Abstract
In a newly isolated temperature-sensitive lethal Escherichia coli mutant affecting the chaperonin GroEL, we observed wholesale aggregation of newly translated proteins. After temperature shift, transcription, translation, and growth slowed over two to three generations, accompanied by filamentation and accretion (in ≈2% of cells) of paracrystalline arrays containing mutant chaperonin complex. A biochemically isolated inclusion body fraction contained the collective of abundant proteins of the bacterial cytoplasm as determined by SDS/PAGE and proteolysis/MS analyses. Pulse–chase experiments revealed that newly made proteins, but not preexistent ones, were recruited to this insoluble fraction. Although aggregation of “stringent” GroEL/GroES-dependent substrates may secondarily produce an “avalanche” of aggregation, the observations raise the possibility, supported by in vitro refolding experiments, that the widespread aggregation reflects that GroEL function supports the proper folding of a majority of newly translated polypeptides, not just the limited number indicated by interaction studies and in vitro experiments.
Keywords: chaperone, misfolding, protein folding
More than 30 years ago, the first genetic experiments were carried out that identified the groE operon of Escherichia coli as involved in macromolecular synthesis, observing it to be required for production of λ and T4 virus phage particles (1, 2). Already in those first experiments it was recognized that groE mutations had effects on host cell growth in the absence of phage infection, implicating this operon in host cell metabolism. Indeed, some years later the groE operon was shown to be essential for E. coli viability (3). At about the same time, studies of a GroEL homologue in mitochondria, Hsp60, showed an involvement in assisting the folding of newly imported polypeptides to their native form (4, 5). The role of providing kinetic assistance to polypeptide folding was further established shortly thereafter by reconstitution in vitro of refolding by the purified groE gene products, the 800-kDa double-ring chaperonin GroEL and the 70-kDa single-ring cochaperonin GroES (6, 7).
Although the polypeptide binding and release actions of the GroEL system are generally understood, a test in vivo of the effect of severe conditional GroEL deficiency on protein metabolism has not been accessible to date. One would expect that many cellular pathways might be affected, based on studies in recent years indicating that ≈10% of bacterial proteins, participating in a host of pathways, interact with GroEL in the cell (8, 9). In particular, ≈12 such interacting proteins have been shown in vitro to be completely dependent on GroEL/GroES to reach the native state, and several of these proteins carry out essential cellular functions, thus providing some explanation for why groE is essential for cell growth. Such proteins require not only the binding function of GroEL, which forestalls misfolding and aggregation, but also the action of folding inside a hydrophilic, confined, cis ternary GroEL/GroES complex (10–13). But interaction and in vitro reconstitution studies cannot uncover the primary effects of shutoff of chaperonin function in the intact physiological system. Here we have assessed this state using a newly derived severe temperature-sensitive lethal GroEL-deficient strain of E. coli that arrests growth in liquid culture at the nonpermissive temperature. Global aggregation of newly translated proteins is observed, raising a question as to whether GroEL may play a more general role than previously thought. Supporting this possibility are observations presented here that two large monomeric proteins are assisted to the native state in vitro by GroEL alone, apparently employing binding and release from open rings.
Results
Isolation and Growth Properties of Temperature-Sensitive Lethal GroEL 461 Mutant.
We previously examined a GroEL mutant E. coli strain with a relatively mild growth phenotype, showing temperature sensitivity on solid media but only slowed growth in liquid medium, without arrest (14). The mild phenotype was a function of this strain carrying both a temperature-sensitive GroEL allele, on a plasmid, and wild-type GroEL, in the bacterial chromosome. Even though the wild-type chromosomal GroEL was regulated by a lac promoter rather than the native one, there was still residual expression of wild-type GroEL in the absence of isopropyl β-d-thiogalactoside to a level 5–10% of normal, reducing the severity of the plasmid-directed mutant phenotype. To produce a more severe phenotype, we placed the same mutant allele of GroEL, E461K, into a strain where the chromosomal GroEL was disrupted, by employing a plasmid shuffling strategy, exchanging an ara-E461K expression plasmid for an ara-wild-type one (15, 16) (Fig. 1A and Fig. 6, which is published as supporting information on the PNAS web site).
Fig. 1.
Construction and temperature-sensitive behavior of the GroEL-deficient 461 strain. (A) Genetic configuration of strains and growth in liquid LB/ampicillin/0.02% arabinose of pBAD-EL/AI90, which serves as the wild-type (wt) strain, and the derived 461 cells. “Shift” indicates the point when the cultures were moved from 20°C to 37°C. Note that the wild-type cells exhibit a doubling time of ≈30–45 min, slightly slower than nonmanipulated wild-type E. coli strains. Note also that the same growth curves were obtained in the absence of ampicillin (data not shown). (B) Immuno-EM of paracrystalline inclusion in a 461 cell. (C) Early onset of deficient function of GroEL in 461 cells after temperature shift. The stringent GroEL substrate protein Rubisco fails to reach a soluble form in 461 early after shift to 37°C (third and fourth lanes). By contrast, ≈30% of newly translated Rubisco reaches the soluble fraction in wild-type cells (first and second lanes). T7-Rubisco-His6, encoded by a second plasmid introduced into wild-type and 461 cells, was induced 15 min after temperature shift by infection with a CE6 λ phage bearing the T7 RNA polymerase gene. Cells were simultaneously radiolabeled with 35S-Translabel. After 1 h, cells were harvested and separated into soluble and insoluble fractions as described in Inclusion Body Preparation in Materials and Methods to assess the relative amounts of newly synthesized soluble, presumably native Rubisco-His6 and insoluble, presumably misfolded/aggregated Rubisco-His6. Rubisco-His6 was collected from the respective fractions by Talon affinity chromatography. Identical portions of eluted material were then analyzed by SDS/PAGE.
E461K changes a residue at the interface between GroEL rings, causing a shift of register between the rings, such that subunits form 1:1 contacts across the interface instead of 1:2 contacts (17). This misalignment is associated with loss of allosteric communication both within and between rings. ATP cooperativity, positive within rings and negative between them (18), is abolished. In vitro, the purified 461 complex binds nonnative proteins but is unable to fold them at 37°C (as compared with 23°C). In particular, 461 complex fails to release subunits of substrate, behaving as a polypeptide “trap” (see Supporting Discussion in Supporting Text, which is published as supporting information on the PNAS web site).
In vivo, the newly isolated 461 strain was temperature-sensitive on both solid and liquid media. As expected for temperature-sensitive behavior due to GroEL deficiency, growth at 37°C was rescued if 461 cells were transformed with a second plasmid expressing wild-type GroEL and GroES at physiological levels. By contrast, a similar plasmid deleted of GroEL, directing expression of only GroES, failed to rescue at 37°C. In liquid LB media after shift to 37°C, the 461 strain exhibited growth arrest occurring over a period of 2–3 h (Fig. 1A). A similar pattern of arrested growth was observed regardless of the amount of arabinose used to regulate expression (0.001–0.1%). A concentration of 0.02% arabinose was selected for general use because plating efficiency of both wild-type and 461 cells was observed to be the most uniform in this amount of inducer. This concentration of arabinose produces a level of GroEL (equal in wild-type and 461 cells both before shift and for 2–3 h afterward) that is ≈5-fold greater than that of nonmanipulated E. coli strains at 37°C.
The 461 culture was still viable at 3–4 h after temperature shift, at which time growth had ceased: when the culture was downshifted to room temperature and plated, colonies were formed. By 12 h, however, the culture was inviable; no colonies were obtained after downshift. Concerning morphology, as was observed previously for GroEL mutant strains, cells became progressively filamentous after temperature shift. A few percent of the cells also developed paracrystalline inclusions, shown by immuno-EM to contain the mutant GroEL itself (Fig. 1B and unpublished observations).
Immediate Onset of GroEL-Deficient Phenotype.
The normal growth behavior of 461 cells immediately after temperature shift raised the question of whether the 461 GroEL is functionally deficient early after temperature shift or whether it requires a period to become defective in protein folding. To address GroEL function, the well studied GroEL-dependent substrate protein Rubisco from Rhodospirillum rubrum was expressed from a T7-regulated coding sequence on a second plasmid, and its solubility was assessed as a measure of whether GroEL function was present. Cells were shifted to 37°C for 15 min, and Rubisco synthesis was then initiated by infection with λ phage bearing the T7 polymerase gene. At the same time, the cells were 35S-radiolabeled. After an hour, the cells were harvested and fractionated into soluble and insoluble fractions, and the Rubisco, bearing a His6 tag, was collected from each (the latter under denaturing conditions) by using Talon affinity resin. The recovered material was analyzed by SDS/PAGE (Fig. 1C). In wild-type cells ≈30% of the newly made Rubisco was found in the soluble fraction, presumably in native form, whereas in the 461 cells no soluble Rubisco was detected. We conclude that the mutant GroEL in 461 cells is defective in folding function even at early time after shift. The phenotypic effects observed thus follow from an early loss of GroEL function.
Transcription and Translation Continue in Mutant Cells at Early Times After Temperature Shift but Are then Strongly Reduced.
Considering that 461 cultures continued to grow at early time after temperature shift, with advancing optical density and increasing length of filamenting bacteria, it seemed likely that protein translation was not arrested despite dysfunction of GroEL. To formally measure translation, pulse-radiolabeling was carried out, comparing equivalent amounts of wild-type and mutant cells directly solubilized at various times after temperature shift (Fig. 2). Rates of translation in 461 and wild-type cells were observed to be equal at 15 and 45 min after shift, but at 2, 3, and 4 h the rate of translation became reduced in the mutant, corresponding with the slowed growth of the culture.
Fig. 2.
Translation in the mutant cells after temperature shift is initially the same as wild type but progressively slows. Additionally, MetE and GroEL are strongly expressed after shift (see text). For the translation experiment, wild-type or 461 cells grown in LB/ampicillin/arabinose at 20°C to the same OD (0.04) were shifted to 37°C, and at the indicated time points 0.4 ml of cells was labeled with 100 μCi of 35S-Translabel for 10 min. Cells were immediately recovered by centrifugation and directly solubilized in SDS sample buffer. The solubilized material from an identical OD equivalent of cells for each strain was analyzed by SDS/PAGE and autoradiographed. Note that wild-type cells were not harvested at 4 h after shift because they had progressed beyond an OD of 1.5.
Overall, the pattern of translated proteins observed for wild-type and 461 cells was remarkably similar. An interesting and reproducible difference was observed, however: the 85-kDa B12-independent methionine biosynthetic enzyme MetE was induced in the mutant cells, observable by 45 min after shift (Fig. 2). This protein was also readily visible in Coomassie-stained gels of the 461 cells after shift (e.g., Fig. 3). Notably, it had been observed to be induced in the milder 461 mutant characterized earlier (14). In addition to induction of MetE, we observed here that, at later times, GroEL (461) itself was strongly translated (Fig. 2), as if more strongly arabinose-induced; consistent with this possibility, at late times after temperature shift a greater level of GroEL was observed in mutant cells than in wild-type cells.
Fig. 3.
A large number of protein species are present in inclusion bodies prepared from 461 cells but not wild-type cells or GroE plasmid-rescued 461 cells. (A) Cell fractionation was carried out as described in Materials and Methods by using lysozyme treatment, sonication, Triton X-100 solubilization, and centrifugation. A Coomassie-stained SDS/PAGE gel shows proteins from soluble (S) and inclusion body (P) fractions prepared from equal OD amounts of cells of wild type harvested in log phase and 461 harvested after 3 h at 37°C (see Supporting Text). (B) Time course of aggregation in 461 cells. Fractionation of 461 cells at permissive temperature (20°C) and at various times after shift to 37°C into soluble (S) and inclusion body (P) fractions was carried out as described in A. Total protein in the respective fractions was displayed in SDS/PAGE.
Transcription was also examined, measuring [3H]uridine incorporation during 1-min time periods at various times after temperature shift (Fig. 7, which is published as supporting information on the PNAS web site). The transcription rate was affected in a similar fashion to translation, falling to ≈50% during the first hour and then virtually arresting by 3 h.
Inclusion Body Fractionation of 461 Cells Reveals a Large Amount of Insoluble Protein and a Large Number of Different Species.
To evaluate the fate of proteins at a biochemical level, standard inclusion body preparations were carried out both on wild-type cells during log-phase growth and on 461 mutant cells at various times after temperature shift. Lysozyme treatment, sonication, and Triton X-100 extraction were carried out on equal amounts of cells. There was an immediately noticeable difference in the size of the inclusion body pellets from wild-type and mutant cells: starting with 10 OD units of cells, a barely visible pellet was obtained from wild-type cells, whereas a dense white precipitate (amounting to >50 μg of total protein, nearly equal to the amount of soluble protein) was recovered from an identical amount of mutant cells harvested 3 h after temperature shift. These observations were confirmed when SDS/PAGE analysis was carried out on equal portions of the wild-type and mutant soluble and inclusion body fractions (Fig. 3A, lanes 1–4). Virtually no protein was present in the inclusion fraction from the wild-type cells, as compared with a large cohort of proteins in the equivalent insoluble fraction of the mutant cells. Indeed, for the mutant, the percentage of total protein in the insoluble fraction increased progressively after shift to 37°C, such that, at 3 h after shift, it was nearly equal to that in the soluble fraction, suggesting that as much as 30–40% of the protein in the 461 cells had lodged in the inclusion body fraction (Fig. 3B). Particularly surprising was that the pattern of species present in the insoluble fraction of 461 cells was similar to that in the soluble fraction of both 461 and wild-type cells. Because only 10–20% of E. coli proteins have been shown to interact stably with the chaperonin in vitro, this suggested that aggregation was occurring as a wholesale process, without regard to whether any particular protein was a bona fide GroEL substrate.
When the same type of fractionation was carried out on 461 cells growing at 20°C, a similar pattern was already visible, albeit at a much reduced level: here only ≈10% of total protein was insoluble (Fig. 3B). Of further note, the 461 mutant GroEL itself remained largely soluble as a double ring after temperature shift (Fig. 3 and Fig. 8, which is published as supporting information on the PNAS web site), indicating that it does not, for example, comprise the major component of the insoluble fraction. In addition, diminishing the level of the mutant 461 chaperonin by shifting cells to glucose for several generations before temperature shift and thereafter produced the same striking aggregation (Fig. 9, which is published as supporting information on the PNAS web site). Finally, 461 cells rescued with a GroES/GroEL-encoding plasmid were examined 3 h after shift to 37°C. At this time point the rescued cells, which grow somewhat more slowly than wild-type cells, have entered log phase growth as compared with arrest of nonrescued cells. The rescued cells exhibited the same fractionation behavior as wild-type cells, with almost all protein in the soluble fraction (Fig. 3A, lanes 5 and 6).
Proteomic Analysis of the Inclusion Body Fraction Reveals both Known GroEL Substrates and Other Abundant Proteins.
To determine the identity of both soluble and insoluble proteins in 461 cells, we carried out proteolysis and MS on the soluble fraction and on the urea-solubilized inclusion bodies using the multidimensional protein identification protocol (MudPIT; ref. 19). In agreement with the SDS/PAGE analysis, the proteomic analysis identified in both fractions a similar set of ≈300 proteins (Table 1, which is published as supporting information on the PNAS web site). Most of the species were cytoplasmic proteins, many known to be highly abundant, but a few periplasmic (secretory) proteins were also observed (for example, plasmid-encoded β-lactamase was present, but Western blot analysis revealed ≈90% to be soluble and processed, presumably in the periplasm; data not shown).
Only some of the cytoplasmic proteins in the inclusion body fraction were recognizable as bona fide GroEL substrate proteins based on previous in vitro validation studies, including MetF, MetK, GatY, and DapA (9). Also, a number of proteins previously identified as either GroEL/ATP-assisted or reversibly bound to GroEL were detected in the inclusion body fraction. The inclusion bodies also contained a large number of abundant cytoplasmic proteins that have not been observed to interact with GroEL. Several of these were overexpressed, purified, and tested in vitro for their dependence on the GroEL system for folding. EF-G, FtsZ, AlaRS, and acyl CoA synthetase were found to be GroEL/ATP-assisted proteins, whereas LysS appeared to be reversibly GroEL-interacting (data not shown). Notably, EF-G, AlaRS, and acyl CoA synthetase are of substantial size relative to the GroEL cavity, measuring 77, 96, and 77 kDa, respectively. In the case of the abundant tubulin-like FtsZ protein, we note that FtsZ-deficient cells exhibit a filamentous cell morphology resembling that observed here (20).
MetE.
MetE, the 85-kDa protein highly induced in the 461 cells after temperature shift, was studied further. Such strong overexpression of MetE was surprising, because cells growing in the presence of a rich LB medium containing methionine would normally repress the methionine biosynthetic genes (21). However, GroEL/GroES has been shown to be absolutely required for MetK folding (9), and its misfolding in the setting of GroEL deficiency would lead to decreased levels of S-adenosylmethionine, which functions as a corepressor with the MetJ protein of the metE operon. Thus, defective MetK and resulting decreased S-adenosyl methionine would result in derepression of metE (see Supporting Discussion in Supporting Text).
Because MetE was induced to such a great extent in the 461 cells, we assayed both its in vitro and in vivo behavior in relation to GroEL/GroES. After acid-induced unfolding, purified MetE was unable to spontaneously refold, whereas 30–50% activity was recovered when GroEL was present in 2-fold molar excess at the time of neutralization. Interestingly, this recovery was not dependent on either ATP or GroES/ATP (data not shown). In vivo, MetE has been found to be particularly susceptible to oxidation, which correlates with the observation in vitro that oxidized glutathione can glutathionylate the protein and induce a conformational change concomitant with enzyme inactivation. Intriguingly, GroEL has been found to specifically associate with the glutathionylated form of the protein (22). Here we assayed the oxidation status of MetE from 461 cells using in vivo thiol trapping. We observed, after cell fractionation, that MetE in the soluble fraction was reduced, but, by contrast, MetE present in the inclusion body fraction was oxidized (Fig. 4A). This finding suggests that, in the absence of GroEL function, newly translated MetE may be vulnerable to misfolding and oxidation.
Fig. 4.
MetE is oxidized in the insoluble fraction of 461 cells as revealed by in vivo thiol trapping (A), and aconitase can be refolded in vitro by GroEL alone (B). (A) Thiol trapping experiments were performed to ascertain the oxidation status of MetE in vivo in 461 cells (wild type does not express MetE in LB medium). Cell fractions were examined by isoelectric focusing and Western blotting (22) (see Supporting Methods in Supporting Text). The first two lanes show controls with purified MetE exposed in vitro to reducing (first lane) and oxidizing (second lane) conditions, then alkylated in the same order as for the cells; the third lane shows the supernatant fraction of alkylated 461 cells; and the fourth lane shows the inclusion body fraction of alkylated 461 cells (20-fold more total protein loaded vs. the third lane). Note that if oxidation or alkylation of MetE within inclusion bodies were different from that of the purified glutathionylated protein, then multiple bands with various isoelectric points would be expected; however, the close correspondence between the in vitro and in vivo thiol trapping samples strongly suggests that MetE is oxidized in the inclusion body fraction of the 461 cells similarly to the purified protein. (B) E. coli and yeast mitochondrial aconitase refolding in vitro: the E. coli enzyme is assisted by GroEL alone whereas the yeast enzyme requires both GroEL and GroES. Acid-unfolded aconitase B was diluted into neutralizing buffer containing the respective components, and refolding was carried out for 1 h. Iron–sulfur cluster formation was then carried out, followed by enzyme assay, as described in ref. 23. Activity is expressed as the percentage recovery of the input material.
Aconitase.
Aconitase B, a 93-kDa monomeric iron–sulfur cluster-containing enzyme, was also studied in vitro. As with MetE, the presence of GroEL alone had a major effect on recovery of native active enzyme after dilution from denaturant, enhancing recovery from 10% in a spontaneous reaction to 60% in the presence of GroEL or a single-ring version, SR1, regardless of the presence of GroES and ATP (Fig. 4B). This finding contrasted with the yeast mitochondrial aconitase (85 kDa), which stringently requires GroES binding in trans to direct release and folding (23).
Newly Translated Proteins, Not Preexistent Ones, Are Incorporated into the Insoluble Fraction.
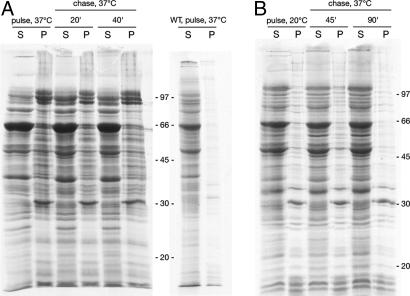
To address more directly whether newly made proteins as opposed to preexistent ones were becoming insoluble, we carried out a pulse–chase radiolabeling experiment on 461 cells, radiolabeling for 10 min at 2 h after temperature shift and chasing for either 20 or 40 min in the presence of chloramphenicol to block further translation. The cells were separated into soluble and inclusion body fractions at the end of the labeling period and after the two times of chase. The results were unambiguous (Fig. 5A). Already by the end of the 10-min labeling period, a substantial fraction, amounting to nearly 50% of newly made protein, was found in the insoluble fraction (Fig. 5A). By contrast, only a few species were found in the insoluble fraction of wild-type cells (Fig. 5A Right). Apparently, in 461 cells, newly translated proteins in nonnative form were at high risk of becoming incorporated into the inclusion bodies. Consistent with such behavior, when shorter labeling times (1 or 5 min) were used there was still a substantial fraction of newly made protein lodging in the insoluble fraction (Fig. 10, which is published as supporting information on the PNAS web site). Strikingly, the pattern of soluble and insoluble proteins at the two times of chase remained identical to that of the pulse period. In particular, there was no further increase in the amount of insoluble protein during the chase, indicating that soluble radiolabeled proteins were not becoming progressively recruited to the insoluble fraction. This finding likely indicates that, once newly translated proteins reached native form, they were no longer at risk for becoming incorporated into the inclusion bodies.
Fig. 5.
Newly translated proteins, not preexistent ones, are subject to misfolding and aggregation in 461 cells. Shown are pulse–chase analyses measuring the partitioning of 35S-labeled newly synthesized proteins into soluble and insoluble fractions. (A) Pulse-labeling of 461 (Left) and wild-type (Right) cells at 37°C followed by chase of 461 cells at 37°C in the presence of 200 μg/ml chloramphenicol to block further translation. (B) Pulse-labeling of 461 cells at 20°C followed by chase in chloramphenicol at 37°C. Thirty-milliliter cultures of 461 cells growing at 20°C (at an OD of ≈0.5) or that had been shifted to 37°C for 2 h were labeled for 20 and 10 min, respectively, with 3 mCi of 35S-Translabel. At the various time points, 10-ml aliquots were removed from the culture and fractionated as in Materials and Methods.
An additional pulse–chase study, carrying out radiolabeling at permissive temperature and then shifting to 37°C in the presence of chloramphenicol, was also performed (Fig. 5B). Here, by the end of the pulse period, a small percentage of proteins was found in the insoluble fraction (≈10%), corresponding to the fractionation behavior of total proteins (Fig. 3B). As with labeling at 37°C, this fractionation pattern was preserved after shift to 37°C and chase for 45 min and 90 min, indicating here also that only newly made proteins are at risk for misfolding and aggregation, whereas already-folded native proteins are not subject to recruitment into the aggregates.
Discussion
Wholesale Aggregation of the Collective of Newly Made Proteins in GroEL-Deficient Cells: Is GroEL Interacting with Most Newly Translated Proteins?
The analysis here of insoluble proteins after temperature shift of GroEL-deficient 461 cells, both in SDS/PAGE and by proteomic study, revealed that a majority of the abundant (observable) bacterial cytoplasmic proteins were present. The gel analyses indicated that the percentage of any given species in the insoluble fraction increased with time: on average ≈10% was insoluble at permissive temperature (20°C), but after 3 h at 37°C, a time when the cells halted further growth, as much as 30–40% was insoluble. How could the general cohort of newly translated proteins wind up in the insoluble fraction if, as is currently postulated, only ≈10% of these are bona fide substrates? Three possible models come to mind: (i) The cytoplasm of the GroEL-deficient cells has reached a physical state where any newly translated protein departing the ribosome can be recruited into an aggregate while it is in the nonnative state (this might be triggered by the misfolding and aggregation of the collective of stringent GroEL substrates); (ii) GroEL deficiency affects another component that plays a more general role in the proper folding of all proteins departing the ribosome; (iii) GroEL itself is involved with folding a far broader collective of proteins than has been recognized by studies to date.
The first of these models seems tenuous given the observation that, even at permissive temperature, there is substantial aggregation (≈10%) of many of the newly translated proteins (Fig. 4), but no altered physical state is observed by EM at 20°C (data not shown). On the other hand, we cannot formally exclude an “avalanche” model, in which stringent GroEL substrates fail to fold and aggregate, providing a multiplicity of nonnative surfaces that interact with and entrain other newly translated protein species.
The second model invokes a new component that would, by definition, need to have a role broader than that defined to date for GroEL. Although the abundance of such a component would not necessarily have to be as great, a null allele would presumably present a phenotype akin to that observed here. We are at a loss to enumerate such a component. For example, although the chaperone trigger factor interacts at the ribosome with most newly translated proteins, it is not essential for cell viability, and wholesale aggregation does not occur in the setting of its deletion (24). DnaK (Hsp70) likewise can be deleted at intermediate temperatures without such an effect (25). Only when both trigger factor and DnaK are deleted is there a general aggregation phenotype similar to but quantitatively less severe than that observed here (24). There is, however, no evidence that GroEL is required for the ongoing function of either trigger factor or DnaK. Interestingly, however, it has recently been observed that the growth defect and wholesale aggregation of DnaK/trigger factor-deficient cells can be rescued by strong overexpression of GroEL/GroES, indicating that the chaperonin system can in fact act on the broad range of substrates, of all molecular sizes, acted on by DnaK and trigger factor (26). Likewise, severe aggregation in RpoH mutants, affecting the heat shock transcription factor σ32, has been rescued by strong overexpression of GroEL/GroES (27).
These latter observations lead to consideration of the third possibility that, in vivo, the interaction of GroEL with newly translated proteins is more general than in vitro reconstitution and coimmunoprecipitation studies have reflected, such that most proteins interact with GroEL in some way to achieve normal folding. In the case of the reconstitution studies, a nonnative protein's ability to bind to GroEL has typically been measured after its dilution from denaturant, necessitating relatively stable physical association with GroEL. For example, in one commonly used assay the nonnative substrate must remain stably associated with the chaperonin during gel filtration, coeluting with it at 800 kDa, to be considered as binding to it. Similarly, in coimmunoprecipitation studies, substrates must remain stably associated with GroEL through steps of cell breakage, immune capture, and washing. It seems conceivable that a wider collective of proteins than these assays have detected may form transient and low-affinity interactions in vivo with GroEL that nonetheless forestall them from aggregation and direct them toward proper folding. For example, the ATP-independent behavior observed here in vitro of two large monomeric enzymes, MetE (85 kDa) and aconitase (93 kDa), may reflect this.
Flux Considerations.
Is there sufficient GroEL, at ≈1–2 μM concentration in the bacterial cytoplasm, to accommodate interaction with the large collective of proteins emerging from a concentration of ribosomes that is 10-fold greater? An earlier theoretical calculation suggested that folding of only a few percent of total newly synthesized protein could be managed (28), but the assumption was made that the folding rate at GroEL would be similar to that of stringent substrates in vitro (0.07–1.1 min−1). These rates reflect a GroEL/GroES reaction cycle of ≈5–10 sec and a requirement for multiple cycles. In contrast, for substrates employing transient association with an open GroEL ring to prevent misfolding, only a single round of binding and release from an open ring may be required, with a release rate that might be well above 1 sec−1 (i.e., 60 min−1). For example, studies of the small protein lysozyme showed that binding and release by GroEL, without ATP or GroES, could accelerate its already rapid folding, involving an off-rate of >2 sec−1 (29). Thus, interaction with the global collective of newly translated proteins seems conceivable. Further studies in vivo may be able to directly observe the spectrum of interacting proteins.
Materials and Methods
Plasmids and Strains.
A pBAD-EL derivative bearing the E461K codon change was produced by PCR mutagenesis and propagation in DH5α at 20°C. After plasmid shuffling to introduce this plasmid in place of pBAD-EL in AI90 cells (Fig. 6), the plasmid was directly recovered from the clonally purified 461 strain grown at 20°C, and the GroEL coding region was sequenced. The original E461K codon change was observed, but an additional change was also present, V417G, confirmed by MS analysis of purified 461 protein. The double-substituted protein was purified and characterized in vitro; its ATPase and refolding behavior were observed to be identical to those of the singly substituted E461K.
Inclusion Body Preparation.
Five OD650 units of bacterial cells were resuspended in 400 μl of 50 mM Tris (pH 8.0)/1 mM EDTA (hypotonic buffer), brought to 0.5 mg/ml lysozyme, and incubated for 5 min. MgCl2 was added to 5 mM, and the mixture was treated with 30 units of DNase I for 5 min. Sonication was then carried out at 4°C. Triton X-100 was added to 0.5%, and the sample was centrifuged at 4°C for 15 min at 14,000 × g. The pellet fraction was washed once with 500 μl of buffer A and solubilized with 450 μl of 10 M urea/100 mM Tris (pH 7.4)/1 mM Tris(2-carboxyethyl)phosphine at 20°C for 30 min.
Rubisco Expression in Wild-Type and 461 Cells.
Wild-type and 461 transformants carrying a second pBR322-based plasmid bearing T7-regulated Rubisco-His6 and a chloramphenicol drug marker were grown at 20°C to an OD of ≈0.5 in media containing 0.2% maltose. Five milliliters of each culture was shifted to 37°C for 15 min, then simultaneously infected with 0.5 ml of CE6 bacteriophage containing a T7 RNA polymerase gene (3.4 × 1010 pfu/ml; Novagen, La Jolla, CA) and radiolabeled with 0.5 mCi (1 Ci = 37 GBq) of 35S-Translabel (GE Healthcare, Piscataway, NJ). After 1 h the cultures were separated as described in Inclusion Body Preparation into soluble and insoluble fractions. Unlabeled Rubisco-His6 (40 μg) was added to each fraction as a carrier, and the respective fractions were incubated with 50 μl of Talon resin (BD Biosciences, Palo Alto, CA) to recover the His-tagged Rubisco. The recovered material was fractionated in SDS/PAGE, and the radiolabeled products were quantitated by using a PhosphorImager (GE Healthcare).
EM and Proteomic Studies.
See Supporting Methods in Supporting Text.
Supplementary Material
Acknowledgments
We thank Pete Lund for the starting E. coli strain; Robert Watson and Jayesh Patel for help with phase contrast imaging; Kirk Beebe and Paul Schimmel for help with tRNA synthetase studies; and Debbie Fass, Jorgé Galan, Rick Lifton, and Brooks Low for valuable discussions. This work was supported by grants from the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
References
- 1.Georgopoulos CP, Hendrix RW, Kaiser AD. Nat New Biol. 1972;239:38–41. doi: 10.1038/newbio239038a0. [DOI] [PubMed] [Google Scholar]
- 2.Takano T, Kakefuda T. Nat New Biol. 1972;239:34–37. doi: 10.1038/newbio239034a0. [DOI] [PubMed] [Google Scholar]
- 3.Fayet O, Ziegelhofer T, Georgopoulos C. J Bacteriol. 1989;171:1379–1385. doi: 10.1128/jb.171.3.1379-1385.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheng M-Y, Hartl FU, Martin J, Pollock RA, Kalousek F, Neupert W, Hallberg EM, Hallberg RL, Horwich AL. Nature. 1989;337:620–625. doi: 10.1038/337620a0. [DOI] [PubMed] [Google Scholar]
- 5.Ostermann J, Horwich AL, Neupert W, Hartl FU. Nature. 1989;341:125–130. doi: 10.1038/341125a0. [DOI] [PubMed] [Google Scholar]
- 6.Goloubinoff P, Christeller JT, Gatenby AA, Lorimer GH. Nature. 1989;342:884–889. doi: 10.1038/342884a0. [DOI] [PubMed] [Google Scholar]
- 7.Martin J, Langer T, Boteva R, Schramel A, Horwich AL, Hartl FU. Nature. 1991;352:36–42. doi: 10.1038/352036a0. [DOI] [PubMed] [Google Scholar]
- 8.Houry WA, Frishman D, Eckerskorn C, Lottspeich F, Hartl FU. Nature. 1999;402:147–154. doi: 10.1038/45977. [DOI] [PubMed] [Google Scholar]
- 9.Kerner MJ, Naylor DJ, Ishihama Y, Maier T, Chang H-C, Sines AP, Georgopoulos CP, Frishman D, Hayer-Hartl M, Mann M, Hartl FU. Cell. 2005;122:209–220. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 10.Xu Z, Horwich AL, Sigler PB. Nature. 1997;388:741–751. doi: 10.1038/41944. [DOI] [PubMed] [Google Scholar]
- 11.Thirumalai D, Klimov DK, Lorimer GH. Proc Natl Acad Sci USA. 2003;100:11195–11197. doi: 10.1073/pnas.2035072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horwich AL, Farr GW, Fenton WA. Chem Rev. 2006;106:1917–1930. doi: 10.1021/cr040435v. [DOI] [PubMed] [Google Scholar]
- 13.Brinker A, Pfeifer G, Kerner MJ, Naylor DJ, Hartl FU, Hayer-Hartl M. Cell. 2001;107:223–233. doi: 10.1016/s0092-8674(01)00517-7. [DOI] [PubMed] [Google Scholar]
- 14.Horwich AL, Low KB, Fenton WA, Hirshfield IN, Furtak K. Cell. 1993;74:909–917. doi: 10.1016/0092-8674(93)90470-b. [DOI] [PubMed] [Google Scholar]
- 15.Quandt J, Hynes MF. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 16.Ivic A, Olden D, Wallington EJ, Lund PA. Gene. 1997;194:1–8. doi: 10.1016/s0378-1119(97)00087-5. [DOI] [PubMed] [Google Scholar]
- 17.Sewell BT, Best RB, Chen S, Roseman AM, Farr GW, Horwich AL, Saibil HR. Nat Struct Mol Biol. 2004;11:1128–1133. doi: 10.1038/nsmb844. [DOI] [PubMed] [Google Scholar]
- 18.Yifrach O, Horovitz A. Biochemistry. 1995;34:5303–5308. doi: 10.1021/bi00016a001. [DOI] [PubMed] [Google Scholar]
- 19.Washburn MP, Wolters D, Yates JR., III Nat Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 20.Addinall SG, Bi E, Lutkenhaus J. J Bacteriol. 1996;178:3877–3884. doi: 10.1128/jb.178.13.3877-3884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hondorp ER, Matthews RG. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Böck A, Curtiss R III, Kaper JB, Neidhardt FC, Nyström T, Rudd KE, Squires CL, editors. Washington, DC: Am Soc Microbiol; 2006. Chap 3.6.1.7. [Google Scholar]
- 22.Hondorp ER, Matthews RG. PLoS Biol. 2004;11:1738–1753. doi: 10.1371/journal.pbio.0020336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaudhuri TK, Farr GW, Fenton WA, Rospert S, Horwich AL. Cell. 2001;107:235–246. doi: 10.1016/s0092-8674(01)00523-2. [DOI] [PubMed] [Google Scholar]
- 24.Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B. Nature. 1999;400:693–696. doi: 10.1038/23301. [DOI] [PubMed] [Google Scholar]
- 25.Hesterkamp T, Bukau B. EMBO J. 1998;17:4818–4828. doi: 10.1093/emboj/17.16.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vorderwülbecke S, Kramer G, Merz F, Kurz TA, Rauch T, Zachmann-Brand B, Bukau B, Deuerling E. FEBS Lett. 2004;559:181–187. doi: 10.1016/S0014-5793(04)00052-3. [DOI] [PubMed] [Google Scholar]
- 27.Gragerov A, Nudler E, Komissarova N, Gaitanaris GA, Gottesman ME, Nikiforov V. Proc Natl Acad Sci USA. 1992;89:10341–10344. doi: 10.1073/pnas.89.21.10341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lorimer GH. FASEB J. 1996;10:5–9. doi: 10.1096/fasebj.10.1.8566548. [DOI] [PubMed] [Google Scholar]
- 29.Coyle JE, Texter FL, Ashcroft AE, Masselos D, Robinson CV, Radford SE. Nat Struct Biol. 1999;6:683–690. doi: 10.1038/10735. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.