became predominant in these mouse brains, we examined the localization of LC3 in control and mutant mouse brains using an anti-LC3 antibody. In control mouse brains, diffuse and/or fibrillary staining of LC3 was detectable in the dendrites and perikarya of cerebral cortical neurons (Figure 5A, inset), as has been described previously for rat cerebellum.37 In dendrites of CD−/− neurons at P23, the staining patterns were similar to those of the control neurons, whereas intense granular staining of LC3 was detected in the perikarya of neurons in the cerebral cortex of CD−/− mice (Figure 5B, inset).
Figure 5.
Immunohistochemical staining of LC3 in brain tissues of CD−/− and CB−/−CL−/− mice and their littermate controls. A and B: Cerebral cortical regions of CD+/− (A) and CD−/− (B) mouse brains at P24. Positive staining of LC3 appears fibrillary in dendrites of pyramidal neurons and diffuse in their perikarya (A, inset). Its staining is also fibrillary in dendrites of the neurons but appears granular in their perikarya (B, inset). C and D: Purkinje cells in the cerebellum of CB+/−CL+/− (C) and CB−/−CL−/− (D) mice at P12. Fibrillary staining of LC3 is demonstrated in dendrites of Purkinje cells from both control and mutant mice (C, D), whereas its staining is relatively weak and diffuse in the perikarya of CB+/−CL+/− control neurons (C, inset) but intense and granular in those of CB−/−CL−/− neurons (D, inset). E and F: Nerve fibers in the corpus callosum of CB+/−CL+/− (E) and CB−/−CL−/− (F) mice at P12. No clear-cut staining for LC3 can be seen in the control axons (E), whereas intensely stained dots are accumulated in the mutant axons. Scale bars: 50 μm (A–F); 20 μm (insets in A–D).
In CB−/−CL−/− mice, immunopositive granules for LC3 were abundantly demonstrated in the perikarya of Purkinje cells in the cerebellum (Figure 5D, inset) and cerebral cortical neurons (data not shown), whereas diffuse and fibrillary staining was distinct in the perikarya and dendrites of Purkinje cells (Figure 5C, inset) and cerebral cortical neurons of CB+/−CL+/− mice. Because autophagosome-like vacuoles were abundantly shown in axons in the corpus callosum of CB−/−CL−/− mouse brains (Figure 2, D and E), immunostaining for LC3 was applied to the region; numerous LC3-positive granules were demonstrated in the CB−/−CL−/− axons (Figure 5F), but no positive staining of LC3 was detected in the axons of the control corpus callosum (Figure 5E).
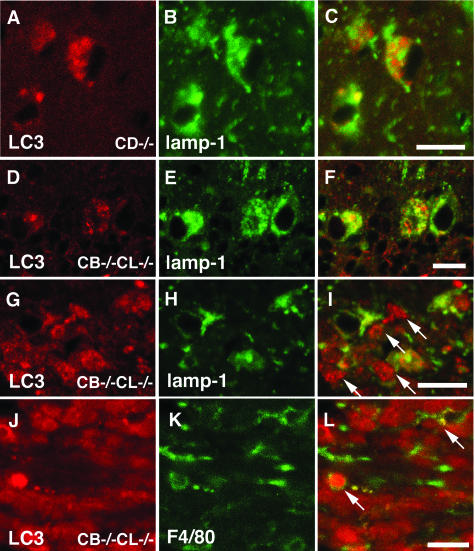
We further examined whether these LC3-positive granules in the neuronal perikarya of CB−/−CL−/− neurons also possess lysosomal characteristics by double-immuno-fluorescent staining for LC3 and lamp1, an abundant lysosomal integral membrane protein. Even in the CD−/− mouse brains at P23, it was possible to find cerebral cortical regions that did not emit autofluorescence. As shown in Figure 6, A–F, some LC3-positive granules were also stained for lamp1 in the cytoplasm of cerebral cortical neurons and cerebellar Purkinje cells in CD−/− and CB−/−CL−/− mice, but a considerable number of LC3-positive granules, which were relatively large in size and intensity, were distinct from lamp1-positive granules, indicating that these granules were nascent, because they are free from any lysosomal characteristics. These results suggest that the number of LC-3-immunopositive autophagosomes/autophagolysosomes is increased in neuronal cells of CD−/− and CB−/−CL−/− mouse brains.
Figure 6.
Double-immunofluorescent staining of LC3 (A, D, G, J; red color) with lamp1 (B, E, H; green color) in cerebral cortical neurons (A–C) of CD−/− mice at P23, cerebellar Purkinje cells (D–F), and the corpus callosum (G–I) of the CB−/−CL−/− mice at P13 or with F4/80 (J, green color) in the corpus callosum (J–L) of the CB−/−CL−/− mice at P13. A–F: A considerable number of LC3-positive granules are also stained for lamp1, but certain numbers of LC3-positive granules (arrows), which are large in size and stain intensely, are distinct from lamp1 (C and F, overlay). G–I: LC3-positive staining in the corpus callosum (arrows) is primarily distinct from lamp1 staining (I, overlay). J–L: F4/80-positive microglial cells possess LC3-positive staining (arrows) (L, overlay). Scale bars, 15 μm.
In addition to the neuronal perikarya, the accumulation of autophagosome-like structures was shown in myelinated and unmyelinated axons and microglia-like cells in the corpus callosum of the mouse brain (Figure 2, D and E), double-immunofluorescent staining for LC3 and lamp1 or F4/80 was performed in the region. Most LC3-positive structures were negative for lamp1 in the corpus callosum (Figure 6, G–I), whereas F4/80-positive microglial cells occasionally enwrapped LC3-positive granules (Figure 6, J–L). These data suggest that accumulated vacuolar structures in axons of the corpus callosum of the mutant mouse brains are nascent autophagosomes and microglial cells invaded into the region possess heterophagosomes containing autophagosomes.
LC3 Is Localized on the Isolation Membrane of Autophagosomes in Neurons of CD−/− and CB−/−CL−/− Mouse Brains
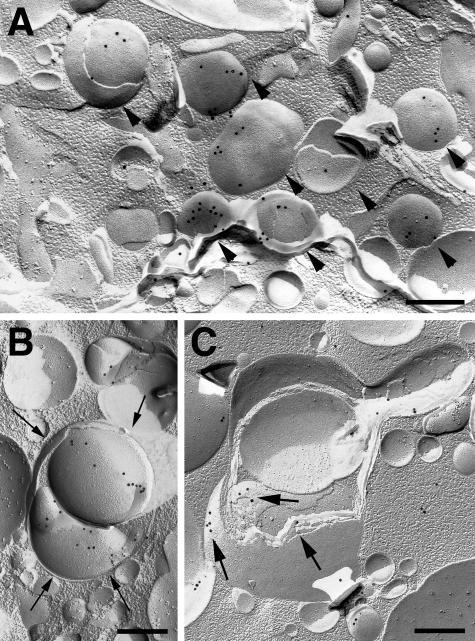
Because LC3-immunopositive granules were abundant in neurons of mutant mouse brains, we further examined the precise localization of LC3 by immunoelectron microscopy. We first attempted to use immunoelectron microscopy using postembedding and cryothin section immunogold methods, but were not able to realize positive signals. We therefore used the SDS-FRL method that was originally developed to study the two-dimensional distribution of integral membrane proteins and phospholipids in biomembranes, using weakly fixed materials.25 As shown in Figure 7, A–C, specific gold particles indicative of LC3 were clearly demonstrated on the membranes of perikaryal granules in anterior spinal neurons and cerebellar Purkinje cells of CD−/− mice. The membranes of the granules that were labeled with gold particles for LC3 possessed almost no intermembrane particles (Figure 7A). In some cases, granular structures with membranes labeled by the gold particles were further enwrapped by membrane structures (Figure 7B), while positive deposits were found on membranes of multilamellar bodies (Figure 7C). Considering the fact that granules with negligible numbers of intermembrane particles and multilayered vacuoles are typical of autophagosomes when observed by freeze-fracture electron microscopy, these data suggest that LC3, probably the LC3-II form, is specifically localized on the isolation membrane of autophagosomes in mutant mouse neurons.
Figure 7.
Immunocytochemical freeze-fracture replica images of an anterior spinal neuron (A) and cerebellar Purkinje cells (B, C) of CD−/− mice at P20 incubated with an anti-LC3 antibody. A: Immunogold labeling is observed on the membrane-bound organelles accumulated in the cytoplasm of the anterior neuron (arrowheads). B and C: In the perikarya of Purkinje cells, positive signals are well localized on membranes of two granules that are further encircled by membrane structures (thin arrows) (B), while they are also detected on multilamellar membranes (thick arrows). Scale bars, 0.25 μm.
GFP-LC3 Is Converted into LC3-II and Labels Accumulated Autophagosomes in GFP-LC3/CD−/− Mouse Brains
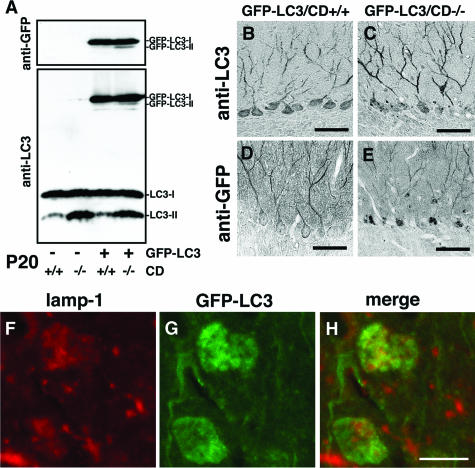
Using GFP-LC3 transgenic mice,23 we generated CD−/− mice expressing GFP-LC3 to further confirm the accumulation of autophagosomes in neurons of CD−/− mouse brains. Like in CD+/− and CD−/− mouse brains, the LC3-I form was a major form in GFP-LC3/CD+/+ mouse brains, while the LC3-II form became increased in GFP-LC3/CD−/− mouse brains (Figure 8A). The ratios of protein amounts of LC3-II to LC3-I were examined, and found to be similar between CD+/− and GFP-LC3/CD+/+ mouse brains or CD−/− and GFP-LC3/CD−/− mouse brains. In the case of GFP-LC3, the amount of GFP-LC3-I was much larger than that of GFP-LC3-II in both GFP-LC3/CD+/+ and GFP-LC3/CD−/− mouse brains. However, GFP-LC3-II distinctly appeared in GFP-LC3/CD−/− brains, but only faintly in GFP-LC3/CD+/+ brains (Figure 8A).
Figure 8.
Expression of GFP-LC3 in brains of GFP-LC3/CD−/− mice. A: Molecular forms of GFP-LC3 in brains of GFP-LC3/CD+/+ and GFP-LC3/CD−/− mice at P20. GFP-LC3-I is a major form in both GFP-LC3/CD+/+ and GFP-LC3/CD−/− brains, whereas GFP-LC3-II appears faintly in GFP-LC3/CD+/+ brains and distinctly in GFP-LC3/CD−/− brains when immunostained with either anti-GFP or anti-LC3. Moreover, the quantitative relationship of LC3-II to LC3-I in brains of CD+/+ and CD−/− mice does not differ depending on GFP-LC3 expression. B–E: Immunohistochemical images of GFP-LC3 and LC3 (B and C), or GFP-LC3 (D and E) in cerebellar Purkinje cells of GFP-LC3/CD+/+ (B and D) and GFP-LC3/CD−/− (C and E) mice at P20. Immunoreactivity for LC3-GFP and LC3, that was immunoreacted with anti-LC3, is more distinct and intense in dendrites and bodies of Purkinje cells than that for GFP-LC3 that was immunostained with anti-GFP. Diffuse or fibrillary staining for GFP-LC3 and LC3, or GFP-LC3 is clear in dendrites and bodies of Purkinje cells from a GFP-LC3/CD+/+ mouse, whereas granular staining for them (spheroids, arrows) is observed not only in Purkinje cell bodies but also in their dendrites from a GFP-LC3/CD−/− mouse. F–H: Double-fluorescence images of lamp1 (red color, F) and GFP-LC3 (green, G) in cerebral cortical neurons of GFP-LC3/CD−/− brains at P20. Lamp1 immunoreactivity is often localized in GFP-LC3-positive granules, but a considerable number of GFP-LC3-positive granules are distinct from lamp1 immunoreactivity (H). Scale bars: 50 μm (A–E); 10 μm (F–H).
When immunostained with either anti-LC3 or anti-GFP, intense fibrillary staining was detected in dendrites of cerebellar Purkinje cells in both GFP-LC3/CD+/+ and GFP-LC3/CD−/− mice (Figure 8, B–E), while granular staining appeared in the perinuclear region of the cells in GFP-LC3/CD−/− mice (Figure 8, C and E). The positive staining in both GFP-LC3/CD+/+ and GFP-LC3/CD−/− mouse brains, however, was much more intense when stained with anti-LC3 than that when stained with anti-GFP (Figure 8, B–E), because anti-LC3 recognizes both endogenous LC-3 and GFP-LC3. In some cases, granular immunodeposits for LC3/GFP-LC3 were detected in dendrites of GFP-LC3/CD−/− Purkinje cells (Figure 8, C and E). To confirm the relationship between lysosomes and autophagosomes, GFP-LC3/CD−/− mouse cortical neurons were stained for lamp1, and found that lamp1 immunoreactivity was often localized in GFP-LC3-positive granules, but a considerable number of GFP-LC3-positive granules were free from lamp1 immunoreactivity (Figure 8, F–H). These data confirmed the enormous accumulation of autophagosomes in neurons of CD−/− mouse brains.
LC3 in Peripheral Tissues of CD−/− and CB−/−CL−/− Mice
The accumulation of lysosome-like structures immuno-positive for subunit c has been previously shown in peripheral tissue cells of CD−/− mice, including hepatocytes and cardiac muscles.11 We therefore examined the protein amounts of LC3-I and LC3-II in the liver and heart of both CD+/− and −/− mice, and the localization of LC3 in these tissue cells.
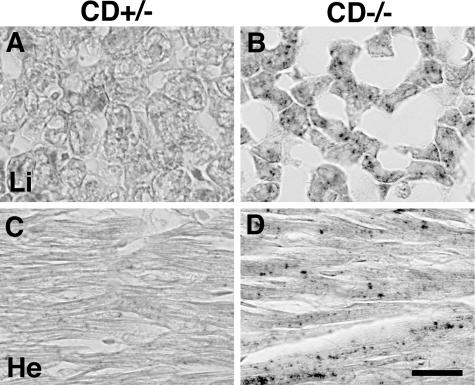
Different from brain tissues, LC3-I was only weakly detected in hepatic (Figure 9, A and B) and cardiac (Figure 8, C and D) tissues from CD+/− mice, and almost absent in those from CD−/− mice. On the contrary, the amounts of LC3-II were high in hepatic (Figure 9, A and B) and cardiac (Figure 9, C and D) tissues of CD−/− mice even at P15, compared to those of CD+/− mice, respectively. The LC3-II form was even more elevated in the CD−/− tissues at P22 than in the CD+/− tissues. Immunohistochemical observations showed that granular staining of LC3 was distinctly detected in hepatocytes and cardiac muscle cells of CD−/− mice, but not in those of CD+/− mice (Figure 10, A–D).
Figure 9.
Expression of LC3 in peripheral tissues of CD−/− (A–E) and CB−/−CL−/− (F, G) mice and their littermate controls (CD+/−, CB+/−CL+/−). A and B: Molecular forms of LC3 in the liver of CD+/− and CD−/− mice at P15 and P22 (A), and densities of LC3-II (B). C and D: Molecular forms of LC3 in the heart of CD+/− and CD−/− mice at P15 and P22 (C), and densities of LC3-II (D). E and F: Molecular forms of LC3 in the liver and heart of CB+/−CL+/− and CB−/−CL−/− mice at P13 (E), and densities of LC3-II (F).
Figure 10.
Immunostaining of LC3 in hepatic (A, B) and cardiac (C, D) tissues of CD+/− (A, C) and CD−/− (B, D) mice at P23. Punctate staining for LC3 is distinct in hepatocytes and cardiac muscular cells of CD−/− mice. Scale bar, 20 μm.
In addition to peripheral tissues from CD−/− mice, the molecular forms of LC3 were also examined in cardiac and hepatic tissues from CB−/−CL−/− and CB+/−CL+/− mice at P13. Like CD−/− mouse tissues, the amounts of LC3-II were significantly high in both cardiac and hepatic tissues from the CB−/−CL−/− mice, compared to those from CB+/−CL+/− mice (Figure 9, E and F).
Discussion
The main findings of the present study using electron microscopic, laser scanning microscopic, and biochemical techniques included the following: 1) many AVs and GROD-like inclusions occupied neuronal perikarya at terminal stages of CD−/− and CB−/−CL−/− mouse brains; 2) like CD−/− brains, neurons and microglial cells in CB−/−CL−/− brains showed positive staining of subunit c and autofluorescence; 3) AVs in which GROD-like inclusions, together with part of the cytoplasm, were encircled by double-membrane saccules were often observed in these cathepsin-deficient neurons; 4) massive staining for LC3 was intensely observed in neuronal perikarya and dendrites of these mutant brains; 5) in axons of the corpus callosum of these mutant mice, LC3-positive granules were not co-localized with lamp1, while these AVs were primarily encircled by double-membrane saccules; 6) the ratio of the amounts of LC3-II to LC3-I became distinctly high at the terminal stages of these mutant mice by Western blotting, while no clear changes were detected in the amounts of LC3 mRNA; and 7) LC3 molecules were clearly localized on the membrane of autophagosomes when examined by the SDS-FRL method. Based on these data, neuropathological features of these mutant mice are summarized in Table 1.
Autophagy Is Involved in the Accumulation of Lysosomal Compartments in Neurons of CD−/− and CB−/−CL−/− Mouse Brains
The present study showed that LC3-II predominated in CD−/− and CB−/−CL−/− mouse brains at their terminal stages and LC3-immunopositive AVs accumulated in the perikarya of cathepsin-deficient neurons. Until recently, no study has shown the presence of endogenous LC3 on the membrane of autophagosomes in tissue cells by electron microscopy using the immunogold method, except for the present SDS-FRL method. Indeed, numerous autophagosomes, showing an LC3 signal on their outer membrane, were present in the neuronal perikarya when brain tissue was analyzed by the SDS-FRL method. Because LC3-II is present on the isolation membrane of autophagosomes,14 these autophagosomes are likely to be nascent (AVi). This notion is also supported by the fact that LC3-positive granules in the perikarya of mutant neurons were often distinct from lamp1-positive lysosomes.
It has been shown that autophagy participates in neurodegenerative disorders such as Huntington’s disease,38–40 Alzheimer’s disease,41–43 Parkinson’s disease,44 and transmitted spongiform encephalopathy.45–47 The involvement of autophagy has also been demonstrated in lysosomal storage disease such as Pompe’s disease.48,49 The cerebral cortical neurons of CD−/− mouse brains show a new form of lysosomal accumulation disease with a phenotype resembling NCL (Batten disease).11,12,21 The present morphometric study on these CD−/− neurons revealed that lysosomal compartments including GROD-like inclusions and fingerprint profiles, which are typical hallmarks in NCL neurons, and AVs occupied nearly 42% of the perikaryal volume at P23. The volume density of GROD-like inclusions rapidly increased from P15 to P23, corresponding well with increases in volume densities of AVi and AVd, while nearly half of AVi contained GROD-like inclusions in CD−/− neurons at P23. In cerebral cortical neurons of CB−/−CL−/− brains at P13, GROD-like inclusions and AVs were found to occupy nearly 30% of the perikaryal volume, while ∼75% of AVi contained GROD-like inclusions. These data strongly support the view that autophagy is involved in the accumulation of lysosomal compartments in CD−/− and CB−/−CL−/− neurons; in particular, GROD-like inclusions are likely to be derived from AVi through AVd.
It has previously been shown that neuropathological features of CB−/−CL−/− mice resemble a lysosomal storage disorder that is distinct from classical NCL and subunit c is not found in accumulated lysosomes of the mutant neurons.13 However, the present study demonstrated that ultrastructural and morphometric features of CB−/−CL−/− neurons that possessed numerous AVs and GROD-like inclusions were very similar to those of CD−/− neurons. Moreover, immunoreactivity for subunit c and autofluorescence were detected in neurons and F4/80-immunopositive microglial cells of CB−/−CL−/− brains, while neuronal cells in these mutant mouse brains were intensely stained for periodic acid-Schiff. It has been shown that administration of cysteine proteinase inhibitors such as leupeptin and E64 induces the accumulation of lysosomes with ceroid-lipofuscin in brain cells of young rats.50 At present, it remains unknown whether subunit c is one of common substrates of cathepsins B and L. However, because CB and CL are major cysteine proteinases in lysosomes of neurons and microglial cells,26 it is reasonable to assume that the loss of these two proteinases in part suppresses the degradation of subunit c in lysosomes of neurons and microglial cells. From these results it seems likely that the pathological configuration of CB−/−CL−/− mice is very similar to that of NCLs.
Moreover, neuropathological features were primarily similar between CD−/− and CB−/−CL−/− mouse brains. In particular, the volume density of GROD-like inclusions was very similar between CD−/− mice at P23 and CB−/−CL−/− mice at P13, indicating that neuropathological damages were much severer in CB−/−CL−/− brains than in CD−/− brains when considering age difference between the two mutant mice. However, fingerprint profiles were not observed in CB−/−CL−/− mouse neurons but in CD−/− mouse neurons. Such a difference may be attributed to the difference in accumulated/undegraded substances in lysosomal compartments between the two mutant mouse neurons.
No accumulation of typical storage lysosomes has been reported in either CB−/− and CL−/− mouse tissue cells, respectively.18,20 This indicates that CB and CL, respectively, are dispensable in lysosomal proteolysis. In CB−/−CL−/− mouse CNS neurons, however, some common substrates of CB and CL, although unknown at present, are not degraded by other lysosomal proteinases, resulting in the accumulation of GROD-like inclusions and AVs in the neurons. It has been shown that the amount of the LC3-II form is increased in cultured cells treated with chloroquine, which raises pH in lysosomes,51 this indicating that autophagy is enhanced in these cells. These lines support the view that the presence of undigested substrates in lysosomes such as subunit c in CD−/− mice and unknown substrates in CB−/−CL−/− mice induces autophagy. In other words, the presence of GROD-like inclusions and AVs with undigested materials such as GROD-like inclusions may be a potent inducer of autophagy in neuronal cells. Until recently, it remains primarily unknown what signaling is essential for autophagosome formation. It has recently been shown that in conditional Atg7-knockout mice in which autophagy is absent specifically in the liver, numerous ubiquitinated aggregates are detected in the cytosol of hepatocytes with the presence of functional proteasomes.52 This suggests that protein ubiquitination may serve as a signal to the autophagic process in addition to the proteasomes pathway.52 The presence of ubiquitin-positive inclusion bodies is one of the common pathological characteristics of neurodegenerative diseases including lysosomal storage disorders.53,54 Therefore, it may be worthwhile to analyze the signaling pathway of autophagosome formation by use of cathepsin-deficient mice in which autophagosomes are abundantly present.
The present RT-PCR study demonstrated that the amounts of the LC3 gene transcript did not vary between cathepsin-deficient and control brains at various stages examined, indicating that changes in amounts of LC3 molecules depend primarily on the posttranslational events. In intact brains, LC3-I was a major form at each stage examined, while the amounts of LC3-II were larger in CD−/− brains at P8, P15, and P23, and in CB−/−CL−/− brains at P13 than those of LC3-I, respectively. It has recently been shown using HeLa cells and primary hepatocytes that, during starvation-induced autophagy, the significant amount of LC3-II is degraded by lysosomal proteinases and increased when inhibited by cysteine and aspartic proteinase inhibitors.55,56 In other words, LC3 is likely to be a substrate of CD and a common substrate of CB and CL. Moreover, the increase in the LC3 level by such proteinase inhibitors does not occur under nutrient-rich conditions.55,56 From these lines of evidence, it is possible to assume that the significant increases in amounts of LC3-II in cathepsin-deficient brains at each stage are attributed to the fact that autophagy is facilitated and the degradation of LC3 in autophagolysosomes is in part suppressed.
The Accumulation of Nascent AVs in Axons of the Corpus Callosum Is One of the Pathological Findings of CD−/− and CB−/−CL−/− Mouse Brains
Under various pathological conditions, vacuolar structures with double and single membranes, multilamellar bodies, and dense bodies have been demonstrated to appear in axons of CNS tissues and cultured neurons.47,57–60 In addition to these ultrastructural analyses and monodansylcadaverine staining for the demonstration of AVs in axons of CNS tissues, the present study showed that AVs found in axons of CD−/− and CB−/−CL−/− mouse brains were immunopositive for LC3 and negative for lamp1 by double immunostaining and primarily encircled by ER-like membrane saccules. These data indicate the followings: 1) there are machineries required for autophagosome formation in axons of brain tissue, 2) AVs in axons of the corpus callosum in CD−/− and CB−/−CL−/− brains are primarily nascent, and 3) the accumulation of AVs in the axons that may correspond well with meganeurites/spheroids could be one of pathological findings in these mutant mouse brains resembling NCLs. These AVs formed in distal axons usually undergo regulated transport back to the cell body, although it remains unknown how and where such nascent AVs in the axons are acidified and the degradation starts. However, the present results showing enormous accumulation of AVs in axons of the mutant neurons suggest that retrograde axonal transport is impeded in these mutant neurons.
Moreover, the present immunohistochemical study on LC3 demonstrated that the accumulation of AVs was also found in dendrites of the cerebellar and cerebral cortical neurons from the cathepsin-deficient mouse brains. These accumulated AVs did not differ from those in the perikarya of neurons from the mutant mouse brains when analyzed by electron microscopy (Supplemental Figure 2 at http://ajp.amjpathol.org). Recently, autophagosomes/AVi, multivesicular bodies, multilamellar bodies, and cathepsin-containing autophagolysosomes have been shown to be the predominant organelles and accumulate in large number in dystrophic neurites of brains from Alzheimer’s disease.61 Considering the fact that various types of AVs including cathepsin-positive autophagolysosomes are present in dystrophic neurites,61 such AVs detected in the case of AD may be different from those in axons of cathepsin-deficient brains and, if anything, resemble those in dendrites of the mutant mouse brains. Although the acidification and transport system may be dependent primarily on disease situations, the accumulation of AVs in the cases of cathepsin-deficient brains is apt to occur earlier and more frequently in axons than in dendrites.
The Conversion of LC3-I to LC3-II and Positive Staining of LC3 Are Also Excellent Markers for Induction of Autophagy in Brains
The behavior of LC3-II has been defined in murine embryonic stem cells,62 the soleus muscles of chloroquine-treated rats,51 in rat podocytes,56 and in embryonic tissue cells of lamp1 and lamp2 double-deficient mice.63 Based on experiments using transgenic mice systemically expressing LC3 fused to green fluorescent protein (GFP-LC3), it has been proposed that the regulation of autophagy is organ-dependent and the role of autophagy is not restricted to the starvation response.23 In most peripheral tissue cells, autophagy is distinctly induced under fasting conditions, whereas starvation does not induce autophagy in brain tissue.23 Although the question of why autophagy is not induced in brain tissue after starvation has not yet been answered, it is likely that ischemia,26 proteinase inhibitors such as leupeptin,60 and lysosomal proteinase deficiencies, as shown in the present study, enhance autophagy activity in CNS neurons. In fact, in GFP-LC3-expressing CD−/− mouse brains, GFP-LC3 was converted from GFP-LC3-I into GFP-LC3-II without interfering the conversion of endogenous LC3-I to LC3-II. More importantly, the accumulation of autophagosomes in CD−/− mouse brains, evidenced by electron microscopy and LC3 immunocytochemistry, was confirmed also by the finding that GFP-LC3-positive granules were accumulated in neuronal perikarya and dendrites of GFP-LC3/CD−/− mouse brains.
Collectively, the current data demonstrate that accumulated lysosomal structures with undigested substrates in neurons of CD−/− or CB−/−CL−/− mouse brains are primarily LC3-immunopositive, suggesting that these structures are derived from autophagosomes and also that autophagy is highly induced in these mutant mouse brains. These lines of evidence suggest that the activation of autophagy plays a pivotal role in the accumulation of lysosomal structures in CNS neurons of mouse models for NCL (Batten disease)/lysosomal storage disorders.
Supplementary Material
Footnotes
Address reprint requests to Yasuo Uchiyama, Department of Cell Biology and Neurosciences, Osaka University Graduate School of Medicine, 2-2 Yamadaoka, Suita, Osaka, Japan. E-mail: uchiyama@anat1.med.osaka-u.ac.jp.
Supported by the Ministry of Education, Science, Sports, and Culture, Japan [grants-in-aid for Creative Scientific Research (16GS0315 to Y.U.) and for Encouragement of Young Scientists (16790125 to M.S. and 17790141 to M.K.)]; the Fukushima Society for the Promotion of Medicine (to M.K.); the Kazato Research Foundation (to M.K.); and the Deutsche Forschungsgemeinschaft.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vittorini S, Paradiso C, Donati A, Cavallini G, Masini M, Gori Z, Pollera M, Bergamini E. The age-related accumulation of protein carbonyl in rat liver correlates with the age-related decline in liver proteolytic activities. J Gerontol A Biol Sci Med Sci. 1999;54:B318–B323. doi: 10.1093/gerona/54.8.b318. [DOI] [PubMed] [Google Scholar]
- Larsen KE, Sulzer D. Autophagy in neurons: a review. Histol Histopathol. 2002;17:897–908. doi: 10.14670/HH-17.897. [DOI] [PubMed] [Google Scholar]
- Uchiyama Y. Autophagic cell death and its execution by lysosomal cathepsins. Arch Histol Cytol. 2001;64:233–246. doi: 10.1679/aohc.64.233. [DOI] [PubMed] [Google Scholar]
- Yuan J, Lipinski M, Degterev A. Diversity in the mechanisms of neuronal cell death. Neuron. 2003;40:401–413. doi: 10.1016/s0896-6273(03)00601-9. [DOI] [PubMed] [Google Scholar]
- Ohsawa Y, Isahara K, Kanamori S, Shibata M, Kametaka S, Gotow T, Watanabe T, Kominami E, Uchiyama Y. An ultrastructural and immunohistochemical study of PC12 cells during apoptosis induced by serum deprivation with special reference to autophagy and lysosomal cathepsins. Arch Histol Cytol. 1998;61:395–403. doi: 10.1679/aohc.61.395. [DOI] [PubMed] [Google Scholar]
- Hall NA, Lake BD, Dewji NN, Patrick AD. Lysosomal storage of subunit c of mitochondrial ATP synthase in Batten’s disease (ceroid-lipofuscinosis). Biochem J. 1991;275:269–272. doi: 10.1042/bj2750269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kominami E, Ezaki J, Muno D, Ishido K, Ueno T, Wolfe LS. Specific storage of subunit c of mitochondrial ATP synthase in lysosomes of neuronal ceroid lipofuscinosis (Batten’s disease). J Biochem (Tokyo) 1992;111:278–282. doi: 10.1093/oxfordjournals.jbchem.a123749. [DOI] [PubMed] [Google Scholar]
- Palmer DN, Martinus RD, Cooper SM, Midwinter GG, Reid JC, Jolly RD. Ovine ceroid lipofuscinosis. The major lipopigment protein and the lipid-binding subunit of mitochondrial ATP synthase have the same NH2-terminal sequence. J Biol Chem. 1989;264:5736–5740. [PubMed] [Google Scholar]
- Fearnley IM, Walker JE, Martinus RD, Jolly RD, Kirkland KB, Shaw GJ, Palmer DN. The sequence of the major protein stored in ovine ceroid lipofuscinosis is identical with that of the dicyclohexylcarbodiimide-reactive proteolipid of mitochondrial ATP synthase. Biochem J. 1990;268:751–758. doi: 10.1042/bj2680751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike M, Nakanishi H, Saftig P, Ezaki J, Isahara K, Ohsawa Y, Schulz-Schaeffer W, Watanabe T, Waguri S, Kametaka S, Shibata M, Yamamoto K, Kominami E, Peters C, von Figura K, Uchiyama Y. Cathepsin D deficiency induces lysosomal storage with ceroid lipofuscin in mouse CNS neurons. J Neurosci. 2000;20:6898–6906. doi: 10.1523/JNEUROSCI.20-18-06898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H, Zhang J, Koike M, Nishioku T, Okamoto Y, Kominami E, von Figura K, Peters C, Yamamoto K, Saftig P, Uchiyama Y. Involvement of nitric oxide released from microglia-macrophages in pathological changes of cathepsin D-deficient mice. J Neurosci. 2001;21:7526–7533. doi: 10.1523/JNEUROSCI.21-19-07526.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felbor U, Kessler B, Mothes W, Goebel HH, Ploegh HL, Bronson RT, Olsen BR. Neuronal loss and brain atrophy in mice lacking cathepsins B and L. Proc Natl Acad Sci USA. 2002;99:7883–7888. doi: 10.1073/pnas.112632299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsumi Y. Molecular dissection of autophagy: two ubiquitin-like systems. Nat Rev Mol Cell Biol. 2001;2:211–216. doi: 10.1038/35056522. [DOI] [PubMed] [Google Scholar]
- Tanida I, Tanida-Miyake E, Ueno T, Kominami E. The human homolog of Saccharomyces cerevisiae Apg7p is a protein-activating enzyme for multiple substrates including human Apg12p, GATE-16, GABARAP, and MAP-LC3. J Biol Chem. 2001;276:1701–1706. doi: 10.1074/jbc.C000752200. [DOI] [PubMed] [Google Scholar]
- Tanida I, Tanida-Miyake E, Komatsu M, Ueno T, Kominami E. Human Apg3p/Aut1p homologue is an authentic E2 enzyme for multiple substrates, GATE-16, GABARAP, and MAP-LC3, and facilitates the conjugation of hApg12p to hApg5p. J Biol Chem. 2002;277:13739–13744. doi: 10.1074/jbc.M200385200. [DOI] [PubMed] [Google Scholar]
- Deussing J, Roth W, Saftig P, Peters C, Ploegh HL, Villadangos JA. Cathepsins B and D are dispensable for major histocompatibility complex class II-mediated antigen presentation. Proc Natl Acad Sci USA. 1998;95:4516–4521. doi: 10.1073/pnas.95.8.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saftig P, Hetman M, Schmahl W, Weber K, Heine L, Mossmann H, Koster A, Hess B, Evers M, von Figura K. Mice deficient for the lysosomal proteinase cathepsin D exhibit progressive atrophy of the intestinal mucosa and profound destruction of lymphoid cells. EMBO J. 1995;14:3599–3608. doi: 10.1002/j.1460-2075.1995.tb00029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Roth W, Wong P, Nelson A, Farr A, Deussing J, Villadangos JA, Ploegh H, Peters C, Rudensky AY. Cathepsin L: critical role in Ii degradation and CD4 T cell selection in the thymus. Science. 1998;280:450–453. doi: 10.1126/science.280.5362.450. [DOI] [PubMed] [Google Scholar]
- Koike M, Shibata M, Ohsawa Y, Nakanishi H, Koga T, Kametaka S, Waguri S, Momoi T, Kominami E, Peters C, Figura K, Saftig P, Uchiyama Y. Involvement of two different cell death pathways in retinal atrophy of cathepsin D-deficient mice. Mol Cell Neurosci. 2003;22:146–161. doi: 10.1016/s1044-7431(03)00035-6. [DOI] [PubMed] [Google Scholar]
- Saftig P, Peters C, von Figura K, Craessaerts K, Van Leuven F, De Strooper B. Amyloidogenic processing of human amyloid precursor protein in hippocampal neurons devoid of cathepsin D. J Biol Chem. 1996;271:27241–27244. doi: 10.1074/jbc.271.44.27241. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Dono K, Gotoh K, Shibata M, Koike M, Marubashi S, Miyamoto A, Takeda Y, Nagano H, Umeshita K, Uchiyama Y, Monden M. Participation of autophagy in the degeneration process of rat hepatocytes after transplantation following prolonged cold preservation. Arch Histol Cytol. 2005;68:71–80. doi: 10.1679/aohc.68.71. [DOI] [PubMed] [Google Scholar]
- Rash JE, Yasumura T. Direct immunogold labeling of connexins and aquaporin-4 in freeze-fracture replicas of liver, brain, and spinal cord: factors limiting quantitative analysis. Cell Tissue Res. 1999;296:307–321. doi: 10.1007/s004410051291. [DOI] [PubMed] [Google Scholar]
- Nitatori T, Sato N, Waguri S, Karasawa Y, Araki H, Shibanai K, Kominami E, Uchiyama Y. Delayed neuronal death in the CA1 pyramidal cell layer of the gerbil hippocampus following transient ischemia is apoptosis. J Neurosci. 1995;15:1001–1011. doi: 10.1523/JNEUROSCI.15-02-01001.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama Y, Watanabe M. A morphometric study of developing pancreatic acinar cells of rats during prenatal life. Cell Tissue Res. 1984;237:117–122. doi: 10.1007/BF00229206. [DOI] [PubMed] [Google Scholar]
- Liou W, Geuze HJ, Geelen MJ, Slot JW. The autophagic and endocytic pathways converge at the nascent autophagic vacuoles. J Cell Biol. 1997;136:61–70. doi: 10.1083/jcb.136.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn WA., Jr Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J Cell Biol. 1990;110:1923–1933. doi: 10.1083/jcb.110.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y, Guhde G, Suter A, Eskelinen EL, Hartmann D, Lullmann-Rauch R, Janssen PM, Blanz J, von Figura K, Saftig P. Accumulation of autophagic vacuoles and cardiomyopathy in LAMP-2-deficient mice. Nature. 2000;406:902–906. doi: 10.1038/35022595. [DOI] [PubMed] [Google Scholar]
- Haltia M. The neuronal ceroid-lipofuscinoses. J Neuropathol Exp Neurol. 2003;62:1–13. doi: 10.1093/jnen/62.1.1. [DOI] [PubMed] [Google Scholar]
- Koike M, Shibata M, Ohsawa Y, Kametaka S, Waguri S, Kominami E, Uchiyama Y. The expression of tripeptidyl peptidase I in various tissues of rats and mice. Arch Histol Cytol. 2002;65:219–232. doi: 10.1679/aohc.65.219. [DOI] [PubMed] [Google Scholar]
- Fujimoto K. Freeze-fracture replica electron microscopy combined with SDS digestion for cytochemical labeling of integral membrane proteins. Application to the immunogold labeling of intercellular junctional complexes. J Cell Sci. 1995;108:3443–3449. doi: 10.1242/jcs.108.11.3443. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsawa Y, Zhang G, Kametaka S, Shibata M, Koike M, Waguri S, Uchiyama Y. Purification, cDNA cloning, and secretory properties of FLRG protein from PC12 cells and the distribution of FLRG mRNA and protein in rat tissues. Arch Histol Cytol. 2003;66:367–381. doi: 10.1679/aohc.66.367. [DOI] [PubMed] [Google Scholar]
- Dunn WA., Jr Autophagy and related mechanisms of lysosome-mediated protein degradation. Trends Cell Biol. 1994;4:139–143. doi: 10.1016/0962-8924(94)90069-8. [DOI] [PubMed] [Google Scholar]
- Mann SS, Hammarback JA. Gene localization and developmental expression of light chain 3: a common subunit of microtubule-associated protein 1A(MAP1A) and MAP1B. J Neurosci Res. 1996;43:535–544. doi: 10.1002/(SICI)1097-4547(19960301)43:5<535::AID-JNR3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- Roizin L, Stellar S, Willson N, Whittier J, Liu JC. Electron microscope and enzyme studies in cerebral biopsies of Huntington’s chorea. Trans Am Neurol Assoc. 1974;99:240–243. [PubMed] [Google Scholar]
- Tellez-Nagel I, Johnson AB, Terry RD. Studies on brain biopsies of patients with Huntington’s chorea. J Neuropathol Exp Neurol. 1974;33:308–332. doi: 10.1097/00005072-197404000-00008. [DOI] [PubMed] [Google Scholar]
- Kegel KB, Kim M, Sapp E, McIntyre C, Castano JG, Aronin N, DiFiglia M. Huntington expression stimulates endosomal-lysosomal activity, endosome tubulation, and autophagy. J Neurosci. 2000;20:7268–7278. doi: 10.1523/JNEUROSCI.20-19-07268.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cataldo AM, Hamilton DJ, Barnett JL, Paskevich PA, Nixon RA. Properties of the endosomal-lysosomal system in the human central nervous system: disturbances mark most neurons in populations at risk to degenerate in Alzheimer’s disease. J Neurosci. 1996;16:186–199. doi: 10.1523/JNEUROSCI.16-01-00186.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadelmann C, Deckwerth TL, Srinivasan A, Bancher C, Bruck W, Jellinger K, Lassmann H. Activation of caspase-3 in single neurons and autophagic granules of granulovacuolar degeneration in Alzheimer’s disease. Evidence for apoptotic cell death. Am J Pathol. 1999;155:1459–1466. doi: 10.1016/S0002-9440(10)65460-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu WH, Kumar A, Peterhoff C, Shapiro Kulnane L, Uchiyama Y, Lamb BT, Cuervo AM, Nixon RA. Autophagic vacuoles are enriched in amyloid precursor protein-secretase activities: implications for beta-amyloid peptide over-production and localization in Alzheimer’s disease. Int J Biochem Cell Biol. 2004;36:2531–2540. doi: 10.1016/j.biocel.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Anglade P, Vyas S, Javoy-Agid F, Herrero MT, Michel PP, Marquez J, Mouatt-Prigent A, Ruberg M, Hirsch EC, Agid Y. Apoptosis and autophagy in nigral neurons of patients with Parkinson’s disease. Histol Histopathol. 1997;12:25–31. [PubMed] [Google Scholar]
- Boellaard JW, Schlote W, Tateishi J. Neuronal autophagy in experimental Creutzfeldt-Jakob’s disease. Acta Neuropathol (Berl) 1989;78:410–418. doi: 10.1007/BF00688178. [DOI] [PubMed] [Google Scholar]
- Boellaard JW, Kao M, Schlote W, Diringer H. Neuronal autophagy in experimental scrapie. Acta Neuropathol (Berl) 1991;82:225–228. doi: 10.1007/BF00294449. [DOI] [PubMed] [Google Scholar]
- Jeffrey M, Scott JR, Williams A, Fraser H. Ultrastructural features of spongiform encephalopathy transmitted to mice from three species of bovidae. Acta Neuropathol (Berl) 1992;84:559–569. doi: 10.1007/BF00304476. [DOI] [PubMed] [Google Scholar]
- Baudhuin P, Hers HG, Loeb H. An electron microscopic and biochemical study of type Ii glycogenosis. Lab Invest. 1964;13:1139–1152. [PubMed] [Google Scholar]
- Novikoff AB. Lysosomes in nerve cells. Hyden H, editor. Amsterdam: Elsevier Publishing Company; The Neuron. 1967:pp 319–377. [Google Scholar]
- Ivy GO. Protease inhibitors as a model for NCL disease, with special emphasis on the infantile and adult forms. Am J Med Genet. 1992;42:555–560. doi: 10.1002/ajmg.1320420427. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Nakagawa M, Yoshikawa A, Sasagawa N, Yoshimori T, Ohsumi Y, Nishino I, Ishiura S, Nonaka I. The first molecular evidence that autophagy relates rimmed vacuole formation in chloroquine myopathy. J Biochem (Tokyo) 2002;131:647–651. doi: 10.1093/oxfordjournals.jbchem.a003147. [DOI] [PubMed] [Google Scholar]
- Komatsu M, Waguri S, Ueno T, Iwata J, Murata S, Tanida I, Ezaki J, Mizushima N, Ohsumi Y, Uchiyama Y, Kominami E, Tanaka K, Chiba T. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J Cell Biol. 2005;169:425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan SS, Beyreuther K, Schmitt HP. Neuronal ubiquitin and neurofilament expression in different lysosomal storage disorders. Clin Neuropathol. 1992;11:251–255. [PubMed] [Google Scholar]
- Ardley HC, Hung CC, Robinson PA. The aggravating role of the ubiquitin-proteasome system in neurodegeneration. FEBS Lett. 2005;579:571–576. doi: 10.1016/j.febslet.2004.12.058. [DOI] [PubMed] [Google Scholar]
- Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005;1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- Asanuma K, Tanida I, Shirato I, Ueno T, Takahara H, Nishitani T, Kominami E, Tomino Y. MAP-LC3, a promising autophagosomal marker, is processed during the differentiation and recovery of podocytes from PAN nephrosis. FASEB J. 2003;17:1165–1167. doi: 10.1096/fj.02-0580fje. [DOI] [PubMed] [Google Scholar]
- Hollenbeck PJ. Products of endocytosis and autophagy are retrieved from axons by regulated retrograde organelle transport. J Cell Biol. 1993;121:305–315. doi: 10.1083/jcb.121.2.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen KE, Fon EA, Hastings TG, Edwards RH, Sulzer D. Methamphetamine-induced degeneration of dopaminergic neurons involves autophagy and upregulation of dopamine synthesis. J Neurosci. 2002;22:8951–8960. doi: 10.1523/JNEUROSCI.22-20-08951.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WL, Lewis J, Yen SH, Hutton M, Dickson DW. Ultrastructural neuronal pathology in transgenic mice expressing mutant (P301L) human tau. J Neurocytol. 2003;32:1091–1105. doi: 10.1023/B:NEUR.0000021904.61387.95. [DOI] [PubMed] [Google Scholar]
- Bednarski E, Ribak CE, Lynch G. Suppression of cathepsins B and L causes a proliferation of lysosomes and the formation of meganeurites in hippocampus. J Neurosci. 1997;17:4006–4021. doi: 10.1523/JNEUROSCI.17-11-04006.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon RA, Wegiel J, Kumar A, Yu WH, Peterhoff C, Cataldo A, Cuervo AM. Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J Neuropathol Exp Neurol. 2005;64:113–122. doi: 10.1093/jnen/64.2.113. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J Cell Biol. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskelinen EL, Schmidt CK, Neu S, Willenborg M, Fuertes G, Salvador N, Tanaka Y, Lullmann-Rauch R, Hartmann D, Heeren J, von Figura K, Knecht E, Saftig P. Disturbed cholesterol traffic but normal proteolytic function in LAMP-1/LAMP-2 double-deficient fibroblasts. Mol Biol Cell. 2004;15:3132–3145. doi: 10.1091/mbc.E04-02-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.