Abstract
To understand the molecular basis that supports the dynamic gene expression programs unique to T cells, we investigated the genomic landscape of activating histone modifications, including histone H3 K9/K14 diacetylation (H3K9acK14ac), H3 K4 trimethylation (H3K4me3), and the repressive histone modification H3 K27 trimethylation (H3K27me3) in primary human T cells. We show that H3K9acK14ac and H3K4me3 are associated with active genes required for T cell function and development, whereas H3K27me3 is associated with silent genes that are involved in development in other cell types. Unexpectedly, we find that 3,330 gene promoters are associated with all of these histone modifications. The gene expression levels are correlated with both the absolute and relative levels of the activating H3K4me3 and the repressive H3K27me3 modifications. Our data reveal that rapidly inducible genes are associated with the H3 acetylation and H3K4me3 modifications, suggesting they assume a chromatin structure poised for activation. In addition, we identified a subpopulation of chromatin regions that are associated with high levels of H3K4me3 and H3K27me3 but low levels of H3K9acK14ac. Therefore, these regions have a distinctive chromatin modification pattern and thus may represent a distinct class of chromatin domains.
Keywords: chromatin landscape, epigenome, gene expression, genome-wide mapping
Numerous covalent modifications on histones have been discovered, most of which occur at the N-terminal tails of histones (1). Depending on the nature and position of the modification, they are linked to different gene activities. For example, acetylation of histone H3 on lysines 9 and 14 and methylation on lysines 4, 36, and 79 (activating marks) are linked to active transcription, whereas methylation on lysines 9 and 27 (repressive marks) is linked to transcriptional repression (1–3). Histone acetylation and deacetylation are highly dynamic processes that are controlled by the counteracting action of histone acetyltransferases (HATs) and histone deacetylases (HDACs) (4). Histone methylation catalyzed by histone methyltransferases (HMTs) is believed to be a more stable epigenetic mark and can be associated with either gene activation or repression, depending on the nature and position of the modification (5). Activating marks such as histone acetylation and H3 K4 trimethylation (H3K4me3) are colocalized in active chromatin (6–9), whereas repressive marks, including H3 K9 trimethylation (H3K9me3) and H3 K27 trimethylation (H3K27me3), are found in inactive chromatin (10–12). Different hypotheses, including histone code and signaling platform models, have been proposed for mechanisms used in regulation of gene expression by histone modifications (13–16).
Human resting T cells circulating in the bloodstream have a unique gene expression program that distinguishes them from other cell types (17). Activation of T cells upon encountering specific antigens rapidly induces or represses thousands of genes. Accompanying the gene expression changes during T cell activation and differentiation, extensive changes in histone modifications have been detected in several lymphocyte-specific genes (18–24). To understand the relationship between histone modifications and gene expression on a whole-genome scale, we have investigated the landscape of genome-wide histone modifications, including both the activating marks H3K9acK14ac and H3K4me3 and the repressive mark H3K27me3, by using an unbiased genome-wide mapping technique (GMAT) that combines the chromatin immunoprecipitation (ChIP) and serial analysis of gene expression (SAGE) protocols (25, 26). Our data indicate that constitutively expressed or rapidly inducible genes, which are required for T cell development and function, are associated with high levels of H3 acetylation and H3K4me3 signals, whereas permanently silenced genes, which are involved in the development of other cell types, are associated with high levels of H3K27me3 signals. Unexpectedly, we found that not only are the H3K4me3 and H3K27me3 modifications colocalized at thousands of promoters in differentiated human T cells, but also both their absolute and relative levels correlate with the expression levels of these genes. Our data suggest that these histone modifications together may define the chromatin landscape important for gene expression in T cells.
Results
Both Activating and Repressive Epigenetic Marks Are Enriched in the Promoter Region.
GMAT analysis involves sequencing ChIP DNA samples by using the long-serial analysis of gene expression protocol (SAGE), which detects 21-bp sequence tags from each DNA fragment pulled-down by specific antibodies. The detection frequency of each tag represents the level of a particular histone modification at the genomic locus containing the sequence tag. Our previous analysis with GMAT revealed that histone acetylation islands are correlated with transcription/chromatin regulatory elements, including promoters, enhancers, and locus control regions (LCRs) (25). We now have determined the distribution of other important histone modifications, including H3K4me3 and H3K27me3, in the entire genome of human primary T cells. A comprehensive epigenomic map was constructed based on these data (see Fig. 7, which is published as supporting information on the PNAS web site; a searchable and interactive high-resolution map can be found at http://dir.nhlbi.nih.gov/papers/lmi/epigenomes).
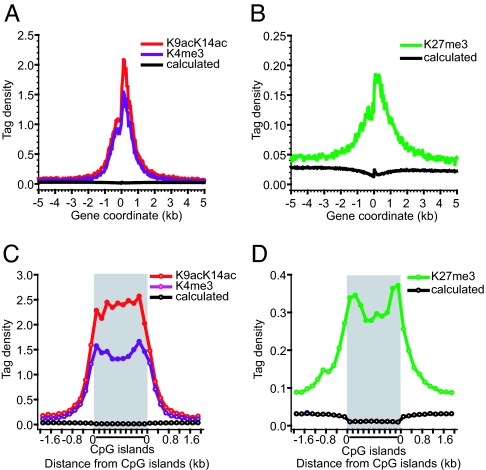
Activating marks, including H3K9acK14ac and H3K4me3, are concentrated in promoter regions in the human genome (7, 9, 25). To compare the distribution between these activating marks to those of repressive marks, we aligned 21,355 annotated genes relative to their transcription start sites (TSSs) and plotted the levels of modification across a 10-kb region. Interestingly, all marks examined were enriched in a 2-kb promoter region surrounding the TSSs (Fig. 1A and B), indicating that the promoter regions are critical target sites for repressive marks as well (Fig. 1B). It is interesting to note that the peak acetylation and H3K4me3 signals appear to occur immediately after the TSSs (Fig. 1A). CpG islands are important regulatory elements of transcription that exist in more than half of the human genes (27). Our previous studies indicated that they are associated with high levels of H3 acetylation (25). We now found that the H3K4me3 and H3K27me3 signals also are enriched in the CpG islands (Fig. 1 C and D). Interestingly, CpG islands outside promoter regions also were modified at a similar level to the promoter-associated CpG islands (data not shown). Analysis with a segment-based χ2 test indicates that the enrichment of all modifications in both the promoter regions and the CpG islands was statistically significant (P < 0.001).
Fig. 1.
Critical transcriptional regulatory elements are targets for both active and repressive epigenetic marks. (A) Here, 10-kb regions of 21,355 genes were aligned relative to their TSSs (x axis). The y axis shows the detected tag density. The black line indicates the calculated value, assuming the tags were detected randomly. H3K9acK14ac and H3K4me3 are represented by red and dark violet lines, respectively. (B) Same as in A, except that H3K27me3 is represented by green. (C) The detected tags within 4,973 CpG islands with sizes of 1–2 kb were counted in a window corresponding to 10% of the CpG island length and normalized to the number of NlaIII sites in that window to yield the tag density. The GMAT tags outside of the CpG islands were similarly counted in a 200-bp window and normalized to the number of NlaIII sites in the window. Each value point represents 10% of the sequence length within the CpG islands and 200 bp outside of the CpG islands. The color codes are the same as in A. (D) Same as in C, except that H3K27me3 is represented by green.
It is noteworthy that the H3K27me3 signals were 8- to 10-fold lower than the H3 acetylation and H3K4me3 signals in the promoter regions, suggesting that H3K27me3 is less enriched in gene promoter regions. Indeed, we found that 34% and 32% of the total H3 acetylation and H3K4me3 tags, respectively, were detected in the 3-kb promoter regions surrounding the TSSs, whereas only 7% of the total H3K27me3 tags were detected in the promoter regions, indicating that the H3K27me3 modification is more spread out in the genome. Because the promoters marked by the H3K27me3 modification are different from the ones marked by the H3K4me3 and H3 acetylation modifications (Fig. 2B), it is not likely that the H3K27me3 enrichment in the promoter regions was caused by antibody cross-reactivity. However, we cannot rule out the possibility that the chromatin regions associated with the H3K27me3 modification may be pulled down less efficiently than the regions modified by H3K4me3 because they might be less accessible.
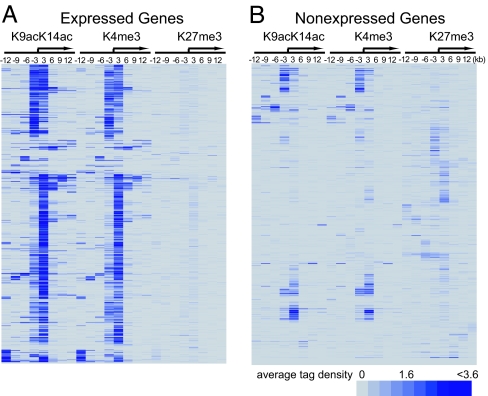
Fig. 2.
H3K4me3 associates with active genes, and H3K27me3 associates with silent genes in T cells. (A) Cluster analysis of H3K9acK14ac, H3K4me3, and H3K27me3 in 1,000 highly active genes. The cluster analysis was performed by using the average tag densities calculated in 3-kb intervals over a total of 24 kb (−12 kb to +12 kb) as shown. A blue band in a segment indicates the detection of the mark, and the relative intensity of band indicates the relative level of the modification. The arrow indicates the TSS. (B) Cluster analysis of H3K9acK14ac, H3K4me3, and H3K27me3 in 1,000 nonexpressed genes, as described in A.
H3K4me3 and H3K27me3 Modifications Associate with Functionally Distinct Genes in T Cells.
To confirm that H3K4me3 associates with active genes whereas H3K27me3 associates with silent genes, we examined the histone modification levels of the 1,000 most active genes and 1,000 silent genes in human T cells (17) by using cluster analysis. We found high levels of both acetylation and K4 methylation in the promoters of almost all of the 1,000 most active genes (t test, P < 0.0001; Fig. 2A). In contrast, the K27 methylation was detected to at least some extent in 76% of the 1,000 silent genes (t test, P = 0.0014; Fig. 2B). Interestingly, many silent promoters were associated with high levels of H3 acetylation and H3 K4 trimethylation (Fig. 2B). Further analysis revealed that many of these promoters are from rapidly inducible genes including NFATC2, AKT2, and RHEB, suggesting that these promoters are in an open chromatin structure and poised for rapid activation.
Next we selected 2,273 and 1,336 genes with high levels of the H3K4me3 and H3K27me3 modifications, respectively, for pathway analysis. The top 10 functional pathways associated with these modifications are shown in Fig. 8A, which is published as supporting information on the PNAS web site. The pathways marked by H3K4me3 signals include T cell receptor (TCR) signaling, Jak-STAT signaling, and cytokine–cytokine receptor interaction pathways (Fig. 8A), which are critically required for T cell development and differentiation. Interestingly, even though the T cells used for the analysis were resting cells, several molecules involved in cell-cycle progression were associated with high levels of H3K4me3 signals (Fig. 8A), suggesting that the cells are prepared to proliferate in response to TCR signaling. In contrast, the pathways marked by H3K27me3 include neuroactive ligand-receptor interaction and axon guidance pathways (Fig. 8B), which are not required for T cell function. Among these genes were transcription factors and cell surface signaling molecules required for development of cells other than T cells, suggesting that they need to be repressed for normal T cell development. We note that many critical transcription factors involved in development also are associated with H3K27me3 in human ES cells (28). Interestingly, only about one-third of K27-methylated regions in the human ES cells overlapped with highly K27-methylated regions in T cells, which mainly contained the genes involved in transcription regulation (data not shown). Many genes encoding cell membrane proteins involved in receptor activity were associated with high levels of H3K27me3 signals in T cells but not in ES cells (data not shown). Our results therefore reveal that the genes involved in T cell development and function are associated with the H3K4me3 modification, whereas the genes involved in development of other cell types are associated with the H3K27me3 modification.
Rapidly Inducible Genes Are Associated with H3 Acetylation and H3K4me3 Modifications.
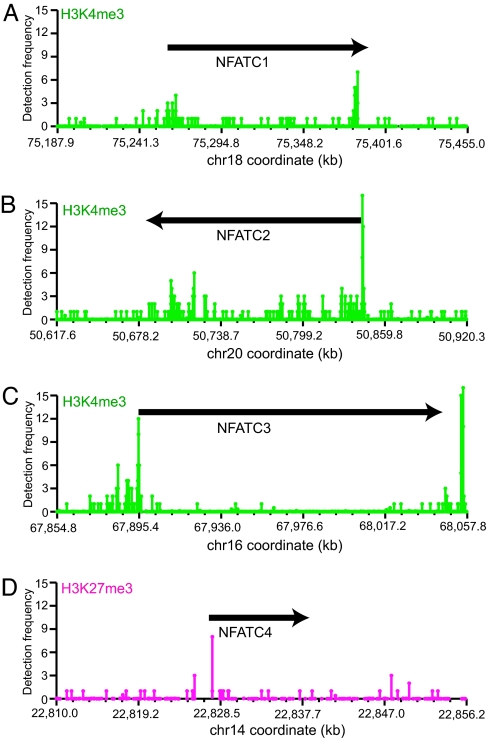
Human genes can be classified into expressed and nonexpressed genes; however, some of the expressed genes are constitutively expressed, whereas others need to be turned off during development or in response to environmental stimuli. Analogously, some of the nonexpressed genes are constitutively silent in certain cell lineages because their ectopic expression might be very toxic, whereas other nonexpressed genes may need to be rapidly turned on during development or in response to stimuli. The complex and diverse patterns of gene expression may be associated with diverse patterns of histone modifications. For example, the constitutively expressed housekeeping β-actin gene exists in a highly open chromatin structure characterized by high levels of H3 acetylation and H3K4me3 throughout the locus (data not shown). The NFATC1, NFATC2, and NFATC3 genes, which are required for T cell activation and differentiation (29, 30), are transcribed only at minimal levels in resting T cells but are rapidly induced by TCR signaling. Interestingly, they were associated with high levels of H3K4me3 signals, especially at their promoter regions (Fig. 3A–C). It also is interesting to note that we detected clusters of H3K4me3 signals near the 3′ ends and in the transcribed regions of these NFAT genes, which may denote regulatory elements for these genes. In contrast, NFATC4, which is not expressed in T cells, is associated with only low levels of H3K4me3 but high levels of H3K27me3 signals (Fig. 3D and data not shown). The high levels of the H3K4me3 signal of the NFATC1, NFATC2, and NFATC3 genes suggest that their minimal levels of transcription in resting T cells may result from a “lack of activation.” When an activator becomes available, as in the case of TCR signaling, their transcription can be rapidly activated. In contrast, the silent NFATC4 and NeuroD genes were associated with high levels of H3K27me3 signals (Fig. 3D and data not shown) to ensure their silent states in T cells. Together, these results are consistent with the notion that the chromatin modified with H3K4me3 and H3 acetylation is competent for transcription, and the chromatin marked with H3K27me3 is incompetent for transcription.
Fig. 3.
The H3K4me3 modification for the TCR-inducible NFATC1 (A), NFATC2 (B), and NFATC3 (C) genes and H3K27me3 modification for the silent and noninducible NFATC4 gene (D) are shown. The positions of these genes are indicated. x axis, chromosome coordinate of the locus. y axis, level of the modification as indicated by the detection frequency of the GMAT tags.
Genome-Wide Colocalization Analysis of the H3K9acK14ac, H3K4me3, and H3K27me3 Modifications.
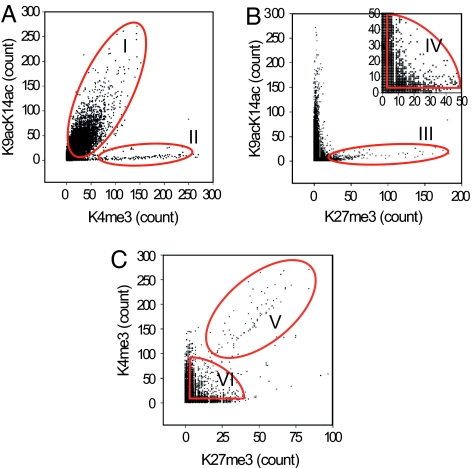
To examine the location relationship between these different histone modifications in an unbiased way for the entire human genome in T cells, we analyzed whole-genome scattergrams of pairwise marks by comparing their average modification levels in 3-kb sequence windows. The results show that ≈90% of the H3K9acK14ac and H3K4me3 signals were colocalized in the human genome (Fig. 4A). Circle I highlighted 14,528 such genomic regions. Interestingly, a distinct population of 357 chromatin regions was associated with high levels of H3K4me3 but with no or low levels of H3K9acK14ac (Fig. 4A, highlighted in circle II). Because these regions are far from any identified genes, it is possible that H3K4me3 has a function in addition to its participation in transcriptional activation of promoters. The scatter analysis between H3K9acK14ac and H3K27me3 indicates that the highly K27-methylated regions did not overlap with the highly acetylated chromatin regions (Fig. 4B, circle III). Circle IV Fig. 4B Inset highlights 8,636 genomic regions with low to moderate levels (3 to 50 detected tag counts) of both H3K9acK14ac and H3K27me3. As we showed previously (25), most tags (even single-copy tags) are true positive signals. Moreover, the data in Fig. 6 A and B also show that this level of modification is well correlated with gene expression. Therefore, the signals in this region are not just noise but true signals. Our data indicate that the H3K9acK14ac and H3K27me3 modifications colocalize in a significant population of chromatin regions. The comparison between H3K4me3 and H3K27me3 revealed extensive colocalization of these two marks in the regions highlighted as V and VI (Fig. 4C). Most of the regions in V are identical with those in region II (Fig. 4A). Therefore, circle VI represents chromatin regions with low to moderate levels of H3K9acK14ac, H3K4me3, and H3K27me3 modifications, many of which are located in gene promoter regions, whereas circle V marks chromatin regions with high levels of H3K4me3 and H3K27me3 but low levels of H3 acetylation signals, which are located far from any known genes. It was recently observed that the H3K4me3 and H3K27me3 modifications colocalize at several genes critical for development in human ES cells, suggesting that the H3K4me3 signal may prime the chromatin structure for the genes to be activated during development (31). However, our discovery of the extensive colocalization of these two marks suggests that these modifications together may define chromatin landscape in a wide range of genomic regions.
Fig. 4.
Genome-wide colocalization analysis of the H3K9acK14ac, H3K4me3, and H3K27me3 modifications. Pairwise scatter analyses of the histone modifications. The histone modifications indicated were analyzed by using GMAT. The number of tags (tag count) detected in 3-kb windows was calculated across the entire human genome. The scatter analyses of the tag counts were performed by comparing two histone modifications in A–C, as indicated. B Inset shows the windows with low to moderate tag numbers between 3 and 50.
Fig. 6.
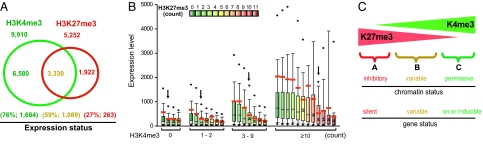
Gene expression levels correlate with both the absolute and the relative levels of the H3K27me3 and H3K4me3 modifications. (A) The Venn diagram depicts the promoters associated with three tags or more of H3K4me3 (green circle) and H3K27me3 (red circle). The percentage of genes expressed and the average expression levels detected by DNA microarrays are indicated in parentheses below the circles. (B) Box-and-whisker plot showing the range of gene expression levels correlated with the histone modifications. The genes are grouped first according to H3K4me3 levels (by the total GMAT tag count in a 3-kb promoter region) and then subgrouped according to H3K27me3 levels (by the total GMAT tag count in a 3-kb promoter region). The y axis represents the arbitrary gene expression levels detected by DNA microarray analysis. The number of H3K27me3 tags corresponding to an ≈50% decrease in expression levels is indicated by the arrow above the column. The boundary of the box closest to 0 indicates the 25th percentile; the line within the box indicates the median; the boundary of the box farthest from 0 indicates the 75th percentile; and whiskers above and below the box indicate the 5th and 95th percentiles, respectively. The outlying points are shown as filled circles. The red thick lines indicate the mean values of each group. ∗ indicates that the decrease in gene expression levels compared with no H3K27me3 tags is significant (P < 0.01). (C) A Model. An inhibitory or permissive chromatin structure is generated by coexistence of the H3K27me3 and H3K4me3 modifications.
Sequential ChIP Assays Confirm the Coexistence of Activating and Repressive Marks at the Same Promoters.
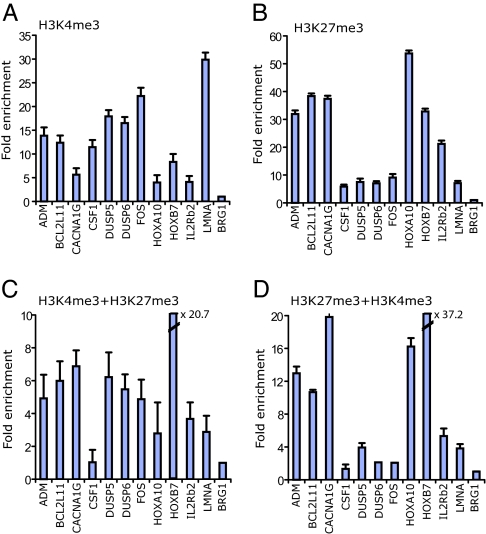
The detection of these apparently opposing modifications at the same genomic loci raises the possibility that their coexistence may be caused by cellular heterogeneity. The CD3+ T cells we used for the genome-wide analysis consisted of ≈33% CD8+ T cells and 67% CD4+ T cells. To confirm that the colocalization of the H3K4me3 and H3K27me3 modifications was not caused by cellular heterogeneity, we analyzed several promoters, which were associated with moderate levels of H3K9acK14ac, H3K4me3, and H3K27me3 modifications in the GMAT libraries (Circles I, IV, and VI in Fig. 4), by using ChIP assays with purified CD4+ T cells that are major components of the CD3+ T cells. Indeed, our data confirmed that most tested promoters were associated with both H3K4me3 and H3K27me3 modifications (Fig. 5A and B). To determine whether these two modifications are located on the same allele and are not independent modifications of the two alleles in CD4+ T cells, we further analyzed these promoters by using sequential ChIP assays. As a positive control, we performed the first ChIP assay by using an anti-H3K4me3 antibody and the second ChIP by using an anti-H3K9acK14ac antibody. This analysis confirmed the colocalization of H3K4me3 and H3K9acK14ac at all of the tested promoters (data not shown). When an anti-H3K27me3 antibody was used for the second ChIP, we detected the colocalization of H3K4me3 and H3K27me3 at all tested promoters except CSF1 (Fig. 5C). Similar results were obtained when the first ChIP was performed by using an anti-H3K27me3 antibody and the second ChIP by using an anti-H3K4me3 antibody (Fig. 5D). These data indicate that these two modifications occur on the same allele. However, our data do not address whether these modifications actually occur on the same nucleosome or on neighboring nucleosomes or whether they are attributable to the cross-linking of distant chromatin fragments in the ChIP assays. At some promoters, it is possible that the detection of these two modifications may be caused by the H3K4me3 modification occurring on one allele and the H3K27me3 modification occurring on the other allele, for example, in a situation of imprinting or monoallelic expression.
Fig. 5.
Colocalization of H3K4me3 and H3K27me3 signals is confirmed by sequential ChIP assays. (A and B) ChIP assays were performed by using anti-H3K4me3 (A) and anti-H3K27me3 (B) antibodies. The fold enrichment of different promoters relative to a BRG1 upstream region was determined by using real-time quantitative PCR. (C and D) Sequential ChIP experiments. (C) H3K4me3 + H3K27me3, sequential ChIP using first the H3K4me3 antibody followed by the H3K27me3 antibody. (D) H3K27me3 + H3K4me3, sequential ChIP using first the H3K27me3 antibody followed by the H3K4me3 antibody. The fold enrichment was determined as described above. The experiments were performed in triplicate.
Gene Expression Levels Correlate with both the Absolute and Relative Levels of the H3K27me3 and H3K4me3 Modifications.
To clarify whether the colocalization of the H3K4me3 and H3K27me3 signals correlates with gene expression, we compared the histone modification patterns with gene expression levels of 16,566 genes, which can be identified as annotated genes from a T cell gene expression database (17). The H3K4me3 signal was detected in 9,910 promoters (60% of all promoters) (Fig. 6A). H3K27me3 was detected in 5,252 promoters (32% of all promoters) (Fig. 6A). Surprisingly, 3,330 promoters were associated with both the activating (H3K4me3) and the repressive (H3K27me3) mark. Because H3K4me3 and H3K9acK14ac were highly correlated at >95% of the promoters, we did not distinguish between the two in the following analysis.
The Venn diagram in Fig. 6A shows three groups of genes based on their H3K4me3 and H3K27me3 modification patterns. The expression levels of each group, which was measured by the average expression levels detected by DNA microarray analysis (17) and the percentage of the genes expressed in the group (250 as a cutoff value for expression), is indicated below the groups. The analysis revealed that ≈76% of the K4-methylated genes were expressed at an average level of 1,684 (arbitrary units), whereas only 27% of the K27-methylated promoters were expressed at a level of only 263. The genes with both the H3K4me3 and H3K27me3 modifications were expressed at intermediate levels (59%; 1,089). To see whether there is a quantitative correlation between gene expression and these two modifications, we used a box-and-whisker plot to show the distribution of gene expression levels relative to the levels of the H3K4me3 and H3K27me3 modifications. The 16,566 genes first were grouped according to the sum of the H3K4me3 tag counts detected in 4-kb promoter regions and then subgrouped according to the sum of the H3K27me3 tag counts in the same regions by using colors from green to red (Fig. 6B). The results indicated that genes with higher levels of the H3K4me3 signal tend to be expressed at higher levels. The presence of H3K27me3 signals tended to correlate with decreased levels of expression in each group. The higher the number of H3K27me3 tags, the lower the expression levels. Interestingly, it seems that the H3K27me3 levels corresponding to a 50% decrease in expression levels depended on the H3K4me3 level. For example, when there were no H3K4me3 signals detected, the presence of one H3K27me3 tag correlated with a 50% decrease in expression (P < 0.01). When there were two tags of H3K4me3, two H3K27me3 tags correlated with a 50% decrease in expression (P < 0.01). When there were >10 H3K4me3 tags, the number of H3K27me3 tags that correlated with a 50% decrease in gene expression increased to 8 (P < 0.01). These data suggest that H3K27me3 acts as a repressive mark, and H3K4me3 correlates with gene activation. Moreover, not only the absolute level of these histone modifications but also the ratios of H3K4me3 to H3K27me3 signals appear to be critical factors associated with chromatin activity.
Discussion
In this study, we have examined and compared the distribution of several critical histone modifications in the entire genome of human T cells. Previous analyses by using ChIP-on-chip assays with tiling arrays covering either chromosomes 21 and 22 or 9,328 1-kb promoters indicate that H3 acetylation and H3K4me3 signals are highly correlated in the promoter regions (7, 9). Our entire genome analysis confirmed the high correlation between the acetylation and H3K4me3 signals in the promoter region. Furthermore, our data indicate that a subpopulation of chromatin regions, which are located far away from any known promoters, are associated with high levels of H3K4me3 and H3K27me3 but low levels of H3K9acK14ac. The histone modifications in these regions differ from both heterochromatin and euchromatin as they are usually described in the literature. A similar case reported in the literature is that centromeric chromatin in Drosophila is associated with H3K4me2 and H3K9me2,3 but depleted of H3K4me3 and H3K9ac (32), indicating that different chromosomal locations may be characterized with different combinations of histone modifications. Therefore, the regions marked by H3K4me3 and H3K27me3 but not H3K9ac may represent chromosomal locations with possibly novel functions.
Correlation of histone modifications with gene expression patterns indicates that H3K4me3 generally is associated with gene expression, whereas H3K27me3 generally is correlated with gene silencing. We find in this study that thousands of promoters are associated with both the H3K4me3 and H3K27me3 modifications in differentiated human T cells, indicating that coexistence of these apparently opposing marks is a widespread phenomenon. Moreover, our data indicate that the levels of gene expression are correlated with both the absolute and relative levels of the H3K4me3 and H3K27me3 signals.
Is histone modification a step of chromatin modification before gene activation or is it simply a consequence of transcriptional activation or repression? Transcriptional activation results in recruitment of histone modification enzymes and, therefore, increased levels of modifications in yeast (33). Indeed, elevated levels of the histone acetylation and H3K4me3 modifications are detected at many promoters and other regulatory regions upon gene activation by TCR signaling (ref. 25 and data not shown). However, we did not detect significant increases in the histone modification levels of many induced genes upon TCR signaling (data not shown), suggesting that histone modifications occur at an earlier step than transcriptional activation for these genes, and they might be a prerequisite for gene activity. We hypothesize that the H3 acetylation and H3K4me3 modifications create a “permissive” chromatin structure (chromatin status C in Fig. 6C), which allows processes such as transcriptional activation and DNA repair. In contrast, the H3K27me3 modification creates an “inhibitory” chromatin structure that does not support active transcription (chromatin status A in Fig. 6C). However, the permissiveness of the chromatin status B is variable and depends on both the absolute and relative levels of these supposedly opposing modifications. Counteracting effects of histone modifications have been demonstrated for histone acetylation and deacetylation in yeast (4) as well as in higher eukaryotes (34). Our findings in this study indicate that the H3K4me3 and H3K27me3 modifications coexist at many genomic regions, and their levels are reciprocally correlated with gene expression levels, suggesting that these two modifications may have counteracting effects in gene expression. Environmental stimuli could shift the equilibrium of these opposing modifications, which would generate a chromatin configuration that may aid or deter gene transcription during T cell development (Fig. 6C). Although we demonstrate the quantitative and reciprocal effects of colocalized activating marks (H3K9acK14ac and H3K4me3) and a repressive mark (H3K27me3) in chromatin activity and gene expression, it is clear that the chromatin status is regulated by many more modifications and regulatory factors in reality. Therefore, it will be necessary to map other critical histone modifications and effector molecules to better understand the mechanisms that control the chromatin function.
Materials and Methods
GMAT Data Analysis.
The GMAT libraries were prepared as described (25). A total of 706,001 and 708,300 21-bp tags were obtained by sequencing 30,720 and 30,336 GMAT clones for H3K4me3 and H3K27me3, respectively. The detection frequency was determined by normalizing the number of times of detection (tag count) to the number of copies of the tag sequence in the genome. The tag density was calculated by dividing the sum of detection frequencies of all tags in a 50-bp window for Fig. 1 and a 3-kb region for Fig. 2 by the number of expected NlaIII sites in the window (25).
The modification levels of 4-kb gene promoters (2 kb upstream and 2 kb downstream of the TSS) were calculated by summing up detection frequencies for all tags detected in the promoters. The promoters with three or more detection frequencies were selected to calculate average expression levels in Fig. 6. All statistical calculations were done by the SAS program. Details can be found in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Sequential ChIP.
Chromatin fractions were prepared as described in ref. 25 by using CD4+ T cells purified from human blood with a CD4+ T cell isolation kit from Miltenyi Biotech (Auburn, CA). The sequential ChIP assays were performed following a published protocol (31). The ChIP or re-ChIP DNA samples were analyzed by real-time PCR using Taqman probes on an ABI 7900HT system (Applied Biosystems, Foster City, CA). The experiments were performed in triplicate.
Supplementary Material
Acknowledgments
We thank Drs. Xin Chen, Tian Chi, Catherine Farrell, Warren J. Leonard, and Carl Wu for critical reading of the manuscript and insightful suggestions. This work was supported by intramural grants to the National Heart, Lung, and Blood Institute, National Institutes of Health.
Abbreviations
- GMAT
genome-wide mapping technique
- ChIP
chromatin immunoprecipitation
- TSS
transcription start site
- TCR
T cell receptor.
Footnotes
The authors declare no conflict of interest.
References
- 1.Berger SL. Curr Opin Genet Dev. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- 2.Wu J, Grunstein M. Trends Biochem Sci. 2000;25:619–623. doi: 10.1016/s0968-0004(00)01718-7. [DOI] [PubMed] [Google Scholar]
- 3.Kornberg RD, Lorch Y. Curr Opin Genet Dev. 1999;9:148–151. doi: 10.1016/S0959-437X(99)80022-7. [DOI] [PubMed] [Google Scholar]
- 4.Kurdistani SK, Grunstein M. Nat Rev Mol Cell Biol. 2003;4:276–284. doi: 10.1038/nrm1075. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Reinberg D. Genes Dev. 2001;15:2343–2360. doi: 10.1101/gad.927301. [DOI] [PubMed] [Google Scholar]
- 6.Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O'Neill LP, Turner BM, Delrow J, et al. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, III, Gingeras TR, et al. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 8.Pokholok DK, Harbison CT, Levine S, Cole M, Hannett NM, Lee TI, Bell GW, Walker K, Rolfe PA, Herbolsheimer E, et al. Cell. 2005;122:517–527. doi: 10.1016/j.cell.2005.06.026. [DOI] [PubMed] [Google Scholar]
- 9.Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B. Nature. 2005;436:876–880. doi: 10.1038/nature03877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobs SA, Taverna SD, Zhang Y, Briggs SD, Li J, Eissenberg JC, Allis CD, Khorasanizadeh S. EMBO J. 2001;20:5232–5241. doi: 10.1093/emboj/20.18.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Noma K, Allis CD, Grewal SI. Science. 2001;293:1150–1155. doi: 10.1126/science.1064150. [DOI] [PubMed] [Google Scholar]
- 12.Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O'Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, Kouzarides T. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- 13.Strahl BD, Allis CD. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 14.Turner BM. BioEssays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 15.Schreiber SL, Bernstein BE. Cell. 2002;111:771–778. doi: 10.1016/s0092-8674(02)01196-0. [DOI] [PubMed] [Google Scholar]
- 16.Henikoff S. Proc Natl Acad Sci USA. 2005;102:5308–5309. doi: 10.1073/pnas.0501853102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su AI, Wiltshire T, Batalov S, Lapp H, Ching KA, Block D, Zhang J, Soden R, Hayakawa M, Kreiman G, et al. Proc Natl Acad Sci USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avni O, Lee D, Macian F, Szabo SJ, Glimcher LH, Rao A. Nat Immunol. 2002;3:643–651. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- 19.Su RC, Brown KE, Saaber S, Fisher AG, Merkenschlager M, Smale ST. Nat Genet. 2004;36:502–506. doi: 10.1038/ng1351. [DOI] [PubMed] [Google Scholar]
- 20.Chi TH, Wan M, Zhao K, Taniuchi I, Chen L, Littman DR, Crabtree GR. Nature. 2002;418:195–199. doi: 10.1038/nature00876. [DOI] [PubMed] [Google Scholar]
- 21.Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae SC, Komori T, Ito Y, Littman DR. Cell. 2002;111:621–633. doi: 10.1016/s0092-8674(02)01111-x. [DOI] [PubMed] [Google Scholar]
- 22.Fields PE, Lee GR, Kim ST, Bartsevich VV, Flavell RA. Immunity. 2004;21:865–876. doi: 10.1016/j.immuni.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 23.Su IH, Basavaraj A, Krutchinsky AN, Hobert O, Ullrich A, Chait BT, Tarakhovsky A. Nat Immunol. 2003;4:124–131. doi: 10.1038/ni876. [DOI] [PubMed] [Google Scholar]
- 24.Smale ST, Fisher AG. Annu Rev Immunol. 2002;20:427–462. doi: 10.1146/annurev.immunol.20.100301.064739. [DOI] [PubMed] [Google Scholar]
- 25.Roh TY, Cuddapah S, Zhao K. Genes Dev. 2005;19:542–552. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roh TY, Ngau WC, Cui K, Landsman D, Zhao K. Nat Biotechnol. 2004;22:1013–1016. doi: 10.1038/nbt990. [DOI] [PubMed] [Google Scholar]
- 27.Larsen F, Gundersen G, Lopez R, Prydz H. Genomics. 1992;13:1095–1107. doi: 10.1016/0888-7543(92)90024-m. [DOI] [PubMed] [Google Scholar]
- 28.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, et al. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hogan PG, Chen L, Nardone J, Rao A. Genes Dev. 2003;17:2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- 30.Graef IA, Chen F, Crabtree GR. Curr Opin Genet Dev. 2001;11:505–512. doi: 10.1016/s0959-437x(00)00225-2. [DOI] [PubMed] [Google Scholar]
- 31.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 32.Sullivan BA, Karpen GH. Nat Struct Mol Biol. 2004;11:1076–1083. doi: 10.1038/nsmb845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng HH, Robert F, Young RA, Struhl K. Mol Cell. 2003;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- 34.Verdin E, Dequiedt F, Kasler HG. Trends Genet. 2003;19:286–293. doi: 10.1016/S0168-9525(03)00073-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.