Abstract
Age-associated differences in estrogen levels critically modify the cutaneous wound healing response. Using a microarray-based approach, we profiled changes in gene expression within the wounds of mice that were wild type or null for the pro-inflammatory cytokine macrophage migration inhibitory factor (MIF) in the presence or absence of estrogen. This experimental design identified more than 600 differentially expressed genes and established MIF as a key player in the wound healing process, regulating many novel repair/inflammation-associated gene targets. Moreover, MIF affected virtually all of the effects of reduced estrogen on wound repair. In humans, serum and wound levels of MIF increased with age and were strongly down-regulated by estrogen in vivo. Estrogen-regulated MIF transcription in vitro via a nuclear factor κB-dependent mechanism. These findings have wide-ranging implications for the many pathophysiological states in which MIF plays an important regulatory role and suggest a potential therapeutic role for MIF in modulating clinical conditions associated with age-related decline in estrogen levels.
Age-related delayed healing, associated with excessive inflammation and matrix degradation, creates a tremendous financial burden to health services worldwide, with costs for largely ineffective treatment exceeding $9 billion per annum in the U.S. We and others have shown that estrogen plays a fundamental role in regulating an appropriate healing response in humans of both sexes and in mice. Because systemic estrogen levels fall with age, precipitously in females and gradually in males, numerous local pro-inflammatory cytokines and growth factors are up-regulated, and the rate of healing declines.1–4 It is important to determine the downstream gene targets of estrogenic action to minimize the adverse systemic side effects (eg, malignancy, thrombosis, etc.) attributed to hormone replacement.
Ovariectomized (OVX) mice, by virtue of negligible systemic estrogen, provide a model of age-associated delayed healing.5 Wounds from these mice take substantially longer to heal and contain significantly increased numbers of inflammatory cells compared with intact mice.1 We recently demonstrated a role for macrophage migration inhibitory factor (MIF) in the delayed healing response of estrogen-depleted mice, because MIF-null mice heal normally in the absence of estrogen, and estrogen strongly down-regulates MIF in vitro and in vivo.6 MIF, a small pro-inflammatory cytokine identified as a central regulator of innate immunity and inflammation,7 has been implicated in a host of disease states including tumorigenesis, arthritis, atherosclerosis, and septic shock8–11 and is associated with tissue injury.12
To further investigate the roles of MIF and estrogen and to dissect their relative contributions to repair/inflammation, gene expression in day 3 cutaneous wounds from four groups of mice were compared using Affymetrix microarrays: MIF-null mice, with and without ovaries (both groups heal normally), and the corresponding MIF wild-type (WT) mice. Day 3 was chosen specifically because at this time point, we have demonstrated an up-regulation of MIF expression in wound tissue and markedly so in OVX mice.6 This approach allows the simultaneous comparison of expression of more than 22,000 probe sets (14,500 genes) in healing wounds from all four mouse groups. Our data suggest that estrogen regulates healing almost exclusively via MIF down-regulation and identifies novel MIF-regulated gene targets and clusters associated with aberrant healing. Moreover, genes involved in estrogen-mediated adverse effects, such as thrombosis and breast cancer, do not involve MIF. Human studies, performed to determine the relevance of our murine findings, showed up-regulation of both systemic and local MIF levels with age, a finding that can be entirely reversed by estrogen treatment. Finally, in vitro studies undertaken to address the mechanism by which estrogen modulates MIF levels demonstrate regulation at the level of transcription via a nuclear factor κB (NF-κB)-dependent mechanism. Taken together, these results suggest that modulation of MIF is a potential therapeutic target to accelerate impaired wound healing.
Materials and Methods
Wounding, Histology, and Immunohistochemistry
Local ethical committee approval was obtained for all human studies. Following informed consent, four healthy, 20- to 39-year-old males and eight health status-defined males (randomized to estrogen or placebo) underwent two 4-mm punch biopsies from the left upper inner arm after local infiltration with 1 ml of 1% lignocaine. The skin was covered by a 5- × 4-cm standardized adhesive hormone replacement patch (Evorel, active patch = 25 μg estradiol/24 hours, or identical placebo patch; Janssen Pharmaceuticals, High Wycombe, UK) through which the two punch biopsies were made. The patch was covered by a Multisorb dry gauze dressing (Smith & Nephew, Hull, UK), both were removed after 24 hours, and wounds were excised at day 7 after wounding. Ten-week-old female mice (MIF wild type, BALB/c, and MIF null, pure BALB/c, >N15) with intact ovaries and 10-week-old mice (MIF wild type, BALB/c, and MIF null, pure BALB/c, >N15) that had undergone ovariectomization 1 month previously were anesthetized and wounded following our established protocol6 (in accordance with home office regulations). Briefly, two equidistant 1-cm-full thickness skin incisional wounds were made through both skin and panniculus carnosus muscle and left to heal by secondary intention. Wounds were excised and bisected at days 3 and 7 after wounding, and one-half of the sample was processed for histology, allowing the midpoint of the wound to be compared between groups. The remaining one-half of each wound was flash frozen and stored at −80°C before RNA/protein extraction. Histological sections were prepared from wound tissue fixed in 10% buffered formal saline and embedded in paraffin. Five-micrometer sections were stained with hematoxylin and eosin or subjected to immunohistochemistry with anti-MIF goat polyclonal antibody (R&D Systems, Abingdon, UK) and biotinylated secondary antibody followed by ABC-peroxidase reagent (Vector Laboratories, Peterborough, UK) with Novared substrate and counterstaining with hematoxylin. Control slides stained with secondary antibody in isolation or control IgG were negative. Total cell numbers and wound area were quantified with Image Pro Plus software as previously described.6
Sample Preparation and Microarray Analysis
Total RNA was isolated from frozen wound tissue by homogenizing in Trizol reagent (Invitrogen, Paisley, UK) following the manufacturer’s instructions. Biotinylated cRNA samples from individual mice were hybridized to mouse 430A oligonucleotide arrays (Affymetrix, Inc., Santa Clara, CA). The balanced experimental design resulted in four conditions with three animals per condition (1 mouse per array). The microarray data were submitted in MIAME (Minimum Information About a Microarray Experiment)-compliant format to the ArrayExpress database (www.mged.org/Workgroups/MIAME/miame.html, accession number e-mexp-209). Our gene enrichment strategy used the following steps. Step 1, normalized array data (RMAExpress13 default settings) were subjected to a present/absent filter (expression value of >100 in at least one array; 58% of 22,627 probe sets passed). Step 2, expression data (log 2) were subjected to a two-factor analysis of variance (Genespring 5.0; Silicon Genetics) and filtered for MIF, OVX, or MIF/OVX interaction P values <0.1. Step 3, A filter was applied for >1.5-fold change between the means of any two sample groups (621 probes sets or 610 individual genes remained). This enriched dataset was segregated into 10 clusters based on similarity of expression profile across the dataset using a k-means clustering algorithm (“Slope” similarity metric “Super Grouper” plugin of maxdView software; see supplementary information online at http://ajp.amjpathol.org). Clustering was performed on the means of each sample condition (log 2) that had been z-transformed (for each probe set, the mean set to 0, SD to 1). Because two pairs of clusters had similar profiles, they were combined to leave the eight clusters presented here. For each cluster, overrepresented gene ontology (GO) groups were identified using the expression analysis systematic explorer (EASE) online tool.14
Quantitative Real-Time Polymerase Chain Reaction (PCR), Comparison with Microarrays, and Statistical Analysis
cDNA was transcribed from 1 μg of wound RNA (Promega RT kit and AMV-reverse transcriptase; Roche, Welwyn Garden City, UK). Quantitative real-time PCR was performed using the SYBR green core kit (Eurogentec, Southampton, UK) following manufacturer’s instructions and an Opticon qPCR thermal cycler (MJ Research). For each primer set an optimal dilution was determined, and melting curves were used to determine product specificity. Each sample was serially diluted over 3 orders of magnitude, and all samples were run on the same 96-well plate. Primer sequences are shown in Supplementary Table 1 (available online at http://ajp.amjpathol.org). Expression ratios were determined relative to a standard sample and normalized using a value derived from four separate control primer sets to 18s rRNA and the housekeeping genes Gapdh, Ywahz, and Hprt. To enable comparison with and validation of microarray data, for each gene, the mean of both real-time ratios and normalized microarray hybridization values was set to 1, and values below 0 were converted into negative-fold change.
Enzyme-Linked Immunosorbent Assay (ELISA) for MIF
ELISA was performed on serum extracted from blood samples collected from 8 premenopausal females, 16 postmenopausal females, and 10 hormone replacement therapy (HRT)-treated postmenopausal females using a coating antibody (monoclonal anti-MIF MAB289; R&D Systems) and a biotinylated anti-human MIF capture antibody (goat anti-human, BAF289; R&D Systems). Human peripheral circulating monocytes were isolated by magnetic cell sorting (Miltenyi Biotech, Bisley, UK) using negative selection to minimize cell activation. Cell viability was determined by Trypan blue. Cells were plated at 12 × 106 cells/ml with 100 μl/well in serum-free phenol red-free medium, treated with or without lipopolysaccharide (LPS; 1 μg/ml) for 6 hours, and washed with phosphate-buffered saline. Cells were then treated with estrogen (1 × 10−10, 1 × 10−9, or 1 × 10−8 mol/L) for 12 hours, and ELISA was performed as above. The NF-κB inhibitor pyrrolidine dithiocarbamate (PDTC) (60 μmol/L; Sigma, Surrey, UK) was co-incubated with LPS for a subgroup of cells.
Transfection of MIF-Promoter Reporter Construct and Luciferase Assay
A 1-kb fragment was derived from the MIF promoter (GenBank AF033192) using 129/SvJ ES DNA as a template. The fragment containing nucleotides 67 to −1029 bp of the MIF gene 5′-flanking sequence was cloned into the pGL3 basic plasmid (Promega, Southampton, UK) with luciferase as a reporter gene. MCF7 (estrogen receptor-positive) cells were transfected with 5 μg DNA/60-mm plate using Superfect (Qiagen, Crawley, UK). In addition, estrogen receptor (ER) negative JEG3 cells were transfected with MIF promoter/reporter and either full-length ERα or ERα lacking the AF-1 transactivation domain. pGL3 basic vector alone was used as a negative control, pGL3 control vector alone was a positive control, and transfection efficiency was determined by PSV βGAL vector (Promega). After 48 hours of incubation, cells were treated with or without LPS (1 μg/ml), estrogen (1 × 10−8 mol/L), and NF-κB inhibitor PDTC (60 μmol/L; Sigma) for 12 hours. Cell lysates were assayed for luciferase or β-galactosidase activity as per manufacturer’s instructions (Promega).
Results
MIF as a Regulator of Wound Healing
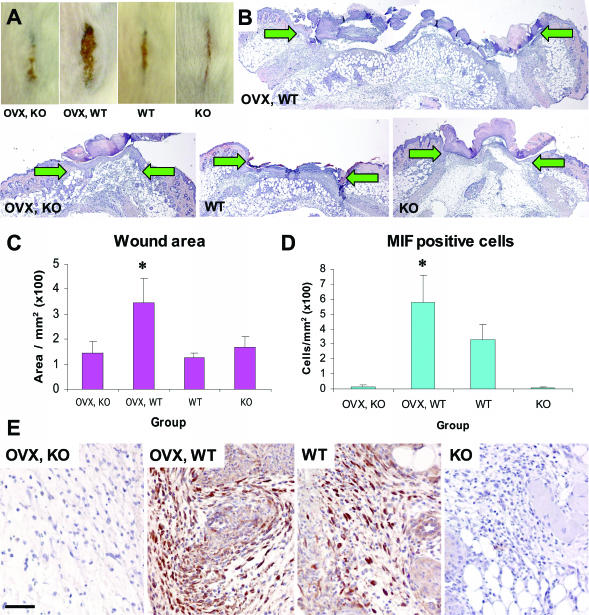
Previous studies have suggested a potential role for MIF in delayed healing in the absence of estrogen.6 To delineate the roles of estrogen and MIF, we used an acute wound healing murine model. As in previous studies,6 delayed wound healing in the absence of circulating estrogen (OVX, MIF wild-type [WT] mice) was confirmed to be associated with increased wound areas (Figure 1, A–C) and dramatically increased numbers of MIF-positive cells within wound tissue (Figure 1, D and E). In contrast, MIF-null mice healed normally regardless of prevailing estrogen levels (Figure 1, B and C; compare OVX, KO with KO). We reasoned that it should be possible to determine which specific genes or groups of genes were involved in these processes and identify the relative effects of MIF and/or estrogen.
Figure 1.
As shown previously MIF is a critical modulator of wound healing, regulated by estrogen in vivo. In mouse, reduced systemic estrogen leads to delayed healing, evident at the gross morphological level (in A, compare OVX, WT with WT) and at the histological level (in B, compare OVX, WT with WT). In the absence of MIF, this effect is abolished (in A, compare OVX, KO with OVX, WT; and in B, compare OVX, KO with OVX, WT). B: Representative hematoxylin and eosin-stained histological sections illustrate the increased wound width (arrows denote the wound edges) and area of wounds (C) from OVX, WT mice compared with other groups; *, t-test P = 0.03. D: Quantification of MIF-positive cells. E: Representative immunohistological MIF localization. D, MIF-null mice lack MIF-positive cells and OVX, WT mouse wounds with reduced systemic estrogen and increased inflammation (A, OVX, WT) have elevated numbers of MIF-positive cells; *, t-test P = 0.0008. E: In intact, WT wounds, MIF localizes predominately to keratinocytes, fibroblasts, and inflammatory cells. In OVX, WT wounds, strong MIF staining is observed in the increased number of inflammatory cells. Bar (E) in A = 5 mm; in B = 370 μm; and in E = 60 μm.
Microarray Analysis of MIF/Estrogen-Regulated Genes
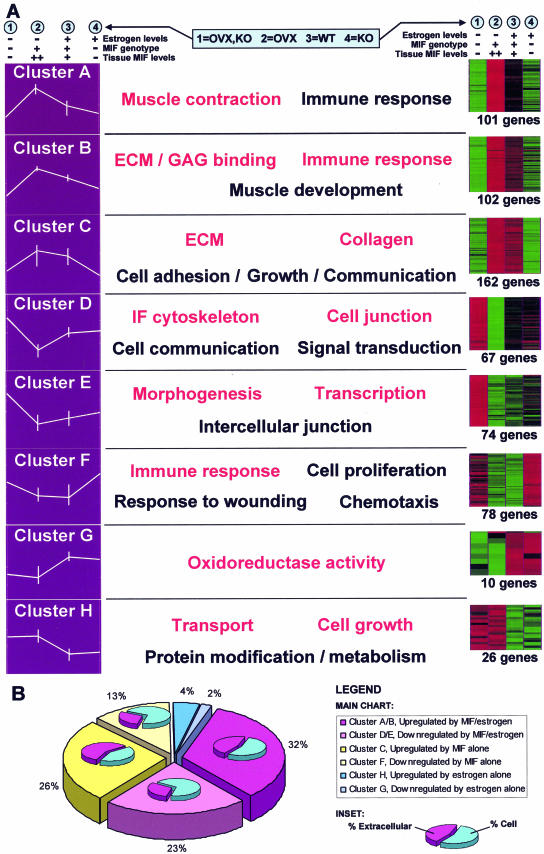
Our experimental strategy was to analyze wound gene expression via microarray analysis using the balanced factorial design shown in Table 1 (one mouse per array). This allowed us to identify genes regulated by either MIF or estrogen (OVX) and crucially to identify an interaction term between the two factors. Microarray analysis used the principles of data mining15 for 1) gene enrichment, 2) clustering, and 3) identification of biologically related gene groups (see Materials and Methods for detailed explanation). Gene enrichment highlighted 610 genes with significant MIF- and/or estrogen-mediated changes in wound tissue expression. Clustering partitioned these into eight gene clusters based on expression profile similarity (Figure 2, Table 2, and Supplementary Table 2, which is available online at http://ajp.amjpathol.org). EASE analysis was used to identify biologically related gene groups based on overrepresentation of GO terms (Figure 2A). These ontology terms indicate biological processes or cellular functions that are specifically altered in the wound environment by MIF and/or estrogen. Single genes belonging to statistically overrepresented GO terms are excellent candidates for mediating the influences of MIF and estrogen on wound healing. The full EASE analysis for each cluster is available in Supplementary Table 3 (available online at http://ajp.amjpathol.org). Astonishingly, few genes were regulated by estrogen alone, with virtually all of the influence of estrogen on gene expression being mediated by MIF (Figure 2B; compare MIF- and MIF/estrogen-regulated genes [94%] with those regulated by estrogen alone [6%]).
Table 1.
Experimental Design
| MIF | Systemic estrogen*
|
|
|---|---|---|
| − | + | |
| − | OVX, KO | INT, KO |
| (Three mice) | (Three mice) | |
| + | OVX, WT | INT, WT |
| (Three mice) | (Three mice) | |
Nonovariectomized mice have intact (INT) ovaries. Hereafter, the INT, KO and INT, WT conditions will be referred to as KO and WT.
Figure 2.
Clusters, profiles, and GO groupings. A: Identified genes were assigned to one of eight distinct clusters using k-means clustering algorithms. On the left, the data for each cluster are represented as a profile of the z-transformed (for each probe set, the mean set to 0 and SD to 1 using maxdView), log (2) values for the mean of each experimental group/condition. Error bars indicate the maximum and minimum values within each group. Displayed on the right is the same z-transformed data shown as an Eisen color plot. The number of genes in each cluster is specified. Red and green indicate positive and negative change from zero, respectively, with color intensity indicating the degree of deviation. The most significantly overrepresented GO terms are shown for the genes within each cluster. Terms in red have the highest statistical significance. B: Graphical representation of the number of genes within each cluster. This highlights the minimal involvement of estrogen alone in healing (blue) with the majority of estrogen’s effects acting through MIF (pink). Subcharts indicate the percentage of extracellular and cellular genes in each cluster. Clusters containing genes up-regulated by MIF levels (clusters A/B and C) are characterized by considerable overrepresentation of extracellular genes.
Table 2.
A Summary of Key Genes from Each Cluster
| Cluster | Gene* | Gene (description) | Gene ontology (GO) group | GO P value† | Max fc/groups‡ | MIF ref.§ |
|---|---|---|---|---|---|---|
| A, Up-regulation by MIF, Ovx interaction | ||||||
| Tnnc | Troponin C, cardiac/slow skeletal | Muscle contraction | 0.00000728 | 7.0/A versus B | x | |
| Tpm3 | Tropomyosin 3 | Muscle contraction | 0.00000728 | 4.1/A versus B | x | |
| Cxcl5 | Chemokine (C-X-C motif) ligand 5 | Immune/defense response | 0.00472 | 5.0/A versus B | √ | |
| C1qb | Complement component 1q, β | Immune/defense response | 0.00472 | 4.9/A versus B | √ | |
| Il6 | Interleukin 6 | Immune/defense response | 0.00472 | 2.9/A versus B | √ | |
| Prg4 | Proteoglycan 4 | Extracellular | 0.000336 | 6.0/A versus B | x | |
| Cd163 | CD163 antigen | Extracellular | 0.000336 | 3.7/A versus B | x | |
| Fbn1 | Fibrillin 1 | Extracellular matrix | 0.000615 | 2.1/A versus B | x | |
| Ets1 | E26 avian leukemia oncogene 1 | Anti-apoptosis/Binding | 0.0256 | 3.1/A versus B | x | |
| B, Up-regulation by MIF, Ovx interaction | ||||||
| Timp1 | Tissue inhibitor of metalloproteinase 1 | Extracellular | 1.74E-09 | 4.1/A versus B | √ | |
| Pdgfra | Platelet derived growth factor receptor, a | Extracellular | 1.74E-09 | 3.7/A versus B | x | |
| Myh6 | Myosin, heavy polypeptide 6, cardiac a | Muscle development | 0.0000391 | 3.7/A versus B | x | |
| Lox | Lysyl oxidase | Oxidoreductase activity | 0.0149 | 3.7/A versus B | x | |
| Cd36 | CD36 antigen | Cell adhesion | 0.00911 | 3.5/A versus B | x | |
| Col4a1 | Procollagen, type IV, α1 | Extracellular matrix | 0.0000266 | 2.9/A versus B | √ | |
| Mfap5 | Microfibrillar associated protein 5 | Extracellular matrix | 0.0000266 | 2.4/A versus B | x | |
| Mmp2 | Matrix metalloproteinase 2 | Extracellular matrix | 0.0000266 | 2.0/A versus B | √ | |
| Vim | Vimentin | Intermediate filament | 0.00457 | 2.5/A versus B | x | |
| C, Up-regulation by MIF only | ||||||
| Mif | Macrophage migration inhibitory factor | Immune response | 0.0205 | 6.6/A versus C | n/a | |
| Col3a1 | Procollagen, type III, α1 | Collagen | 4.8E-15 | 5.9/A versus B | x | |
| Col1a1 | Procollagen, type I, α1 | Collagen | 4.8E-15 | 4.0/B versus C | x | |
| Col1a2 | Procollagen, type I, α2 | Collagen | 4.8E-15 | 3.6/A versus B | x | |
| Lum | Lumican | Extracellular matrix | 5.45E-17 | 3.6/B versus D | x | |
| Fbn1 | Fibrillin 1 | Extracellular matrix | 5.45E-17 | 3.4/B versus D | x | |
| Pcolce | Procollagen C-proteinase enhancer protein | Extracellular | 4.58E-20 | 3.1/A versus B | x | |
| Serpinh1 | Serine proteinase inhibitor clade H 1 | Extracelluar | 4.58E-20 | 2.6/B versus C | x | |
| Ndn | Necdin | Cell growth | 0.0042 | 2.6/A versus B | x | |
| D, Down-regulation by MIF, Ovx interaction | ||||||
| Krt1-15 | Keratin complex 1, acidic, gene 15 | Cytoskeleton | 0.000215 | 2.6/B versus D | x | |
| Wnt4 | Wingless-related MMTV integration site 4 | Cell communication | 0.0111 | 2.4/A versus B | x | |
| Fzd6 | Frizzled 6 | Cell communication | 0.0111 | 2.1/A versus B | x | |
| Notch1 | Notch gene homolog 1 | Cell communication | 0.0111 | 2.0/A versus B | x | |
| Cldn1 | Claudin 1 | Cell junction | 0.00133 | 2.3/A versus B | x | |
| Alox12e | Arachidonate lipoxygenase, epidermal | Oxidoreductase activity | 0.113 | 3.4/A versus B | x | |
| Defb6 | Defensin β6 | Extracellular | 0.121 | 2.7/A versus B | x | |
| E, Down-regulation by MIF, Ovx interaction | ||||||
| Jag1 | Jagged 1 | Morphogenesis | 0.00158 | 4.3/A versus B | x | |
| Dhcr24 | 24-Dehydrocholesterol reductase | Cell communication | 0.0176 | 2.6/A versus C | x | |
| Lhx2 | LIM homeobox protein 2 | Transcription | 0.0176 | 2.3/A versus B | x | |
| Sdc1 | Syndecan 1 | Membrane | 0.00963 | 2.2/A versus B | x | |
| St14 | Suppression of tumorigenicity 14 | Cell migration | 0.0857 | 2.0/A versus B | x | |
| Lad1 | Ladnin 1 | Extracellular matrix | 0.17 | 2.0/A versus B | x | |
| Alcam | Activated leukocyte cell adhesion molecule | Cell adhesion/membrane | 0.00963 | 1.8/A versus B | x | |
| Dsg2 | Desmoglein 2 | Intercellular junction | 0.0109 | 1.5/A versus B | x | |
| F, Down-regulation by MIF only | ||||||
| Thbs1 | Thrombospondin 1 | Extracellular | 0.177 | 3.8/C versus D | x | |
| Gbp1 | Guanylate nucleotide binding protein 1 | Immune response | 1.37E-11 | 3.2/C versus D | x | |
| Ifit1 | Interferon-induced with tetratricopeptide 1 | Immune response | 1.37E-11 | 2.5/B versus D | x | |
| Cxcl10 | Chemokine (C-X-C motif) ligand 10 | Immune response | 1.37E-11 | 2.5/B versus D | x | |
| Cdkn1a | p21 (CDKN1A)-activated kinase 1 | Immune response | 1.37E-11 | 1.7/A versus B | √ | |
| Mmp13 | Matrix metalloproteinase 13 | Extracellular matrix | 0.177 | 2.3/C versus D | √ | |
| Nfkbia | NF-k light chain enhancer in B-cell inhibitor, a | Cell proliferation | 0.00169 | 2.1/A versus B | √ | |
| G, Up-regulation by estrogen | ||||||
| Cyp2g1 | Cytochrome P450, family2, subfamily g,1 | Oxidoreductase activity | 0.00549 | 4.4/B versus D | x | |
| Cyp17a1 | Cytochrome P450, family17, subfamily a,1 | Oxidoreductase activity | 0.00549 | 1.8/B versus D | x | |
| Akr1c18 | Aldo-keto reductase family1, member C18 | Oxidoreductase activity | 0.00549 | 1.6/B versus D | x | |
| Fkbp5 | FK506 binding protein 5 | Chaperone activity/metabolism | 0.136 | 2.0/A versus D | x | |
| H, Down-regulation by estrogen | ||||||
| Cald1 | Caldesmon 1 | Actin binding | 0.285 | 2.5/B versus D | x | |
| Ramp2 | Receptor (calcitonin) activity modifying 2 | Growth/transport | 0.0189 | 2.8/A versus C | x | |
| Snx6 | Sorting nexin 6 | Growth/transport | 0.0189 | 1.8/A versus C | x | |
| Inhbb | Inhibin β-B | Growth/transport | 0.0189 | 1.6/B versus D | √ |
Genes in bold text have been validated by real-time PCR.
EASE-derived (see Materials and Methods) P value for overrepresentation of gene ontology (GO) group (bold indicates significance).
Maximum fold change (fc) between any two experimental groups. In each case, experimental groups compared are defined: A, OVX, KO; B, OVX, WT; C, WT; and D, KO.
Known reference to MIF interaction; √, cited references to MIF regulation; x, novel MIF gene target.
Genes Up-Regulated by MIF with Estrogenic Interaction
In terms of determining genes involved in impaired healing/inflammation, the most important clusters were characterized by MIF regulation with estrogen interaction (Figure 2A, clusters A and B and clusters D and E). Via interaction with estrogen, MIF up-regulated (ie, associated with delayed healing) two clear groups: genes associated with muscle contraction and those involved in immune response or adhesion. MIF down-regulated (ie, associated with accelerated healing) genes involved in virtually all of the major signaling cascades found in the wound environment,16 ie, TLR and NF-κB or IRF, TGF-β and SMAD, PI3K and Akt, and Wnt and Notch, as well as numerous cytoskeletal/epidermal genes (Figure 2A, clusters D and E). Because the experimental protocol used RNA extracted from whole wounds, representing an extremely heterogeneous source of tissues, it is important to note that genes may be appointed to these clusters based on cellular differences between wounds (ie, delayed-healing associated) or inherent differences in RNA expression from a specific lineage (ie, directly regulated by MIF and estrogen interaction).
Clusters A and B comprised genes that have low expression in both MIF-null groups and very high expression in the OVX, WT group (Figure 2; see Table 2 for selected genes), mirroring MIF protein levels in the wound (Figure 1D). Thus genes in this category were associated with delayed healing and increased inflammation. This cluster is the largest and is composed of more than one-third of the genes identified (Figure 2B). Surprisingly, the most significant overrepresentation was for genes in the muscle-associated GO groups or that are muscle related but have yet to be annotated (24 genes or 14%). Tropomyosin 3 (Tpm3) was of particular interest not only in terms of its possible role in fibroblast contraction17 but also because it may act as a pro-inflammatory18 and anti-angiogenic19 agent. Protein localization indicated expression by fibroblasts, inflammatory cells, and blood vessels within the wound but lack of expression in skeletal muscle, confirming that Tpm3 levels were independent of subcutaneous muscle volume retraction (data not shown). A number of skin-expressed genes, such as Atp2a2, Myh7, and Myl3, are involved in cardiac contraction, cardiomyopathy, and fibrosis,20 suggesting a potential role for MIF in these processes. MIF is markedly increased after ischemic myocardial injury,21 as are several other cluster A genes novel to skin or wounds such as Myh6 and Tnnt1, and is involved in destabilization of atherosclerotic plaques.10 Our data suggest that the effects of MIF on such processes may extend beyond its pro-inflammatory role. Other genes included structural extracellular matrix (ECM) proteins (23 genes or 14%), such as fibrillin 1 (Fbn1) and the interacting protein microfibrillar associated protein 5 (Mfap5), and elastic fiber-associated genes, such as lysyl oxidase (Lox). Thrombospondin 2 (Thbs2) and its receptor CD36 were up-regulated by MIF, and both Thbs2 and Fbn1 levels increased in the uninjured skin of aged mice and humans.21,3 Thbs2, an inhibitor of angiogenesis, plays a key role in the organization of granulation tissue architecture. Moreover, Thbs2-null mice display accelerated healing,22 suggesting that abnormal ECM structure may play a role in the impaired healing phenotype observed in our model.
OVX, WT wounds were characterized by excessive inflammation and matrix degradation.1,5 Inflammation-associated genes were prevalent in clusters A/B (77 genes or 41%) and included chemokines, cytokines, and their receptors; ECM-modulating enzymes such as Mmp2 and glucuronidase β (Gusb); and leukocyte/endothelial adhesion molecules. Confinement of Mmp2 to this group (ie, up-regulation in OVX, WT wounds) was of particular interest because its levels increase in normal elderly skin and in age-related impaired acute and chronic wounds.2 Moreover, MIF up-regulates MMP2 levels in prostatic cell lines.23 The pro-inflammatory E26-transformation specific (ETS) family transcription factors Ets1 and Elk3 (involved in LPS-signaling via TLR424) were up-regulated in OVX, WT wounds, as was prostaglandin I2 synthase (Ptgis), an inhibitor of platelet aggregation. Other pro-inflammatory factors included mediators of leukocyte chemotaxis such as Ccr2, and interleukin-2 receptor-γ (Il2rg). This group contained novel MIF/estrogen-regulated macrophage markers, such as Cd163 and macrophage-expressed gene 1 (Mpeg1), and genes associated with adhesion (Icam2, P selectin), rolling (endomucin), and diapedesis (Aoc3). The presence of Il6 in this cluster was of particular note. Interleukin-6 induces Cxcl5, another member of this group that is a potent chemotaxin able to activate inflammatory cells and induce classical pro-inflammatory cytokines.25
Genes Down-Regulated by MIF and Interaction with Estrogen
This group contained genes that have high expression in OVX, MIF-null (OVX, KO) wounds and very low expression in OVX, WT wounds (Figure 2, clusters D and E), representing genes whose down-regulation was associated with impaired healing. In line with the inhibitory effects of estrogen on MIF levels, maximal down-regulation was only seen when levels of systemic estrogen were low, ie, OVX, KO versus OVX, WT wounds. These clusters were enriched with genes that mediate transcription, morphogenesis, and immune response. Multiple genes involved in the induction of apoptosis and G-protein/TGF-β signaling were down-regulated in these impaired healing wounds, and most have been previously associated with invasive carcinomas26,27 but not tissue injury/repair. MIF has been termed a “pro-tumorigenic” factor,27 and in this regard, it is interesting to note that Notch down-regulation predisposes to epidermal carcinomas, suggesting an association between MIF and cutaneous malignancy.28 Numerous genes influencing epithelial cell biology and differentiation were inhibited in this group, including genes known to be modified in epithelializing wounds such as Krt5, Krt14, Notch, Jag1, and Dsg2.29,30 The role of MIF in modulating such genes has never been reported, and because no differences in epithelialization rates were noted between groups, this finding cannot be explained by a temporal delay in expression.
In contrast to the up-regulated gene groups, down-regulated genes included protease inhibitors not previously associated with healing, such as Spint1 and -2, the innate immunity-associated antimicrobial peptide defensin β 6 (Defb6), and anti-inflammatory genes such as Gpx2, previously described only in the gastrointestinal tract. Gpx2 plays a role in suppression of inflammation, and Gpx2-null mice are highly susceptible to bacteria-associated inflammation of the gastrointestinal tract.31 Reduction of this isozyme in the wounds of OVX, WT mice may contribute to the excessive inflammation observed.
Genes Up-Regulated by MIF
This group contains genes that were up-regulated by the presence of MIF (reduced expression in both KO groups) without being influenced by systemic estrogen levels (Figure 2A, cluster C). These genes may be of particular significance in other MIF-regulated processes such as arthritis,9 response to sepsis,11 parasitic infection,32 and renal fibrosis.33 This was the second largest group, containing 162 genes or 26.5% of the total genes identified (Figure 2B). Surprisingly, the most significantly overrepresented GO group was collagen (12 genes or 8%), closely followed by extracellular matrix (83 genes or 51% [71% of annotated genes]). Only a subset of the 31 distinct collagen genes encoded by the mouse genome are expressed in normal skin. Hence, MIF has a profound and previously unappreciated impact on fibroblast collagen synthesis. This cluster contained genes encoding collagen I, III, IV, V, and VI in addition to many basement membrane components such as Nidogen 1 (Nid1), perlecan, laminin 1 B1 subunit (Lamb1-1), and laminin-α 4 (Lama4). This cluster was also enriched with genes encoding small collagen-, proteogycan- and matrix-associated proteins such as dermatopontin (Dpt) and Sparc (both of which interact with TGF-β), biglycan (Bgn), decorin (Dcn), lumican (Lum), and tenascin (Tnc). Up-regulation of these genes may result in fibrosis and abnormal collagen maturation. This cluster was enriched with genes encoding proteolytic enzymes, the most interesting of which was Mmp3, a crucial regulator of matrix turnover.34 Additional proteolytic enzymes included the metalloproteinase Adamts5 and the lysosomal cysteine proteinase cathepsin K (Ctsk). As in the previous cluster, we saw many inflammation-associated genes involved in acute phase response (Orm1), leukocyte-endothelial adhesion (Thy-1), migration (Cxcl12), and diapedesis (Pecam1). Cxcl12 is a novel MIF-regulated cytokine that activates leukocytes, regulates their migration, activates proteases such as MMP9 and is important for homing to areas of ischemic injury.35
Genes Down-Regulated by MIF
These genes had the inverse profile of genes in cluster C (Figure 2A, cluster F). The most significantly overrepresented GO group in this cluster was immune response factors characterized by genes involved in the Th1 response,36 such as Tap1, a macrophage-associated interferon-γ (IFN-γ) and LPS-induced transport protein. Cross-talk between the MIF and IFN pathways is suggested by genes in this cluster, such as Gbp1, an interferon- and LPS-induced guanylate nucleotide-binding protein, interferon-stimulated gene 20 kd (Isg20; a leukocyte-expressed estrogen-responsive37 gene), and Cxcl10 (an IFN-γ-inducible chemokine that is up-regulated through TLR4 and involved in monocyte stimulation). Intriguingly, no association between MIF and these immune regulators has been previously recognized.
MIF appeared to down-regulate a number of cell cycle regulatory genes such as the cyclin-dependent kinase inhibitor p21 (Cdkn1a), a regulator of cell cycle progression also instrumental in the execution of apoptosis after caspase activation,38 and the Jun-B oncogene Schlafen 3 (Slfn3), a negative regulator of cell proliferation that may be differentially expressed in alternatively activated macrophages. This may lead to an anti-apoptotic state in line with the role of MIF in extended inflammation. This cluster also contained a number of extracellular genes, several of which were ECM associated, including Mmp13, a key player in matrix remodeling. An association between MIF and this group of ECM genes has never previously been established and may be of importance in a number of MIF-regulated systems.
Genes Regulated by Estrogen Independent of MIF
Genes up-regulated by estrogen independently of MIF (Figure 2, cluster G) appear to have little impact on repair/inflammation in vivo; however, they may be important in normal skin physiology and could play a role in estrogen-mediated processes in noncutaneous tissues. Surprisingly, few genes were estrogen regulated without MIF interaction. This cluster contained just 10 genes, with only a single GO group being overrepresented: oxidoreductase activity or, more specifically, steroid hormone synthesis. Skin has long been known to contain the enzymes needed for final stage synthesis of potent androgens and estrogens. However, the skin has only recently been identified as a steroidogenic tissue in its own right, shown to contain the full cytochrome P450 system required for the de novo production of sex steroids from cholesterol.39,40 In this study, we found that systemic estrogen deprivation reduced transcription of enzymes necessary for cutaneous synthesis of not only estrogen but other sex hormones, (Cyp17a1, Cyp2g1, and Akr1c18). In rodents, this may be of less relevance in the wound healing response because the adrenal gland does not secrete the potent precursor dehydroepiandrosterone (DHEA); however, in elderly humans in which conversion of precursors, in particular DHEA, determines local tissue estrogen levels, this may be of great importance. Whereas DHEA has been shown to accelerate healing,41 our data suggest that DHEA alone may have little impact on skin physiology in the elderly unless up-regulation of the steroid biosynthetic enzymes occurs in tandem.
Genes independently inhibited by estrogen were confined to the transport/cell growth and metabolism GO groups (26 genes; Figure 2, cluster H). Again, although these genes appear not to be critical to wound repair/inflammation, they may play a role in normal skin homeostasis and in other organs in which estrogen plays a role. Such genes were invariably linked to TGF-β functions/pathways, and the majority of genes in this group are involved in pathways inhibiting cell proliferation, promoting apoptosis, and conferring protection in the pathogenesis of breast and endometrial cancer. This group contained genes not previously associated with estrogenic regulation, such as sorting nexin 6 (Snx6), which associates with the TGF-βR.42 Other specific genes included the estrogen-regulated Pten,43 which regulates cell proliferation, apoptosis, and angiogenesis and is mutated in endometrial carcinoma, and Inhbb, a TGF-β superfamily member involved in cell growth inhibition in human breast cancer cells. Glycoprotein 38 (Gp38), up-regulated by estrogen alone, is able to induce platelet aggregation and thrombosis without the need for plasma components.44 This group contained genes that may be of importance in other systems in which estrogen is involved in coagulation/thrombosis and neoplastic pathways.
Validation
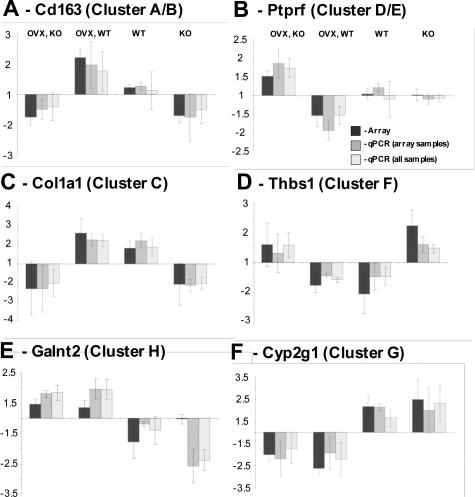
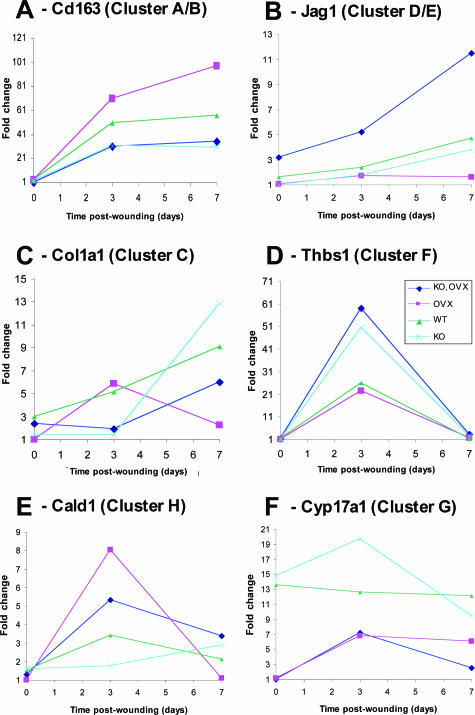
Primers for quantitative real-time PCR were designed to 26 of the 610 significant genes identified in this study (see Materials and Methods). Primer sequences were designed to each gene coding sequence independently of the Affymetrix probe set target region sequence. Hence, our primers and the Affymetrix probes may or may not be directed to the same gene region. In addition, real-time PCR was performed with not only the 12 samples used on the arrays (to enable absolute comparison) but also all 20 samples collected in this study. For all 26 genes, the cluster profile was recapitulated by the real-time results (Figure 3, A–F, and data not shown). Furthermore, in many instances in which statistical significance was not achieved on comparison of two groups from the microarray data, it was achieved when comparing the same two groups with real-time PCR using all 20 samples (data not shown). We examined the expression of selected genes from each cluster over a wound healing time course (Figure 4 and data not shown). The majority of genes were up-regulated on day 3 after wounding (eg, Figure 4D), and in most instances, the day-3 profiles (Figure 3) were maintained (eg, Figure 4A) or expression was down-regulated at day 7 (eg, Figure 4D).
Figure 3.
Validation of array data by quantitative real-time PCR. Six representative comparisons from a total of 26. A: Cd163, a gene up-regulated by MIF with OVX interaction. B: Ptprf, down-regulated by MIF with OVX interaction. C: Col1a1, up-regulated by MIF alone. D: Thbs1, down-regulated by MIF alone. E: Galnt2, down-regulated by estrogen independently of MIF. F: Cyp2g1, up-regulated by estrogen independently of MIF. qPCR (array samples), real-time PCR on the same three mice per group used for arrays; qPCR (array samples) used five mice per group. To enable direct comparison, all three datasets were normalized to give a mean of 1 and values below 1 converted to negative-fold change. The y axis indicates fold change from the mean. Error bars indicate ±SEM.
Figure 4.
Temporal profiles of gene expression during wound healing determined by quantitative real-time PCR. Six representative profiles from a total of 14. A: Cd163, up-regulated by MIF with OVX interaction. B: Jag1, down-regulated by MIF with OVX interaction. C: Col1a1, a gene up-regulated by MIF alone. D: Thbs1, down-regulated by MIF alone. E: Cald1, down-regulated by estrogen independently of MIF. F: Cyp17a1, up-regulated by estrogen independently of MIF.
MIF as a Regulator of Wound Healing in Humans
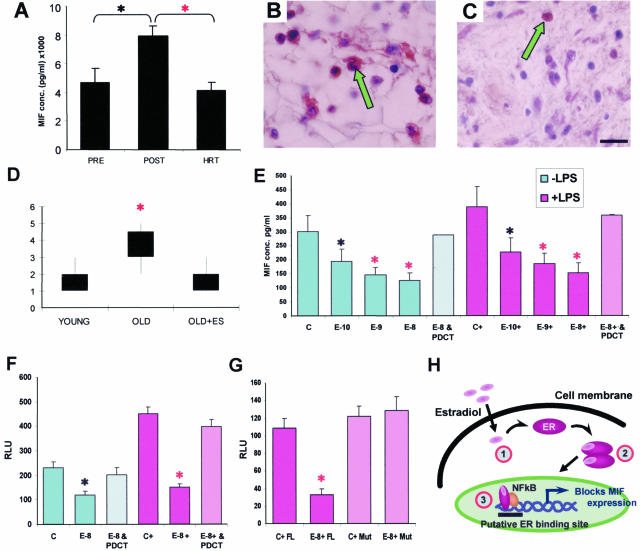
In both ovariectomized mice and postmenopausal women, circulating estrogen was undetectable (data not shown). Having demonstrated inhibition of MIF by estrogen in the mouse and the profound effect of MIF and estrogen on estrogen-depleted murine wound healing, we reasoned that these effects would be recapitulated in age-related delayed human wound healing. In humans, estrogen also down-regulated MIF levels both systemically (Figure 5A) and locally in wound tissue (Figure 5, B–D). In postmenopausal (45- to 60-year-old) women with low systemic estrogen, systemic MIF levels were significantly higher than in younger premenopausal women (Figure 5A). However, in postmenopausal (47- to 60-year-old) women treated with exogenous systemic estrogen (ie, undergoing HRT treatment), MIF levels were markedly reduced and comparable with premenopausal women (Figure 5A). This systemic reduction in MIF levels is especially relevant in terms of response to such conditions as sepsis,11 immune dysregulation, or parasitic infection32 in the elderly. In humans, local MIF levels were also increased in the acute wounds of elderly compared with young subjects (Figure 5D). Acceleration of cutaneous healing by topical estrogen in the elderly was associated with significant reduction in local MIF levels (Figure 5, B, C, and D; compare elderly estrogen treated in C with elderly nontreated in B).
Figure 5.
Mechanism of MIF regulation by estrogen in humans. A: Postmenopausal female subjects (45 to 60 years old with low systemic estrogen) have significantly higher systemic MIF levels than premenopausal subjects (35 to 49 years old). MIF levels in female subjects who were postmenopausal and on HRT (47 to 60 years old) were indistinguishable from the premenopausal group. B: Numerous local MIF-positive cells (arrow) in wounds from aged subjects are dramatically reduced by topical estrogen (C; arrow, compare with B). D: MIF immunostaining scored on a scale of 1 to 5 is elevated in elderly subjects and reversed by topical estrogen. E: Human monocyte MIF production, by both resting (−LPS) and LPS-activated (+LPS) cells, is decreased with concurrent estrogen treatment. E-10, E-9, and E-8 = 10−10, 10−9, and 10−8 mol/L estrogen. In addition, treatment of both resting and LPS-activated estrogen-treated monocytes with the NF-κB inhibitor PDCT (60 μmol/L) significantly reversed this estrogen-dependent reduction in MIF levels. F: Human MCF7 ER+ cells transfected with a MIF-promoter luciferase-reporter construct showed increased activity after LPS activation. Estrogen (10−8 mol/L) inhibited this activity, and the effects of estrogen were blocked by PDCT. G: ER-negative JEG3 cells transfected with MIF promoter/reporter and full-length ERα (FL) showed estrogen-dependent (E-8) down-regulation of MIF promoter activation. When a mutant ERα construct (Mut) lacking the AF-1 domain is instead transfected, estrogen is no longer able to inhibit MIF expression. H: Schematic representation of the role of NF-κB in estrogen receptor-dependent activation of MIF. Step 1, estradiol binds to the ER. Step 2, ER dimerizes. Step 3, ER binds to NF-κB. *, t-test P < 0.05; red asterisk, t-test P < 0.01; Error bars = means ± SEM; n = 10–16 (A), 4 (D), 5 (E–G). Bar (C) in B and C = 9 μm.
Estrogen Regulates MIF at the Level of Transcription
The observation that the impaired wound healing phenotype, resulting from an absence of estrogen, could be rescued in the MIF-null mice, consistent with a dampened inflammatory response, suggested that estrogen directly or indirectly modulated inflammatory cell-derived MIF expression. The observed increase in MIF levels in the absence of estrogen could reflect an overall increase in the number of inflammatory cells (indirect regulation) or signify a cell-specific up-regulation of MIF. In addition, the effects of estrogen on MIF could represent direct transcriptional regulation or could reflect the effects of estrogen on multiple secondary gene products and thus influence MIF indirectly. In concordance with the in vivo up-regulation of MIF levels in acute wound tissue of OVX mice (Figure 1D) and elderly humans (Figure 5D), we showed that estrogen directly inhibited MIF production by both resting and activated human monocytes in a dose-dependent manner (Figure 5E). This statistically significant decrease was, however, abolished when cells were co-treated with the NF-κB inhibitor PDCT (Figure 5E), suggesting a novel role for the well-characterized pro-inflammatory transcription factor NF-κB in estrogen modulation of MIF.
To further investigate the mechanism of MIF inhibition by estrogen, we transfected a MIF promoter-luciferase reporter construct into MCF7 cells (breast adenocarcinoma cell line of epithelial origin). Estrogen treatment of transfected cells resulted in a statistically significant and reproducible reduction in luciferase activity. This inhibitory action of estrogen on LPS-induced promoter activity indicated that the effects of estrogen are directed either genomically or nongenomically via interaction with second messenger systems at the level of transcription (Figure 5F). Again, co-treatment with PDCT reversed the effect of estrogen, indicating involvement of NF-κB. When ER-negative cells (JEG3, placental choriocarcinoma cell line of epithelial origin) were co-transfected with the MIF promoter and ER-α constructs, estrogen no longer inhibited MIF promoter activity when the ER lacked the AF-1 transactivation function (Figure 5G), suggesting a requirement for ER-mediated transcriptional activity and a direct interaction between the ER and the MIF promoter region (summarized in Figure 5H). This has important implications for our microarray findings and supports the concept that where estrogen modulates gene expression, it does so through direct modulation of MIF levels. This is also the first report of a role for the pro-inflammatory transcription factor NF-κB in estrogen-dependent MIF regulation.
Discussion
In the developed world, the majority of women live one-third of their lives in a state of profound estrogenic deprivation. Estrogen is of fundamental importance in the cutaneous wound healing response, accelerating healing in both elderly females and males and in numerous animal models. However, its mechanisms of action are poorly understood.5,45 In reproductive and nonreproductive tissue, estrogen regulates a plethora of interacting gene products with numerous downstream consequences on cell functions. This results in a complex system in which physiological effects are only mimicked by the use of estrogenic ligands (such as in HRT46), leading to adverse effects because of such a generic approach. Recently, the multifunctional cytokine MIF has emerged as a potential candidate mediating the delayed healing and excessive inflammatory response in the absence of estrogen.6 Here, we provide the first demonstration of a direct correlation between age and MIF levels in human acute wounds that systemically corresponds to an age-associated decrease in estrogen levels. Elevated local MIF is associated with an increased inflammatory response and markedly delayed wound healing that is entirely reversed by exogenous estrogen treatment.
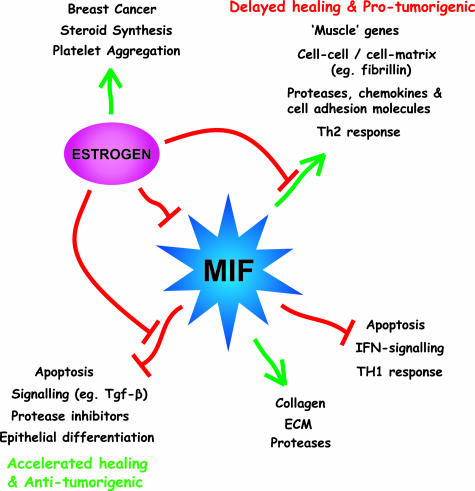
We have used gene array technology to determine the importance of MIF as a regulator of estrogen-mediated healing at the molecular level. Our experimental design has allowed us to identify more than 600 differentially expressed genes that segregate into several distinct clusters corresponding to regulation by MIF and/or estrogen. Different gene ontology groups are specifically overrepresented in each cluster, indicating that MIF and estrogen orchestrate a diverse range of functions within the wound environment. MIF has a direct effect on the expression of genes involved in all aspects of the wound healing process in addition to genes associated with cellular proliferation, differentiation, and apoptosis in a wide range of cell types. In contrast estrogen appears to primarily inhibit MIF, with indirect effects on healing via MIF. Thus, MIF is a key player in the healing wound that regulates many aspects of the impaired healing response when levels of estrogen are reduced (Figure 6). Furthermore, MIF is responsible for mediating most, if not all, of the effects of estrogen-depletion on wound repair. Specific genes can be delineated as showing advantageous effects (estrogen/MIF regulated accelerated healing) versus potential adverse effects (estrogen only-mediated pro-thrombosis and tumorigenesis). Future studies are required to determine the biological significance of such observations.
Figure 6.
A summary of the effects of MIF and estrogen on healing.
This in vivo study assesses changes in gene expression between mouse wounds corresponding to four experimental conditions. There are advantages and disadvantages to this approach. The main limitation is that cellular composition will vary between conditions. An in vitro (homogeneous cell type) experimental design would allow easier assessment of definitive changes in inherent RNA expression. However, findings would be entirely cell type-specific and not directly relevant to healing of a three-dimensional tissue. In contrast the experimental design uses whole-wound tissue and so directly measures in vivo changes in wound gene expression between conditions. Although identified genes provide clear clues to their lineage of origin, a major undertaking, and avenue for future experimentation, will be to delineate the cellular source of differentially expressed genes. We are confident that genes in clusters C, F, G, and H are regulated at the level of inherent tissue expression, because in these clusters, differences are observed despite virtually identical cellular composition (Figure 1). It is possible that differential expression of genes in clusters A, B, D, and E is due to differences in cellular composition, particularly in OVX, WT wounds. We are especially interested in these genes because they represent candidates for clinical manipulation to accelerate healing.
Taken together, our data suggest that impaired healing, in the absence of estrogen via elevated MIF, is associated with dysregulated differentiation, cell contractile machinery, altered signaling and transcription, coupled with a proteolytic and a pro-inflammatory state. MIF, with no estrogenic interaction, and above a threshold level, appears to be involved in the positive regulation of structural ECM-type genes and the negative regulation of specific immune response genes, IFN-regulated genes, and pro-apoptotic genes. Many of these genes are entirely novel and have never been related to the biological functions of MIF. For example, numerous structural collagen genes, including the major types present in normal unwounded skin and in wound healing, appear to be surprisingly and strongly up-regulated by MIF. This observation has major implications for the role of MIF in pro-fibrotic disorders such as renal sclerosis33 and atherosclerosis.10 It is surprising that in light of the number of affected genes in the MIF-null mice, in particular collagen and structural ECM genes, that wound healing is essentially normal. Such data highlight functional redundancy of large gene groups and parallel the findings from specific transgenic models in which compensation by related genes occurs.47,48 It is interesting to note that at day 7 after wounding, Col1 expression is markedly up-regulated in the absence of MIF. In addition, MIF up-regulates specific proteases such as Mmp3 and pro-inflammatory genes such as Cxcl12, which may act to counteract enhanced matrix expression. In this regard, it is important to note that OVX, WT wounds, characterized by very high MIF levels and up-regulated ECM expression, demonstrate a marked decrease in overall matrix deposition,5 suggesting that tissue proteolysis plays a major role.
NF-κB has been implicated as an important regulator of pro-inflammatory responses, which can be inhibited by estrogen in vivo49. In this study, we have clearly demonstrated a role for NF-κB in the inhibition of MIF by estrogen in two separate in vitro systems. In both human monocytes and LPS-activated MCF7 cells, NF-κB inhibition reverses the effects of estrogen on MIF. These findings confirm NF-κB as an integral transcription factor required for the inhibition of MIF promoter activity. Previous reports suggest that estrogen inhibits and TNF-α activates NF-κB, which is responsible for the transcription of a number of pro-inflammatory mediators.50 Our data suggest a mechanism whereby NF-κB itself plays a crucial role in estrogen-mediated inhibition of MIF production. It is possible that the estrogen receptor and NF-κB may not associate directly but instead could compete for limiting amounts of a third factor necessary for MIF promoter activation. An analogous situation has been demonstrated for cell adhesion molecule induction in smooth muscle cells where estrogen receptor-α and RelA (p65 NF-κB) compete for the CREB-binding protein-related protein p300.51
It is noteworthy that several NF-κB regulatory genes were included in those identified by microarray. The IκBα (Nfkbia) gene is down-regulated in the delayed healing of OVX, WT wounds (cluster F), whereas Csnk2a1 is up-regulated in response to reduced estrogen (cluster H; ie, up-regulated in delayed-healing wounds). IκBα and IκBβ sequester NF-κB in the cytoplasm and prevent it from binding to DNA in the nucleus, thereby tightly regulating activation of downstream genes. Recent studies using IκBβ knock-in mice have identified a unique injury-specific role for IκBα in NF-κB-mediated activation of pro-inflammatory responses.52 In contrast, phosphorylation of NF-κB by elevated Csnk2a1 in OVX, WT wounds will potentially enhance expression of pro-inflammatory genes such as MIF. Csnk2a1-mediated phosphorlyation of the p65 subunit of NF-κB enhances inducible nitric-oxide synthase transcription.53 Further elucidation of the role of NF-κB in the regulation of MIF levels will be essential to understanding this complex process.
Our data have wide-ranging implications for the role of MIF in a variety of estrogen-regulated systems, including osteoporosis, atherosclerosis,10 and autoimmune disorders.54 When estrogen levels fall, specific patterns of gene expression are induced by raised MIF levels, leading to altered immune responses, cell differentiation and contractility, reduced signaling intermediates, and impaired healing. We can clearly delineate groups of genes and individual genes that have not previously been associated with wound healing, MIF, or estrogen. Such genes warrant further investigation not only in terms of cutaneous wound healing, but also for all aspects of tissue repair, inflammation, and estrogen-mediated regulation of cell behavior. Moreover, our data suggest that there is a selective advantage in estrogen acting through the modulation of one specific cytokine in nonreproductive tissue. The dramatic inhibition of MIF by estrogen may be of fundamental importance in reproductive functions, because MIF has been implicated in dysregulated fetal tolerance, recognition of paternal antigens, and fetal rejection.55,56 Estrogen-mediated inhibition of MIF leading to accelerated healing may have been actively selected to ensure fast tissue repair after menstruation and parturition or passively co-selected with other advantageous phenomena such as protection of the fetus. That MIF should be the “master regulator” of the effects of estrogen is of importance not only in its clinical implications for wound healing (ie, inhibition of MIF in the elderly as a therapeutic strategy to accelerate healing) but also as a mechanism for other estrogen-mediated actions.
Supplementary Material
Acknowledgments
We thank L. Wardleworth and A. Hayes of the University of Manchester Core Microarray Facility for excellent technical help and advice. We thank Kejian Lei (National Institutes of Health, Bethesda, MD) for the MIF-promoter construct.
Footnotes
Address reprint requests to Professor Gillian S. Ashcroft, Room C2265, Faculty of Life Sciences, University of Manchester, Michael Smith Building, Oxford Road, Manchester M13 9PT, UK. E-mail: gillian.s.ashcroft@manchester.ac.uk.
Supported by grants from The Wellcome Trust and The Health Foundation.
Supplemental material for this article can be found on http://ajp.amjpathol.org.
References
- Ashcroft GS, Horan MA, Ferguson MW. Aging is associated with reduced deposition of specific extracellular matrix components, an upregulation of angiogenesis, and an altered inflammatory response in a murine incisional wound healing model. J Invest Dermatol. 1997;108:430–437. doi: 10.1111/1523-1747.ep12289705. [DOI] [PubMed] [Google Scholar]
- Ashcroft GS, Horan MA, Herrick SE, Tarnuzzer RW, Schultz GS, Ferguson MW. Age-related differences in the temporal and spatial regulation of matrix metalloproteinases (MMPs) in normal skin and acute cutaneous wounds of healthy humans. Cell Tissue Res. 1997;290:581–591. doi: 10.1007/s004410050963. [DOI] [PubMed] [Google Scholar]
- Ashcroft GS, Kielty CM, Horan MA, Ferguson MW. Age-related changes in the temporal and spatial distributions of fibrillin and elastin mRNAs and proteins in acute cutaneous wounds of healthy humans. J Pathol. 1997;183:80–89. doi: 10.1002/(SICI)1096-9896(199709)183:1<80::AID-PATH1104>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- Ashcroft GS, Mills SJ, Ashworth JJ. Ageing and wound healing. Biogerontology. 2002;3:337–345. doi: 10.1023/a:1021399228395. [DOI] [PubMed] [Google Scholar]
- Ashcroft GS, Dodsworth J, van Boxtel E, Tarnuzzer RW, Horan MA, Schultz GS, Ferguson MW. Estrogen accelerates cutaneous wound healing associated with an increase in TGF-beta1 levels. Nat Med. 1997;3:1209–1215. doi: 10.1038/nm1197-1209. [DOI] [PubMed] [Google Scholar]
- Ashcroft GS, Mills SJ, Lei K, Gibbons L, Jeong MJ, Taniguchi M, Burow M, Horan MA, Wahl SM, Nakayama T. Estrogen modulates cutaneous wound healing by downregulating macrophage migration inhibitory factor. J Clin Invest. 2003;111:1309–1318. doi: 10.1172/JCI16288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calandra T, Roger T. Macrophage migration inhibitory factor: a regulator of innate immunity. Nat Rev Immunol. 2003;3:791–800. doi: 10.1038/nri1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson JD, Shoaibi MA, Maestro R, Carnero A, Hannon GJ, Beach DH. A proinflammatory cytokine inhibits p53 tumor suppressor activity. J Exp Med. 1999;190:1375–1382. doi: 10.1084/jem.190.10.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morand EF, Leech M. Macrophage migration inhibitory factor in rheumatoid arthritis. Front Biosci. 2005;10:12–22. doi: 10.2741/1501. [DOI] [PubMed] [Google Scholar]
- Schober A, Bernhagen J, Thiele M, Zeiffer U, Knarren S, Roller M, Bucala R, Weber C. Stabilization of atherosclerotic plaques by blockade of macrophage migration inhibitory factor after vascular injury in apolipoprotein E-deficient mice. Circulation. 2004;109:380–385. doi: 10.1161/01.CIR.0000109201.72441.09. [DOI] [PubMed] [Google Scholar]
- Bernhagen J, Calandra T, Mitchell RA, Martin SB, Tracey KJ, Voelter W, Manogue KR, Cerami A, Bucala R. MIF is a pituitary-derived cytokine that potentiates lethal endotoxaemia. Nature. 1993;365:756–759. doi: 10.1038/365756a0. [DOI] [PubMed] [Google Scholar]
- Yu CM, Lai KW, Chen YX, Huang XR, Lan HY. Expression of macrophage migration inhibitory factor in acute ischemic myocardial injury. J Histochem Cytochem. 2003;51:625–631. doi: 10.1177/002215540305100508. [DOI] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatetsky-Shapiro G, Tamayo P. Microarray data mining: facing the Challenges. SIGKDD Explorations. 2003;5:1–5. [Google Scholar]
- Werner S, Grose R. Regulation of wound healing by growth factors and cytokines. Physiol Rev. 2003;83:835–870. doi: 10.1152/physrev.2003.83.3.835. [DOI] [PubMed] [Google Scholar]
- Tomasek JJ, Gabbiani G, Hinz B, Chaponnier C, Brown RA. Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat Rev Mol Cell Biol. 2002;3:349–363. doi: 10.1038/nrm809. [DOI] [PubMed] [Google Scholar]
- Takubo T, Tatsumi N. Microscopic co-distributions of myosin, actin, alpha-actinin and vinculin in human neutrophils during movement. Haematologia. 1999;29:285–294. [PubMed] [Google Scholar]
- Donate F, Juarez JC, Guan X, Shipulina NV, Plunkett ML, Tel-Tsur Z, Shaw DE, Morgan WT, Mazar AP. Peptides derived from the histidine-proline domain of the histidine-proline-rich glycoprotein bind to tropomyosin and have antiangiogenic and antitumor activities. Cancer Res. 2004;64:5812–5817. doi: 10.1158/0008-5472.CAN-04-0440. [DOI] [PubMed] [Google Scholar]
- Richard P, Charron P, Carrier L, Ledeuil C, Cheav T, Pichereau C, Benaiche A, Isnard R, Dubourg O, Burban M, Gueffet JP, Millaire A, Desnos M, Schwartz K, Hainque B, Komajda M, EUROGENE Heart Failure Project Hypertrophic cardiomyopathy: distribution of disease genes, spectrum of mutations, and implications for a molecular diagnosis strategy. Circulation. 2003;107:2227–2232. doi: 10.1161/01.CIR.0000066323.15244.54. [DOI] [PubMed] [Google Scholar]
- Agah A, Kyriakides TR, Letrondo N, Bjorkblom B, Bornstein P. Thrombospondin 2 levels are increased in aged mice: consequences for cutaneous wound healing and angiogenesis. Matrix Biol. 2004;22:539–547. doi: 10.1016/j.matbio.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Kyriakides TR, Tam JW, Bornstein P. Accelerated wound healing in mice with a disruption of the thrombospondin 2 gene. J Invest Dermatol. 1999;113:782–787. doi: 10.1046/j.1523-1747.1999.00755.x. [DOI] [PubMed] [Google Scholar]
- Meyer-Siegler K. Macrophage migration inhibitory factor increases MMP-2 activity in DU-145 prostate cells. Cytokine. 2000;12:914–921. doi: 10.1006/cyto.2000.0682. [DOI] [PubMed] [Google Scholar]
- Chen YH, Layne MD, Chung SW, Ejima K, Baron RM, Yet SF, Perrella MA. Elk-3 is a transcriptional repressor of nitric-oxide synthase 2. J Biol Chem. 2003;278:39572–39577. doi: 10.1074/jbc.M308179200. [DOI] [PubMed] [Google Scholar]
- McLoughlin RM, Hurst SM, Nowell MA, Harris DA, Horiuchi S, Morgan LW, Wilkinson TS, Yamamoto N, Topley N, Jones SA. Differential regulation of neutrophil-activating chemokines by IL-6 and its soluble receptor isoforms. J Immunol. 2004;172:5676–5683. doi: 10.4049/jimmunol.172.9.5676. [DOI] [PubMed] [Google Scholar]
- Garnis C, Campbell J, Davies JJ, Macaulay C, Lam S, Lam WL. Involvement of multiple developmental genes on chromosome 1p in lung tumorigenesis. Hum Mol Genet. 2005;14:475–482. doi: 10.1093/hmg/ddi043. [DOI] [PubMed] [Google Scholar]
- Mitchell RA. Mechanisms and effectors of MIF-dependent promotion of tumourigenesis. Cell Signal. 2004;16:13–19. doi: 10.1016/j.cellsig.2003.07.002. [DOI] [PubMed] [Google Scholar]
- Thelu J, Rossio P, Favier B. Notch signalling is linked to epidermal cell differentiation level in basal cell carcinoma, psoriasis and wound healing. BMC Dermatol. 2002;2:7. doi: 10.1186/1471-5945-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto A, Martinez-Arias A, Martin P. Mechanisms of epithelial fusion and repair. Nat Cell Biol. 2001;3:E117–E123. doi: 10.1038/35074643. [DOI] [PubMed] [Google Scholar]
- Pedersen TX, Leethanakul C, Patel V, Mitola D, Lund LR, Dano K, Johnsen M, Gutkind JS, Bugge TH. Laser capture microdissection-based in vivo genomic profiling of wound keratinocytes identifies similarities and differences to squamous cell carcinoma. Oncogene. 2003;22:3964–3976. doi: 10.1038/sj.onc.1206614. [DOI] [PubMed] [Google Scholar]
- Esworthy RS, Aranda R, Martin MG, Doroshow JH, Binder SW, Chu FF. Mice with combined disruption of Gpx1 and Gpx2 genes have colitis. Am J Physiol Gastrointest Liver Physiol. 2001;281:G848–G855. doi: 10.1152/ajpgi.2001.281.3.G848. [DOI] [PubMed] [Google Scholar]
- Satoskar AR, Bozza M, Rodriguez Sosa M, Lin G, David JR. Migration-inhibitory factor gene-deficient mice are susceptible to cutaneous Leishmania major infection. Infect Immunol. 2001;69:906–911. doi: 10.1128/IAI.69.2.906-911.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown FG, Nikolic-Paterson DJ, Hill PA, Isbel NM, Dowling J, Metz CM, Atkins RC. Urine macrophage migration inhibitory factor reflects the severity of renal injury in human glomerulonephritis. J Am Soc Nephrol. 2002;13(Suppl 1):S7–S13. [PubMed] [Google Scholar]
- Girard MT, Matsubara M, Kublin C, Tessier MJ, Cintron C, Fini ME. Stromal fibroblasts synthesize collagenase and stromelysin during long-term tissue remodeling. J Cell Sci. 1993;104:1001–1011. doi: 10.1242/jcs.104.4.1001. [DOI] [PubMed] [Google Scholar]
- Ceradini DJ, Kulkarni AR, Callaghan MJ, Tepper OM, Bastidas N, Kleinman ME, Capla JM, Galiano RD, Levine JP, Gurtner GC. Progenitor cell trafficking is regulated by hypoxic gradients through HIF-1 induction of SDF-1. Nat Med. 2004;10:858–864. doi: 10.1038/nm1075. [DOI] [PubMed] [Google Scholar]
- Neurath MF, Finotto S, Glimcher LH. The role of Th1/Th2 polarization in mucosal immunity. Nat Med. 2002;8:567–573. doi: 10.1038/nm0602-567. [DOI] [PubMed] [Google Scholar]
- Pentecost BT. Expression and estrogen regulation of the HEM45 MRNA in human tumor lines and in the rat uterus. J Steroid Biochem Mol Biol. 1998;64:25–33. doi: 10.1016/s0960-0760(97)00140-4. [DOI] [PubMed] [Google Scholar]
- Agrawal S, Agarwal ML, Chatterjee-Kishore M, Stark GR, Chisolm GM. Stat1-dependent, p53-independent expression of p21(waf1) modulates oxysterol-induced apoptosis. Mol Cell Biol. 2002;22:1981–1992. doi: 10.1128/MCB.22.7.1981-1992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiboutot D, Jabara S, McAllister JM, Sivarajah A, Gilliland K, Cong Z, Clawson G. Human skin is a steroidogenic tissue: steroidogenic enzymes and cofactors are expressed in epidermis, normal sebocytes, and an immortalized sebocyte cell line (SEB-1). J Invest Dermatol. 2003;120:905–914. doi: 10.1046/j.1523-1747.2003.12244.x. [DOI] [PubMed] [Google Scholar]
- Slominski A, Zjawiony J, Wortsman J, Semak I, Stewart J, Pisarchik A, Sweatman T, Marcos J, Dunbar C, Tuckey RC. A novel pathway for sequential transformation of 7-dehydrocholesterol and expression of the P450scc system in mammalian skin. Eur J Biochem. 2004;271:4178–4188. doi: 10.1111/j.1432-1033.2004.04356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araneo BA, Ryu SY, Barton S, Daynes RA. Dehydroepiandrosterone reduces progressive dermal ischemia caused by thermal injury. J Surg Res. 1995;59:250–262. doi: 10.1006/jsre.1995.1162. [DOI] [PubMed] [Google Scholar]
- Parks WT, Frank DB, Huff C, Renfrew Haft C, Martin J, Meng X, de Caestecker MP, McNally JG, Reddi A, Taylor SI, Roberts AB, Wang T, Lechleider RJ. Sorting nexin 6, a novel SNX, interacts with the transforming growth factor-beta family of receptor serine-threonine kinases. J Biol Chem. 2001;276:19332–19339. doi: 10.1074/jbc.M100606200. [DOI] [PubMed] [Google Scholar]
- Guzeloglu-Kayisli O, Kayisli UA, Al-Rejjal R, Zheng W, Luleci G, Arici A. Regulation of PTEN (phosphatase and tensin homolog deleted on chromosome 10) expression by estradiol and progesterone in human endometrium. J Clin Endocrinol Metab. 2003;88:5017–5026. doi: 10.1210/jc.2003-030414. [DOI] [PubMed] [Google Scholar]
- Kato Y, Fujita N, Kunita A, Sato S, Kaneko M, Osawa M, Tsuruo T. Molecular identification of Aggrus/T1alpha as a platelet aggregation-inducing factor expressed in colorectal tumors. J Biol Chem. 2003;278:51599–51605. doi: 10.1074/jbc.M309935200. [DOI] [PubMed] [Google Scholar]
- Ashcroft GS, Greenwell-Wild T, Horan MA, Wahl SM, Ferguson MW. Topical estrogen accelerates cutaneous wound healing in aged humans associated with an altered inflammatory response. Am J Pathol. 1999;155:1137–1146. doi: 10.1016/S0002-9440(10)65217-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis DJ, Knauss J, Bilker W. Hormone replacement therapy and prevention of pressure ulcers and venous leg ulcers. Lancet. 2002;359:675–677. doi: 10.1016/S0140-6736(02)07806-6. [DOI] [PubMed] [Google Scholar]
- Coulombe PA, Tong X, Mazzalupo S, Wang Z, Wong P. Great promises yet to be fulfilled: defining keratin intermediate filament function in vivo. Eur J Cell Biol. 2004;83:735–746. doi: 10.1078/0171-9335-00443. [DOI] [PubMed] [Google Scholar]
- Huang X, Griffiths M, Wu J, Farese RV, Jr, Sheppard D. Normal development, wound healing, and adenovirus susceptibility in beta5-deficient mice. Mol Cell Biol. 2000;20:755–759. doi: 10.1128/mcb.20.3.755-759.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans MJ, Eckert A, Lai K, Adelman SJ, Harnish DC. Reciprocal antagonism between estrogen receptor and NF-kappaB activity in vivo. Circ Res. 2001;89:823–830. doi: 10.1161/hh2101.098543. [DOI] [PubMed] [Google Scholar]
- Rollerova E, Urbancikova M. Intracellular estrogen receptors, their characterization and function. Endocr Regul. 2000;34:203–218. [PubMed] [Google Scholar]
- Speir E, Yu ZX, Takeda K, Ferrans VJ, Cannon RO., III Competition for p300 regulates transcription by estrogen receptors and nuclear factor-kappaB in human coronary smooth muscle cells. Circ Res. 2000;87:1006–1011. doi: 10.1161/01.res.87.11.1006. [DOI] [PubMed] [Google Scholar]
- Fan C, Li Q, Zhang Y, Liu X, Luo M, Abbott D, Zhou W, Engelhardt JF. IkappaBalpha and IkappaBbeta possess injury context-specific functions that uniquely influence hepatic NF-kappaB induction and inflammation. J Clin Invest. 2004;113:746–755. doi: 10.1172/JCI17337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantome A, Pance A, Gauthier N, Vandroux D, Chenu J, Solary E, Jeannin JF, Reveneau S. Casein kinase II-mediated phosphorylation of NF-kappaB p65 subunit enhances inducible nitric-oxide synthase gene transcription in vivo. J Biol Chem. 2004;279:23953–23960. doi: 10.1074/jbc.M313731200. [DOI] [PubMed] [Google Scholar]
- Denkinger CM, Denkinger M, Kort JJ, Metz C, Forsthuber TG. In vivo blockade of macrophage migration inhibitory factor ameliorates acute experimental autoimmune encephalomyelitis by impairing the homing of encephalitogenic T cells to the central nervous system. J Immunol. 2003;170:1274–1282. doi: 10.4049/jimmunol.170.3.1274. [DOI] [PubMed] [Google Scholar]
- Pence H, Petty WM, Rocklin RE. Suppression of maternal responsiveness to paternal antigen by maternal plasma. J Immunol. 1975;114:525–528. [PubMed] [Google Scholar]
- Tofoski JG, Gill TJ., III The production of migration inhibitory factor and reproductive capacity in allogeneic pregnancies. Am J Pathol. 1977;88:333–344. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.