Abstract
Oligodendrocytes undergo extensive changes as they differentiate from progenitors into myelinating cells. To better understand the molecular mechanisms underlying this transformation, we performed a comparative analysis using gene expression profiling of A2B5+ oligodendrocyte progenitors and O4+ oligodendrocytes. Cells were sort-purified ex vivo from postnatal rat brain using flow cytometry. Using Affymetrix microarrays, 1707 transcripts were identified with a more than twofold increase in expression in O4+ oligodendrocytes. Many genes required for oligodendrocyte differentiation were upregulated in O4+ oligodendrocytes, including numerous genes encoding myelin proteins. Transcriptional changes included genes required for cell adhesion, actin cytoskeleton regulation, and fatty acid and cholesterol biosynthesis. At the O4+ stage, there was an increase in expression of a novel proline-rich transmembrane protein (Prmp). Localized to the plasma membrane, Prmp displays adhesive properties that may be important for linking the extracellular matrix to the actin cytoskeleton. Together, our results highlight the usefulness of this discovery-driven experimental strategy to identify genes relevant to oligodendrocyte differentiation and myelination.
Keywords: oligodendrocyte, microarray, myelination, adhesion, cytoskeleton, actin
Introduction
Oligodendrocytes are the myelinating cells of the CNS and are responsible for the synthesis of the myelin sheath, which wraps axons and promotes efficient nerve conduction. Oligodendrocyte progenitors arise in multiple locations in the developing brain and migrate throughout the CNS (Kessaris et al., 2006). As development proceeds, oligodendrocyte progenitors find axonal targets and extend multiple branched processes, which initiate the myelination process (Sherman and Brophy, 2005). Identification of the molecular components that are required at the onset of myelination is an important objective toward understanding this essential developmental process.
A number of receptor/ligand interactions between neurons and oligodendrocytes have been identified, including laminin2-integrin (Buttery and ffrench-Constant, 1999) and contactin-neurofascin (Charles et al., 2002). In addition, downstream effectors of oligodendrocyte process extension have been identified. Fyn expression is increased as oligodendrocytes differentiate and is required for process extension (Osterhout et al., 1999). The Rho family of GTPases are also important cytoskeletal regulators and effectors in the integrin/Fyn pathway (Liang et al., 2004). The dramatic changes in oligodendrocyte morphology during differentiation involve cytoskeletal elements including microtubules and microfilaments (Simpson and Armstrong, 1999; Song et al., 2001; Fox et al., 2006). Myelination begins with process extension, which may require the polymerization of actin-rich filopodia and lamellipodia (Kachar et al., 1986; Asou et al., 1995). Both mechanisms are likely required to orchestrate the complex process of extension, branching, and ultimately the wrapping of axons with the myelin sheath. However, the proteins and pathways that link cell adhesion molecules to changes in oligodendrocyte morphology during differentiation remain poorly characterized.
Elucidating these complex mechanisms that mediate oligodendrocyte maturation requires examining stage-specific changes as they differentiate. This approach necessitates the prospective identification and isolation of these cells at early and late stages of oligodendrocyte lineage progression. Oligodendrocyte progenitors can be identified using the A2B5 antibody (Abney et al., 1983), whereas more mature proligodendroblasts and oligodendrocytes are recognized by the O4 antibody (Sommer and Schachner, 1981; Gard and Pfeiffer, 1990). We reasoned that comparing A2B5+ oligodendrocyte progenitors to O4+ oligodendrocytes would be a productive screening method for identification of genes important for myelination and the establishment of axon–glia interactions. Several reports have used microarrays to study oligodendrocyte development (Scarlato et al., 2000; Cohen et al., 2003; Hu et al., 2004). However, these studies were limited by the number of probe sets available on older version microarrays. Microarrays used in combination with fluorescence-activated cell sorting (FACS), is a particularly powerful strategy, because a large number of known and novel genes can be simultaneously assayed in purified populations of cells prospectively isolated from their native microenvironment within the developing brain. In this study, we used Affymetrix rat microarrays to compare gene expression profiles during the transition from A2B5+ oligodendrocyte progenitors to O4+ oligodendrocytes. This analysis identified both genes and pathways that are activated as oligodendrocytes differentiate. Using this data set, we have identified a novel proline-rich membrane protein (Prmp) that may be important for linking the extracellular matrix to the actin cytoskeleton in oligodendrocytes.
Materials and Methods
Cell preparation and sorting.
Whole brains were dissected from one litter of 10 postnatal day 7 male Sprague Dawley rats (Taconic, Hudson, NY) for each biological replicate, and the cerebellum and meninges were removed in 4°C wash buffer containing the following: 1× HBSS, 20 mm HEPES, 0.1% BSA, and 50 μg/ml gentamicin. The time from killing to RNA extraction buffer was minimized to 6 h. Brains were dissociated using gentle trituration, followed by a 10 min papain and a 10 min DNase I digestion. Cell suspensions were passed through a 40 μm cell strainer to remove nondissociated tissue and centrifuged at 900 × g for 10 min. The resuspended cells were then layered onto a 15–40% Percoll gradient and centrifuged at 2000 × g for 15 min at 4°C. Cells at the interface of the 15–40% Percoll layers were collected, washed twice, and resuspended in wash buffer. Oligodendrocyte progenitors were labeled using the A2B5 mouse monoclonal IgM antibody (Eisenbarth et al., 1979), whereas more mature proligodendroblast and oligodendrocytes were labeled with the O4 mouse monoclonal IgM antibody (Sommer and Schachner, 1981; Gard and Pfeiffer, 1990). Both antibodies were produced and purified from hybridoma cultures grown in-house. The O4 labeling was detected using goat anti-mouse IgM conjugated with phycoerythrin (PE) (Caltag, Burlingame, CA). Normal mouse IgM was used to block excess IgM binding sites before detection of A2B5. A2B5 was custom-conjugated with biotin using a standard biotinylation kit (Pierce, Rockford, IL), according to manufacturer's specifications, and visualized with streptavidin conjugated with PE and TxR (Texas Red) tandem dye (Caltag). Labeled cell suspensions were physically sorted directly into collection tubes containing RNAlater (Ambion, Austin, TX) using a FACSVantageSE flow cytometer (Becton Dickinson, Franklin Lakes, NJ), as previously described (Cohen et al., 2003).
RNA extraction/labeling.
Total RNA was extracted using the RNeasy micro kit (Qiagen, Valencia, CA). The quality of total RNA was assessed using a Bioanalyzer microchip (Agilent, Palo Alto, CA). A total of 100 nanograms of total RNA was amplified following the small sample labeling protocol (vII) of Affymetrix. The protocol involves two rounds of reverse transcription and in vitro transcription with the biotin label being incorporated during the second round of in vitro transcription.
Microarray.
Ten micrograms of biotin-labeled and fragmented cRNA was hybridized to Affymetrix Rat Expression 230 Microarrays (Affymetrix, Santa Clara, CA). Five biological replicates for A2B5 and four biological replicates for O4 were hybridized. The microarrays were hybridized, washed and scanned according to Affymetrix standard protocols. The raw data are deposited in the Gene Expression Omnibus accessible at http://www.ncbi.nlm.nih.gov/geo/ (Barrett et al., 2005).
Microarray data analysis.
Robust multiarray analysis was used for preprocessing of the raw CEL files. The data were then imported into Genespring for normalization and data analysis. The data were normalized using Genespring's per chip normalization (normalized to the 50th percentile) and per gene normalization (normalized to the median). A twofold difference in normalized expression value was used to identify differentially regulated transcripts. In addition, a Welch t test was performed without an assumption of equal variances. The Benjamini and Hochberg multiple testing correction was used with a false discovery rate of 0.05. The twofold cutoff and the statistical test were used to obtain the differentially regulated gene lists. Pathway analysis was performed with KEGG (Kanehisa et al., 2006) and Ingenuity (Redwood City, CA) pathway analysis.
Quantitative real-time PCR.
The LightCycler from Roche (Indianapolis, IN) was used to perform relative quantitative real-time PCR (qRT-PCR). cDNA was generated from total RNA isolated from purified oligodendrocyte lineage cells as described above. Primer pairs were chosen to span one intron and yield a ∼200 bp product size. Primer pairs were validated by performing PCR and running the products on an agarose gel to confirm a single band. In addition, melting curves were generated using the Roche LightCycler analysis software to determine whether there were any spurious amplification products. Actin was chosen as a reference gene because it was expressed at similar levels in both A2B5 and O4 according to the microarray data and using qRT-PCR. Relative quantitative RT-PCR was used to determine the fold difference for 11 genes (Pfaffl et al., 2002). In this method, the PCR efficiency is determined for each primer pair and is calculated by the following: efficiency = 10(−1/slope), with the slope being determined using a dilution series and the Roche LightCycler analysis software. The difference in cycle threshold between A2B5 and O4 was normalized by dividing by the difference in the cycle threshold for actin in A2B5 and O4 sorted cDNA.
Subcloning.
rPrmp and rProfilin2 were obtained using PCR on a cDNA template prepared from whole rat brain. The band was gel purified, subcloned into TOPO vector (Invitrogen, Carlsbad, CA), and then amplified using primers tailed with EcoRI. The products were subcloned into pEGFP-N2 (Becton Dickinson) and pTAG5CMyc (Stratagene, La Jolla, CA). All constructs were verified by DNA sequencing and expression was confirmed by Myc and enhanced green fluorescent protein (EGFP) antibody detection in transfected cells.
Immunocytochemistry.
CG4 or COS7 cells were plated and the following day cotransfected with Prmp-wt or Prmp-ct and EGFP expression vectors using Lipofectamine 2000 (Invitrogen). Each of the conditions was processed in parallel to facilitate comparison between the different groups. Twenty-four hours after transfection, cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. Anti-Myc (Sigma, St. Louis, MO) and anti-EGFP antibodies (Invitrogen) were applied to cells at room temperature for 2 h. Cells were washed three times for 5 min in PBS, incubated with 10% normal donkey serum for 30 min, and detected with donkey anti-mouse-Cy3 and donkey anti-rabbit-FITC (Invitrogen) for 1 h. Cells were covered with a glass cover and Vectashield with 4′,6′-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, CA) and observed with an Olympus (Tokyo, Japan) IX70 microscope through a 40× objective with a 0.6 numerical aperture. The images were captured with a Cool Snap digital camera (Photometrics, Tucson, AZ). For the cell process quantitation assay, transfected cells were identified in the red channel, and the longest process was measured from the cell body to the process tip using the EGFP channel. For area measurements in COS7 cells, the measure area function was used in the IPLab software (Scanalytics, Rockville, MD). Two independent experiments were performed with a total of 50 cells counted for each of the three groups.
Primary cultures.
Primary cultures from neonatal rat brains were prepared as described previously (Armstrong, 1998). Briefly, postnatal day 2 rat brains were dissociated, plated in tissue culture flasks, and allowed to grow for 7–10 d. The flasks were placed on a rotary shaker to dislodge immature oligodendrocyte lineage cells, which were then plated onto poly-d-lysine-coated chamber slides and 24-well plates. Oligodendrocyte progenitors were grown in medium containing PDGF (10 ng/ml) and basic fibroblast growth factor (bFGF) (10 ng/ml), and under these conditions most cells maintained a progenitor phenotype. When the progenitors were plated in differentiation medium lacking PDGF and bFGF, the majority of cells differentiated into mature oligodendrocytes over 72 h in culture.
Western blotting.
HEK293 cells were plated onto poly-d-lysine-coated 35 mm dishes and the following day transfected with Prmp-wt-EGFP, Prmp-ct-EGFP, or EGFP. Twenty-four hours after transfection, lysates were prepared with extraction buffer containing 1% Triton X-100 and protease inhibitors. Cells were disrupted with a tissue grinder and incubated for 60 min at 4 or 37°C. The soluble and insoluble fractions were separated with centrifugation at 12,000 × g for 10 min. The insoluble pellet was solubilized in 2% SDS and 8 m urea. The fractions were separated with SDS-PAGE, and then transferred to polyvinylidene difluoride membranes (Invitrogen). The membranes were blocked with 3% casein and probed with anti-EGFP. The proteins were detected with anti-rabbit IgG conjugated to HRP using the Western Breeze detection kit (Invitrogen).
Cell adhesion assay.
HEK293 cells were plated into 12-well plates and the following day transfected with Prmp-wt-EGFP, Prmp-ct-EGFP, or EGFP. Twenty-four hours after transfection, wells were gently washed with PBS, and loosely adherent cells were collected by centrifugation. The cells were counted using a hemocytometer. Two wells per condition were counted, and five independent replicates for each condition were performed.
Results
Purification of oligodendrocytes
Oligodendrocyte lineage cells were isolated directly from postnatal day 7 rat brains using a positive-selection FACS strategy. This strategy incorporated the initial identification of oligodendroglial cells using early and later stage-specific surface markers (A2B5, O4), which was followed by the physical separation of these cells into homogeneous populations, as illustrated in Figure 1, A and B. Using this sorting strategy, neuronal progenitors that can express low levels of A2B5 were excluded from oligodendroglial progenitors by selectively gating on cells expressing mid to high levels of A2B5 (Maric et al., 2003; Maric and Barker, 2004). This experimental design does not discriminate between early proligodendroblasts, which are O4+ and GalC negative, and more mature oligodendrocytes, which are O4+ and GalC positive. In this study, we refer to both of these stages as O4+ cells. However, to determine what percentage of the O4+ cells were GalC positive, we performed an analytical flow cytometry experiment using O4 and GalC antibodies. We found that 62% of the O4+ cells were GalC negative and 38% of the O4+ cells were GalC positive. Oligodendrocyte lineage cells were sorted directly into RNAlater (Ambion) to prevent RNA degradation, thus preserving their RNA expression state as close as possible to that existing in vivo. The quality of the RNA was assessed with the Bioanalyzer (Agilent), and only samples with visible ribosomal bands were used for labeling (Fig. 1C). As a control for the antibody selection strategy and cell purity obtained by FACS, we plated FACS-purified single A2B5+ cells into oligodendrocyte differentiation medium and cultured them for 6 d. After this period, most cells exhibited characteristic oligodendrocyte morphology and ∼70% acquired O1 immunoreactivity (Fig. 1D), demonstrating that A2B5+ cells retained their intrinsic potential to differentiate into mature oligodendrocytes in vitro.
Figure 1.
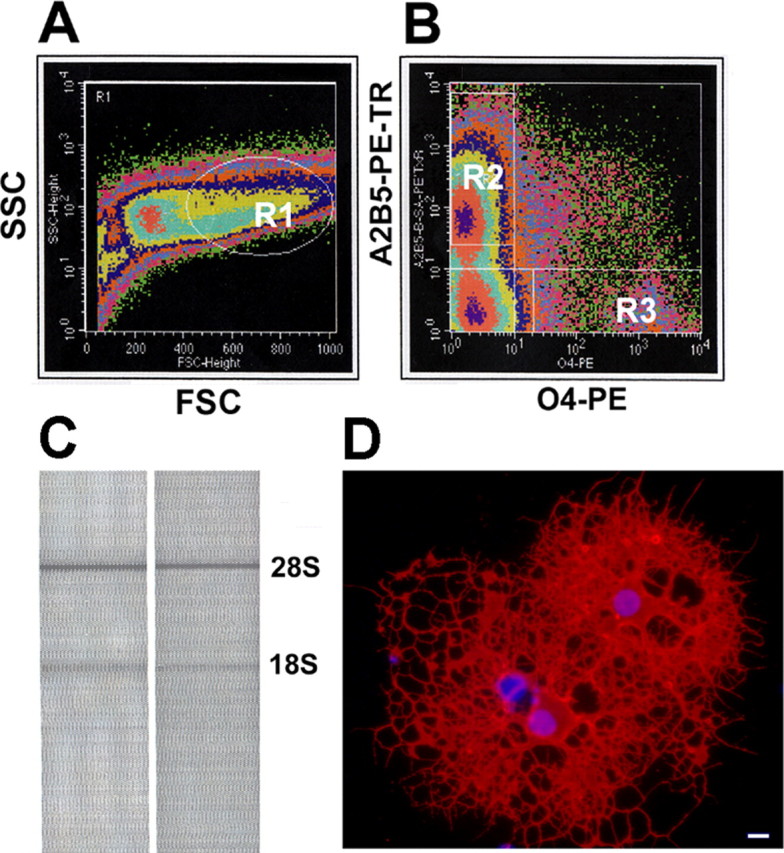
Oligodendrocytes were purified directly from the whole brain of postnatal day 7 rats using flow cytometry. A, FACS density scatter plot in pseudocolor shows forward scatter (FSC) versus side scatter (SSC) properties used to identify viable cells (gate R1). B, A2B5 and O4 immunofluorescence signals were used in combination with gate R1 to sort-purify homogeneous populations of vital cells at early and later stages of oligodendrocyte differentiation. Gates were set to collect single A2B5+ progenitors expressing middle to high levels of A2B5 (top left quadrant, R2) and single O4+ oligodendrocytes (bottom right quadrant, R3). C, Agilent Bioanalyzer analysis of total RNA extracted from oligodendrocytes isolated by FACS demonstrates the intact 28S and 18S rRNA bands. D, Sorted A2B5+ cells cultured for 6 d differentiated into mature oligodendrocytes, as evidenced by strong immunoreactivity with the O1 antibody (red fluorescence). Scale bar, 10 μm.
Gene ontology and pathway analysis
After hybridization and scanning, the microarray data were filtered to obtain a list of genes differentially expressed between A2B5+ and O4+ cells. A twofold cutoff was used in combination with a t test to eliminate transcripts with high variance between the biological replicates. In this study, we focused on transcripts that were upregulated in O4+ cells. When genes expressed in O4+ cells were sorted by normalized expression values, the three genes with the highest normalized expression value were myelin oligodendrocyte basic protein, juxtanodin, and myelin basic protein. Each of these genes is expressed in differentiated oligodendrocytes, thus helping to validate the methods used in this study. There were 1707 transcripts upregulated more than twofold in O4+ cells, whereas 1610 other transcripts were downregulated more than twofold in these cells (Fig. 2A). In assessing the global characteristics of the data, Venn diagrams were used to compare the number of genes present and absent in both A2B5+ and O4+ populations. Using the Affymetrix MAS 5.0 present calls, A2B5+ cells expressed 52% of all genes in four of five replicates, whereas O4+ cells expressed 48% of all genes in three of four replicates. Of the genes called present, 82% were detected in both A2B5+ and O4+ cells (Fig. 2B).
Figure 2.
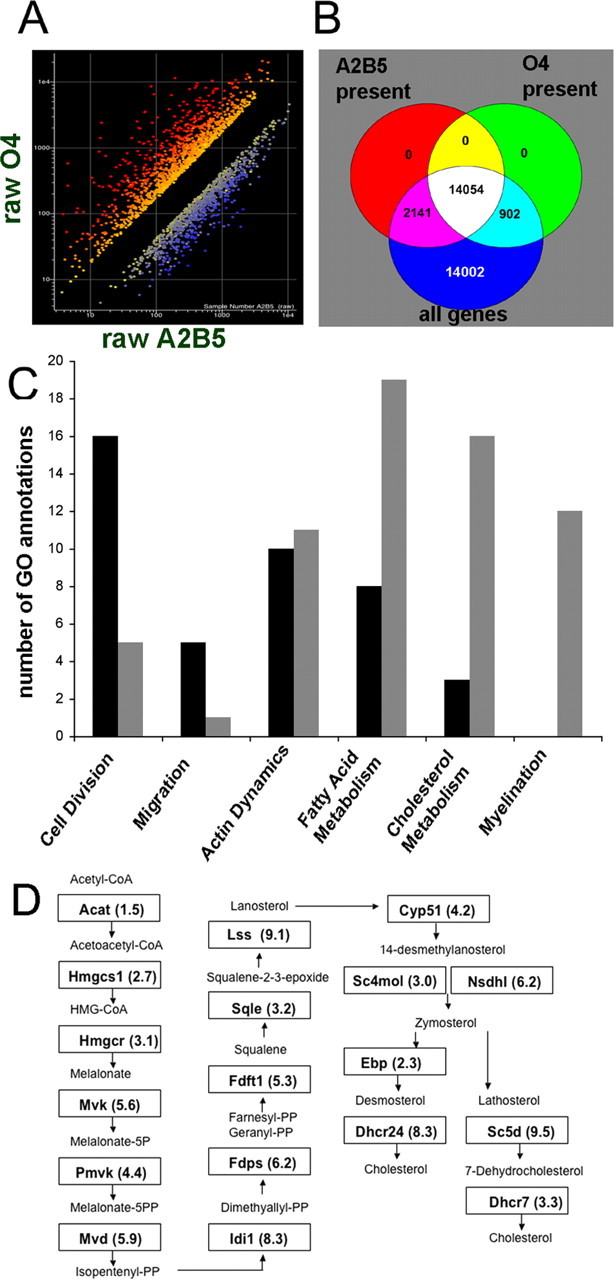
Transcripts upregulated in O4+ cells include many genes known to be involved with myelination. A, Microarray expression values from the Affymetrix rat 230 chip. A, Genes upregulated more than twofold in O4+ cells are shown in red, and genes downregulated more than twofold are shown in blue. B, Venn diagram illustrating the number of genes that were called present in four of five A2B5+ replicates and three of four O4+ replicates. C, Affymetrix GO browser was used to assign biological process terms for transcripts that were either downregulated (black bars) or upregulated (gray bars) in O4+ cells for the following GO designations: cell division, 51301; migration, 16477; actin dynamics, 30036; fatty acid metabolism, 6631, 6633, 6635; cholesterol metabolism, 6695, 8203, 45540; myelination, 42552. D, Genes in the cholesterol biosynthesis pathway with the designated gene symbol and fold upregulation in O4+ cells are shown in their respective boxes.
The Affymetrix Gene Ontology (GO) browser was used to organize the differentially regulated gene lists into functional categories using biological process designations. The number of transcripts was graphed from GO categories relevant to oligodendrocyte development (Fig. 2C). The results showed that cell division and migration designations each contained a greater number of transcripts that were downregulated more than twofold in O4+ cells (black bars), whereas fatty acid, cholesterol, and myelination designations each contained a greater number of transcripts that were upregulated more than twofold (gray bars). We next identified the probe sets for each of the 18 transcripts encoding proteins in the cholesterol biosynthetic pathway (Kanehisa et al., 2006), and found that all 18 transcripts were detected in O4+ cells and 17 of 18 genes in this pathway were upregulated more than twofold in O4+ cells (Fig. 2D). GO analysis also identified15 transcripts that were upregulated more than twofold in O4+ cells annotated with fatty acid metabolism including fatty acid synthase and fatty acid elongase 2 (Table 1).
Table 1.
Transcripts upregulated more than twofold in O4+ cells annotated with the GO terms: fatty acid metabolism (6631) and fatty acid biosynthesis (6633)
| Gene | Fold change | GenBank ID | Gene name |
|---|---|---|---|
| Acac | 2.3 | BI296153 | Acetyl-coenzyme A carboxylase |
| Acly | 3.3 | NM_016987 | ATP citrate lyase |
| Echs1 | 3.5 | NM_078623 | Enoyl coenzyme A hydratase, short chain 1 |
| Faah | 11.3 | NM_024132 | Fatty acid amide hydrolase |
| Facl2 | 4.9 | BI277523 | Fatty acid coenzyme A ligase, long chain 2 |
| Facl3 | 3.7 | NM_057107 | Fatty acid coenzyme A ligase, long chain 3 |
| rELO2 | 7.9 | AI235528 | Fatty acid elongase 2 |
| Fasn | 4.6 | AI179334 | Fatty acid synthase |
| Hadhsc | 2.3 | AA799574 | l -3-Hydroxyacyl-coenzyme A dehydrogenase, short chain |
| Nrbf1 | 2.3 | NM_017209 | Nuclear receptor binding factor 1 |
| 2.0 | AI406826 | Similar to carnitine palmitoyltransferase I (LOC308579) | |
| 19.8 | AW527783 | Similar to fatty acid hydroxylase (LOC307853), mRNA | |
| 3.2 | BI286134 | Similar to fatty acid transport protein 4 (LOC311839) | |
| Slc25a1 | 3.4 | NM_017307 | Solute carrier family 25, member 1 |
| 7.2 | AI009805 | Strong similarity to elongation of very long chain fatty acids |
Actin regulation and cell adhesion genes
The GO analysis identified a number of transcripts encoding proteins involved with the regulation of the actin cytoskeleton and cell adhesion. The KEGG actin polymerization pathway was used to categorize transcripts upregulated in O4+ cells that encode actin polymerization proteins (Kanehisa et al., 2006). Microarray analysis identified the presence of the transcripts encoding proteins involved with the stress fiber, filopodia, and lamellipodia pathways (Table 2). In addition, there was a total of 42 transcripts that encode cell adhesion proteins upregulated in O4+ cells including several transcripts that encode proteins known to be involved with oligodendrocyte adhesion such as myelin-associated glycoprotein, claudin11, and Neurofascin (Tables 3 and 4).
Table 2.
Transcripts detected in O4+ cells found in the KEGG regulation of actin cytoskeleton pathway
| Gene | Fold change | GenBank ID | Gene name |
|---|---|---|---|
| Stress fiber | |||
| RhoN | 8.3 | AI136709 | Similar to RhoN |
| Rgnef | 5.8 | BM386483 | Similar to Rho-guanine nucleotide exchange factor |
| Chn2 | 15.0 | BE120930 | Chimaerin 2 (Rho Gap3) |
| Rock | 1.4 | BF556405 | Rho-associated coiled-coil forming kinase 2 |
| Pip5K2a | 1.8 | AI407351 | Phosphatidylinositol-4-phosphate 5-kinase, type II |
| Gsn | 3.3 | BI285576 | Similair to gelsolin |
| Actn4 | 1.5 | NM_031675 | Actinin α 4 |
| Csrp1 | 5.9 | NM_017148 | Cysteine and glycine-rich protein 1 |
| Tmod2 | 5.8 | BF285511 | Tropomodulin 2 |
| Myo1c | 14.9 | BI279786 | Strong similarity to protein myosin I myr 4–rat |
| Pfn2 | 2.2 | NM_030873 | Profiln II |
| Filopodia | |||
| Cdc42 | 1.3 | AA925473 | Cell division cycle 42 |
| NWASP | 1.2 | BM388523 | Neural Wiskott–Aldrich syndrome protein |
| Arpc1a | 3.0 | NM_031146 | Actin-related protein complex subunt 1a |
| Lamellipodia | |||
| Rac | 1.6 | AA799542 | Rho family, small GTP binding protein Rac1 |
| Pak | 3.2 | NM_019210 | (CDKN1A)-activated kinase 3 |
| Abi1 | 1.0 | NM_024397 | Eps8 binding protein (e3B1) |
| Wasf1 | 7.3 | BI295427 | Similar to WASP family 1 (LOC294568), mRNA |
| Arpc1a | 3.0 | NM_031146 | Actin-related protein complex subunit 1a |
| Pfn2 | 2.2 | NM_030873 | Profilin II |
| Tmsb4 | 0.8 | BG668902 | Thymosin β -4, Y isoform |
Transcripts upregulated more than twofold in O4+ cells are identified in bold and include transcripts encoding proteins required for filopodia, lamellipodia, and stress fiber formation.
Table 3.
Transcripts upregulated more than twofold in O4+ cells identified with the GO term adhesion (7155)
| Gene | Fold change | GenBank ID | Gene name |
|---|---|---|---|
| Adam23 | 2.1 | AI535213 | A disintegrin and metalloprotease domain 23 |
| App | 2.5 | BM986220 | Amyloid β (A4) precursor protein |
| Bcan | 2.5 | U37142 | Brevican |
| Cdh13 | 5.2 | BI282750 | Cadherin 13 |
| Cdh20 | 4.9 | BF419648 | Cadherin 20 (predicted) |
| Cd9 | 2.9 | AI227627 | CD9 antigen |
| Cspg2 | 7.3 | AF084544 | Chondroitin sulfate proteoglycan 2 |
| Cldn11 | 24.5 | NM_053457 | Claudin 11 |
| Cntn1 | 13.9 | NM_057118 | Contactin 1 |
| Ddr1 | 2.6 | L26525 | Discoidin domain receptor family, member 1 |
| 5.8 | BM386842 | Elastin microfibril interfacer 2 (predicted) | |
| Fez1 | 4.2 | NM_031066 | Fasciculation and elongation protein ζ 1 (zygin I) |
| Gp1bb | 9.9 | BM387903 | Glycoprotein Ib, β polypeptide |
| Gpc1 | 4.4 | NM_030828 | Glypican 1 |
| Hapln2 | 139.9 | AI145465 | Hyaluronan and proteoglycan link protein 2 |
| Igsf4a | 2.5 | BE117767 | Immunoglobulin superfamily, member 4A (predicted) |
| Itgb4 | 2.4 | NM_013180 | Integrin β 4 |
| Lsamp | 6.4 | U31554 | Limbic system-associated membrane protein |
| Mcam | 3.1 | BI277043 | Melanoma cell adhesion molecule |
| Pcdh21 | 3.8 | NM_053572 | MT-protocadherin |
| Mpdz | 2.5 | NM_019196 | Multiple PDZ domain protein |
| Mag | 29.2 | NM_017190 | Myelin-associated glycoprotein |
| Neo1 | 4.6 | U68726 | Neogenin |
| Nfasc | 2.5 | BI285436 | Neurofascin |
| Ninj2 | 61.1 | NM_021595 | Ninjurin 2 |
| Opcml | 8.6 | NM_053848 | Opioid-binding protein/cell adhesion molecule-like |
| Pkp4 | 2.7 | BE113437 | Plakophilin 4 (predicted) |
| Col2a1 | 8.0 | AF305418 | Procollagen, type II, α 1 |
| Col9a1 | 3.0 | BM388861 | Procollagen, type IX, α 1 (predicted) |
| Col9a2 | 2.6 | BM384121 | Procollagen, type IX, α 2 (predicted) |
| Col9a3 | 8.9 | BM389001 | Procollagen, type IX, α 3 (predicted) |
| Col11a1 | 2.8 | BM389291 | Procollagen, type XI, α 1 |
| Col11a2 | 6.0 | BM391350 | Procollagen, type XI, α 2 |
| 2.6 | AW918040 | Protocadherin 17 (predicted) | |
| 5.5 | BF415436 | Protocadherin 9 | |
| Spock1 | 2.1 | AI406342 | Sparc/osteonectin, cwcv kazal-like proteoglycan 1 |
| Tnr | 19.1 | NM_013045 | Tenascin R |
| Tyro3 | 11.1 | NM_017092 | TYRO3 protein tyrosine kinase 3 |
Table 4.
Transcripts upregulated more than twofold in O4+ cells identified with the GO term myelination (42552) and with PubMed literature searching for proteins expressed in differentiated oligodendrocytes
| Gene | Fold change | GenBank ID | Gene name |
|---|---|---|---|
| Aspa | 4.6 | NM_024399 | Aspartoacylase |
| Atrn | 4.9 | NM_031351 | Attractin |
| Casr | 58.8 | NM_016996 | Calcium-sensing receptor |
| Cldn11 | 24.5 | NM_053457 | Claudin 11 |
| Cnp1 | 26.4 | M18630 | Cyclic nucleotide phosphodiesterase 1 |
| Tpo1 | 5.1 | NM_133395 | Developmentally regulated protein TPO1 |
| Enpp2 | 5.3 | NM_057104 | Ectonucleotide pyrophosphatase 2 |
| Fyn | 9.4 | NM_012755 | Fyn proto-oncogene |
| Gjb1 | 21.0 | NM_017251 | Gap junction membrane channel protein β 1 |
| Klk6 | 34.0 | NM_019175 | Kallikrein 6 |
| Mal | 124.0 | NM_012798 | Myelin and lymphocyte protein |
| Mbp | 40.5 | NM_017026 | Myelin basic protein |
| Mog | 48.0 | NM_022668 | Myelin oligodendrocyte glycoprotein |
| Mag | 29.2 | NM_017190 | Myelin-associated glycoprotein |
| Mobp | 205.5 | X90402 | Myelin-associated oligodendrocytic basic protein |
| Omg | 10.1 | AA964584 | Oligodendrocyte myelin glycoprotein |
| Z49858 | 11.7 | NM_022533 | Plasmolipin |
| Plp | 4.3 | NM_030990 | Proteolipid protein |
| Nkx6.2 | 27.3 | BF400968 | Similar to homeotic protein Gtx—mouse |
| Edg8 | 61.9 | AF115249 | Sphingosine 1-phosphate receptor |
| Sox10 | 25.7 | NM_019193 | SRY-box containing gene 10 |
| Tspan2 | 18.5 | AI228231 | Tetraspan 2 |
| Tf | 7.6 | AA945178 | Transferrin |
| Ugt8 | 41.5 | L21698 | UDP-glucuronosyltransferase 8 |
| Zfp111 | 6.4 | NM_133323 | Zinc finger protein 111 |
Microarray validation
GO analysis and literature searching identified 25 genes upregulated in O4+ cells that are annotated with the GO term myelination or are reported in the literature to play a role in oligodendrocyte differentiation (Table 4). Additionally, qRT-PCR was performed on a subset of transcripts upregulated in O4+ cells. The 11 transcripts chosen from different functional categories displayed fold changes ranging from a high magnitude to near the twofold cutoff. All 11 genes chosen for validation gave similar results as the microarrays. In some cases, the magnitude of the fold difference was different, but in each case the direction of the fold change and the twofold cutoff of the qRT-PCR agreed with the microarray data (Fig. 3A).
Figure 3.
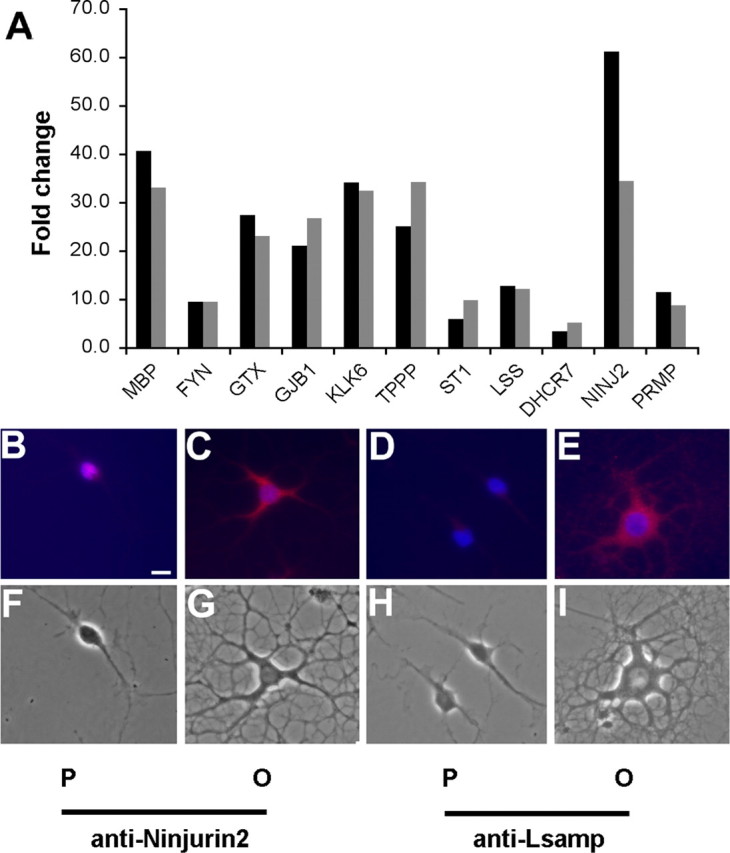
Transcripts identified as differentially regulated by microarray analysis were validated by qRT-PCR and immunocytochemistry. A, Relative qRT-PCR (gray bars) was performed on RNA isolated by FACS, and compared with the data obtained with microarrays (black bars). Summary of qRT-PCR data on 11 transcripts including several genes known to be upregulated during oligodendrocyte differentiation (Mbp, Fyn, Nkx6.2, and Gjb1) and transcripts involved with cholesterol biosynthesis including Osc and Dhcr7. B–I, Immunocytochemistry using anti-Ninjurin2 (B, C, F, G) and Lsamp (D, E, H, I) was used to validate the array data at the protein level. Primary oligodendrocyte cultures were grown in the presence of PDGF and bFGF to maintain their immature progenitor state (P) (B, D) or in the absence of these growth factors to allow differentiation into oligodendrocytes (O) (C, E). Strong signal was detected in the cell body and processes of differentiated oligodendrocytes incubated with anti-Ninjurin2 (C, G), whereas little Ninjurin2 staining was observed in oligodendrocyte progenitors (B, F). Strong signal was also detected in the cell body and processes of differentiated oligodendrocytes incubated with anti-Lsamp (E, I), whereas little signal was observed in oligodendrocyte progenitors (D, H). DAPI staining of nuclei is blue. Scale bar, 10 μm.
To validate the microarray data at the protein level, we obtained antibodies that are specific for Ninjurin2 and limbic system-associated membrane protein (Lsamp) both of which were upregulated in O4+ cells. An oligodendrocyte primary culture system was used that allows the control of differentiation by the addition of PDGF and bFGF. In cultures maintained in PDGF and bFGF, most cells are A2B5+, maintain a bipolar morphology, and continue to proliferate. In the absence of growth factors, the oligodendrocyte progenitors differentiate into mature oligodendrocytes. Immunocytochemistry was used with anti-Ninjurin2 (Araki and Milbrandt, 2000) and anti-Lsamp (Levitt, 1984) to label progenitor and differentiated cultures. Progenitor cultures incubated with anti-Ninjurin2 appeared to have detectable protein expression in the cell body (Fig. 3B). However, in differentiated cultures, there was strong expression of this protein in the cell body and in the major cell processes (Fig. 3C). A similar pattern was seen with anti-Lsamp, with weak signal in the progenitor cultures (Fig. 3D) and strong expression in the cell body and the major processes of differentiated cells (Fig. 3E).
Identification of Prmp
We hypothesized that novel genes encoding proteins required for myelination could be identified from the list of transcripts upregulated in O4+ cells. We searched this gene list for transcripts encoding novel proteins with putative transmembrane domains and identified four candidate genes, which were subcloned into expression vectors. One of these novel transcripts encodes a protein, which we have named Prmp. The Prmp gene is found in the human (TMEM58), mouse (Tmem58), and rat (similar to Scotin) databases. Prmp is expressed in both oligodendrocyte progenitors and O4+ oligodendrocytes, and there is a 10-fold increase in Prmp expression in O4+ cells (Fig. 3A).
Prmp is a predicted single pass transmembrane protein with an extracellular N terminus and cytoplasmic C terminus (Krogh et al., 2001). Prmp is highly conserved between human, rat, and mouse (Fig. 4A). Prmp has weak homology to the gene that encodes Scotin, which is a proapoptotic endoplasmic reticulum-resident protein (Bourdon et al., 2002). Prmp contains a predicted signal peptide between amino acids 1 and 27, a region of E-cadherin homology between amino acids 42 and 52, a transmembrane domain between amino acids 86 and 108, and a proline-rich domain between amino acids 131 and 195 (Fig. 4A). There is also a predicted N-glycosylation site at amino acid 38. The C-terminal proline-rich domain contains a predicted RhoA effector conserved domain (Marchler-Bauer and Bryant, 2004). Members containing this domain include diaphanous-related formin-1 of the formin family of proteins, which are important actin cytoskeleton regulators.
Figure 4.
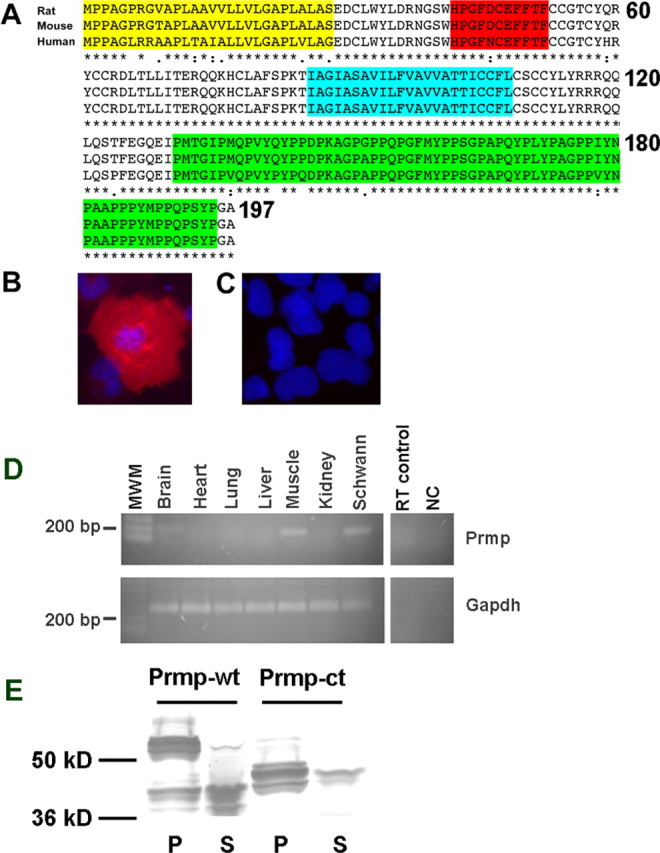
Prmp is a highly conserved detergent-insoluble transmembrane protein. A, Amino acid alignment of rat, mouse, and human orthologs of Prmp. Prmp is a predicted single pass transmembrane protein with an N-terminal extracellular domain containing a predicted signal peptide (yellow) and a region of homology with E-cadherin (red). The predicted transmembrane domain is highlighted in blue. The C terminus contains a proline-rich domain highlighted in green. COS7 cells transfected with C-terminal Myc-tagged Prmp were fixed and permeabilized (B) or surface-stained live with anti-Myc (C). C-terminally tagged Prmp was only detected after permeabilization. DAPI staining of nuclei is blue. D, RT-PCR was performed on various adult mouse tissues. Prmp was detected in the brain, skeletal muscle, and Schwann cells. MWM, Molecular weight marker; NC, negative control; RT, no reverse transcriptase control. E, HEK293 cells were transfected with an EGFP-tagged Prmp expression vector, and 24 h after transfection, lysates were prepared with a 1% Triton X-100 extraction buffer at 4°C. Both the full-length Prmp-wt and Prmp-ct deletion mutant were found predominantly in the detergent-insoluble fraction (P). Only a small percentage of the transfected full-length Prmp-wt or Prmp-ct was recovered in the detergent-soluble fraction (S).
The predicted topology of Prmp was tested using immunocytochemistry on fixed/permeabilized and live-cell surface staining. A C-terminal Myc epitope-tagged Prmp expression vector was transfected into COS7 cells. The results revealed strong Myc expression after these cells were fixed and permeabilized (Fig. 4B), whereas there was no detectable Prmp expression in the live surface staining conditions (Fig. 4C).
RT-PCR was used to determine the tissue expression profile of Prmp. Prmp mRNA was detected in brain, skeletal muscle, and Schwann cell cultures (Fig. 4D). In the CNS, the oligodendrocyte lineage expression of Prmp mRNA was identified in the microarray data set and confirmed using RT-PCR with oligodendrocytes purified by flow cytometry (Fig. 3A).
Prmp partitioned in the detergent-insoluble fraction, consistent with a possible close association with the cytoskeleton. An EGFP C-terminal Prmp expression vector was transfected into HEK293 cells, and lysates were prepared with 1% Triton-X100 at 4°C. Nearly all of the full-length Prmp-wt was retained in the detergent-insoluble fraction (Fig. 4E). We obtained similar results with lysates prepared at 37°C (data not shown). Prmp-wt migrates slightly larger than the predicted 48.4 kDa, and there appear to be multiple forms, possibly because of N-glycosylation. The soluble fraction contains an apparent truncated version of Prmp that corresponds to the approximate size of the cytoplasmic domain plus the EGFP fusion (36.7 kDa). A Prmp deletion mutant lacking the cytoplasmic domain (Prmp-ct) is also predominately retained in the detergent-insoluble fraction, indicating that the proline-rich C terminus does not confer detergent insolubility.
Immunocytochemistry performed on HEK293 cells localized Prmp to the plasma membrane, with some Prmp protein appearing to be concentrated at cell–cell junctions (Fig. 5A). Double labeling with phalloidin indicates that Prmp appears to be colocalized with actin at cell–cell junctions. A Prmp deletion mutant lacking the cytoplasmic domain also appears to be localized to cell–cell junctions (Fig. 5B). The homology of the proline-rich domain of Prmp to the formin family members like diaphanous-related formin 1 predicted a possible interaction with proteins that regulate actin dynamics such as Profilin2. To test this hypothesis, Profilin2 was transfected into HEK293 cells. The results show that Profilin2 is localized to the plasma membrane and is colocalized with actin (Fig. 5C). Cotransfection of Profilin2 and Prmp reveal that both Prmp and Profilin2 appear to be colocalized at cell–cell junctions (Fig. 5E). Control experiments done with cotransfection of Prmp and EGFP did not reveal any strong overlap at cell–cell junctions (Fig. 5D).
Figure 5.
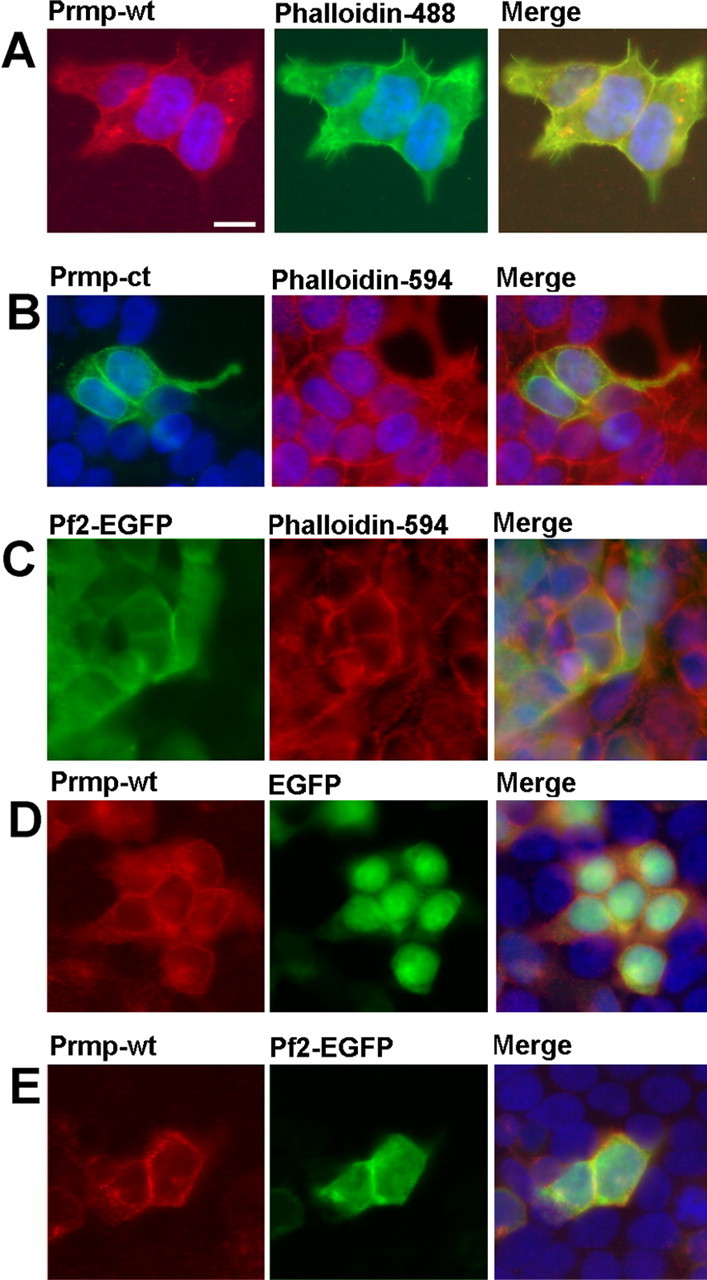
Prmp is localized to the plasma membrane in HEK293 cells, and colocalizes with actin and Profilin2. A, Myc-tagged Prmp-wt was transfected into HEK293 cells. At 24 h after transfection, the cells were fixed and detected with anti-Myc (red) and phalloidin 488 (green). B, EGFP-tagged Prmp-ct was transfected into HEK293 cells and detected with anti-EGFP (green) and phalloidin 594 (red). C, An EGFP-tagged Profilin2 was transfected into HEK293 cells and detected with anti-EGFP (green) and phalloidin-594 (red). D, Control EGFP transfected HEK293 cells demonstrate little colocalization of Prmp and EGFP. E, Prmp tagged with Myc was cotransfected with Profilin2 tagged with EGFP, and detected with anti-Myc (red) and anti-EGFP (green). Prmp and Profilin2 appear to colocalize at cell–cell junctions. DAPI staining of nuclei is blue. Scale bar, 10 μm.
Prmp functional characterization
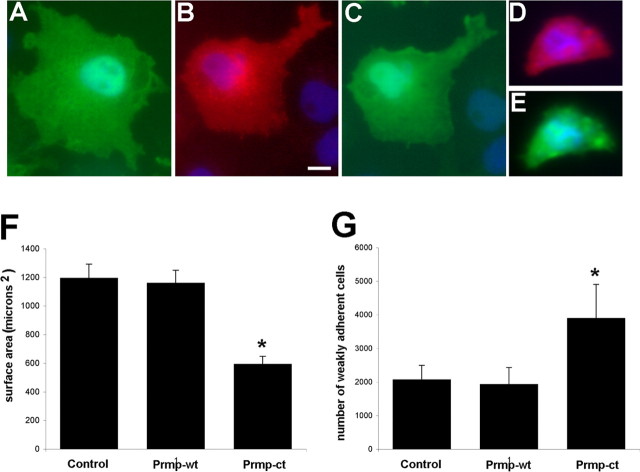
To assess the function of Prmp, the proline-rich domain was deleted. A C-terminal deletion mutant (Prmp-ct) was constructed that lacks the entire cytoplasmic domain of Prmp. When Prmp-wt was transfected into COS7 cells (Fig. 6B,C), there was no apparent effect on morphology when compared with control transfected cells (Fig. 6A). However, in COS7 cells transfected with Prmp-ct (Fig. 6D,E), the cells had a dramatic reduction in the extent of membrane spreading. Nearly all cells that had been transfected with Prmp-ct appeared to be reduced in size, and many were much smaller, rounded-up cells. Quantifying these data indicated that transfection of Prmp-ct reduced membrane spreading by nearly twofold compared with control and Prmp-wt (Fig. 6F). The inhibition of membrane spreading is similar to phenotypes induced by a constitutively active RhoA signaling pathway (Khyrul et al., 2004). We tested whether the RhoA inhibitor Y-27632 could rescue the Prmp-ct inhibition. However, we did not observe any rescue of Prmp-ct transfected cells treated with Y-27632 (data not shown). To determine whether the Prmp-ct deletion mutant was affecting the survival of the transfected cells, a cell viability assay was performed. Twenty-four hours after transfection, cells were trypsinized, labeled with propidium iodide and Hoechst dyes, and analyzed with flow cytometry. Control transfected cells were 97.9% viable, Prmp-wt cells were 98.9% viable, and Prmp-ct transfected cells were 98.7% viable.
Figure 6.
Deletion of the C terminus of Prmp reduces membrane spreading of COS7 cells. A, COS7 cells were transfected with control EGFP and detected with anti-EGFP. The EGFP channel was used to measure membrane spreading. COS7 cells were cotransfected with Prmp-wt (red; B), and EGFP (green; C). COS7 cells were cotransfected with a C-terminal deletion mutant (Prmp-ct; D) and (EGFP; E). The corresponding EGFP channel in E demonstrates the reduced size of COS7 cells that have been transfected with Prmp-ct. DAPI staining of nuclei is blue. Scale bar, 10 μm. F, Quantitation of Prmp transfection in COS7 cells. The total area was measured using the EGFP channel and IPLab software. Fifty cells from two independent experiments were measured for each group. Error bar represents SEM. *p < 0.01, Student's t test (Prmp-wt compared with Prmp-ct). G, Cell adhesion assays were performed on HEK293 cells transfected with EGFP control, Prmp-wt-EGFP, and Prmp-ct-EGFP. Twenty-four hours after transfection, wells were gently washed and loosely adherent cells were counted with hemacytometer. Two wells per condition were counted, and five independent replicates were performed. Error bars represent the SEM. *p < 0.05, Student's t test (Prmp-wt compared with Prmp-ct).
Cell adhesion assays were performed to test whether the reduction in membrane formation had any effect on adhesion. There was no difference in the number of adherent cells comparing Prmp-wt to control transfected cells. However, when cells were transfected with Prmp-ct, there was a twofold increase in the number of loosely adherent cells (Fig. 6G). Homotypic cell adhesion assays were also performed, but we did not detect any evidence for Prmp acting as a homotypic cell adhesion protein (data not shown).
To determine whether Prmp had similar effects on a more relevant cell type, the above transfection experiments were repeated using CG4 cells. CG4 cells are an oligodendrocyte cell line that is typically used as an in vitro model for oligodendrocyte differentiation. When CG4 cells were transfected with Prmp, Prmp-wt could be observed localized to the cell body and the processes, but Prmp-wt had no apparent effect on morphology of the transfected cells (Fig. 7A,B). However, the morphology of Prmp-ct transfected CG4 cells was strongly affected, because these cells were unable to normally extend processes (Fig. 7C,D) when compared with control transfected cells (Fig. 7E).
Figure 7.
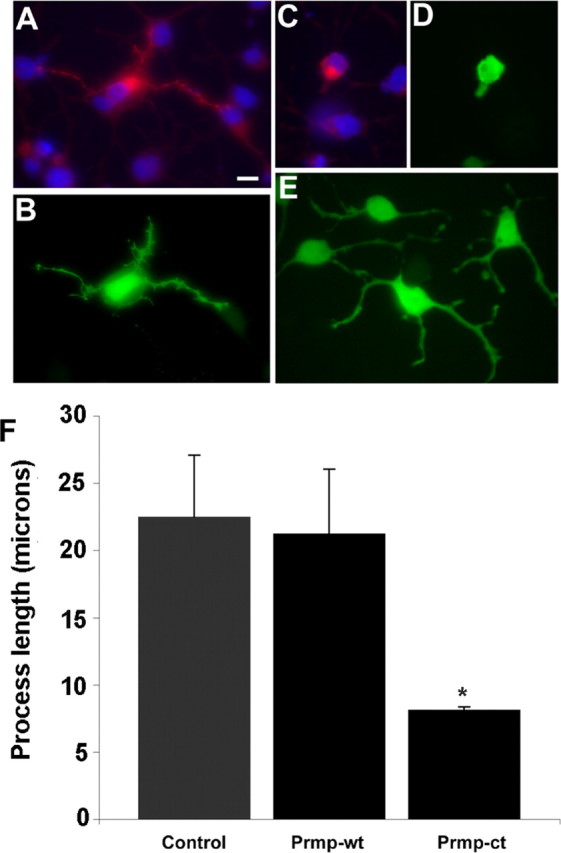
Deletion of Prmp C terminus reduces process length of CG4 cells. CG4 cells were transfected with an expression vector containing Prmp tagged with Myc (A). Cotransfection with EGFP was used to measure the length of the longest process (B). A C-terminal deletion mutant (Prmp-ct) was transfected into CG4 cells (C). The corresponding EGFP channel in C demonstrates the severe reduction in process length of CG4 cells that have been transfected with Prmp-ct (D). E, Control EGFP transfected CG4 cells. DAPI staining of nuclei is blue. Scale bar, 10 μm. F, Quantitation of process length after Prmp transfection in CG4 cells. The longest process was measured from the cell body to the end of the process using the EGFP channel and IPLab software. Fifty cells from two independent experiments were measured for each group. Error bars represent SEM. *p < 0.01, Student's t test (Prmp-wt compared with Prmp-ct).
Discussion
In this study, we sought to identify changes in gene expression during the transition from A2B5+ oligodendrocyte progenitors into O4+ oligodendrocytes, and to identify molecular components that are required at the onset of myelination. We identified a number of genes that are upregulated as oligodendrocytes differentiate, including transcripts encoding myelin proteins, fatty acid and cholesterol biosynthesis enzymes. Analysis of this data set demonstrates that at the O4+ stage of oligodendrocyte development, cells are preparing to myelinate with the production of enzymes and proteins required for synthesizing the myelin sheath. The microarray data were validated by multiple approaches, including qRT-PCR, immunocytochemistry, and published expression data. The most compelling validation comes from the sheer number of genes known to be involved in myelination that were identified as upregulated in O4+ cells. We found 25 genes encoding transcripts that have a myelin term assigned by GO or were found by PubMed searches to be involved with oligodendrocyte differentiation. In addition, many of these genes exhibited very high fold increases compared with their expression levels in A2B5+ progenitor cells (Table 4), and most were found in the top 1% of normalized expression values of all the genes on the microarray. As an additional validation, we compared the microarray data to published oligodendrocyte myelin proteomic data, which lists 98 proteins detected in mature myelin (Taylor et al., 2004). The microarrays detected as present in O4+ cells 94% of the transcripts that encode proteins identified by proteomic methods in myelin. Additionally, 10% of transcripts were identified as upregulated in O4+ cells that encode proteins identified in mature myelin. The correlation between the two data sets provides additional validation of this study, and improves confidence given to predictions about the translation of mRNA into protein.
Analysis of the transcripts detected in each population of cells indicated that many of the genes required in mature oligodendrocytes were already transcriptionally active in A2B5+ oligodendrocyte progenitors. Of the 1707 transcripts on the list of genes upregulated in O4+ cells, 76% were detected as present in A2B5+ cells. The other 24% of the genes upregulated in O4+ cells that were not detected in A2B5+ cells did not appear to be of any particular class, and included myelin proteins such as myelin basic protein, transcription factors such Nkx6.2, and cholesterol biosynthetic enzymes such as lanosterol synthase.
Cholesterol is a major constituent of the myelin sheath and cholesterol biosynthesis has been shown to take place locally in the brain (Jurevics and Morell, 1995). In addition, cholesterol biosynthesis within the oligodendrocyte lineage is required for proper myelination (Saher et al., 2005). Gene ontologies and pathway analysis of our microarray data set identified the cholesterol biosynthetic pathway as being activated in O4+ cells. Seventeen of 18 genes required to synthesize cholesterol were upregulated more than twofold in O4+ cells. Sterol response elements have been identified in cholesterol gene promoters, and sterol response element binding proteins (SREBPs) have been shown to regulate cholesterol gene expression in Schwann cells (Leblanc et al., 2005). SREBP2 was upregulated 2.5-fold in O4+ cells, suggesting that SREBP2 may play a role in the coordinate regulation of cholesterol biosynthetic genes in oligodendrocytes. In addition, the cholesterol biosynthesis pathway was previously reported to be activated in Schwann cell development and injury models (Nagarajan et al., 2002; Verheijen et al., 2003). Together, these data confirm the importance of cholesterol biosynthesis within the oligodendrocyte lineage, and highlight the ability of microarrays to detect pathways that have been activated during development.
The dramatic changes in morphology that take place as oligodendrocytes differentiate are indicative of major alterations in the cytoskeleton. We detected changes in transcripts important in several different aspects of actin cytoskeleton regulation, including proteins such as neuronal Wiskott–Aldrich syndrome protein (N-WASP) and WASP-family verprolin-homologous protein 1 (WAVE1), which in other systems are effector proteins important for the formation of filopodia and lamellipodia (Table 2). Both filopodia and lamellipodia formation have been reported to occur as oligodendrocytes extend processes and initiate contact with axons (Kachar et al., 1986; Asou et al., 1995). In addition, WAVE1 was recently reported to be required for proper CNS myelination (Kim et al., 2006). Previous reports have identified RhoA and RhoGAP as regulators of oligodendrocyte differentiation, and it was shown that downregulation of RhoA activity by RhoGAP was required for process extension (Wolf et al., 2003; Liang et al., 2004). In this study, microarray analysis detected the presence of RhoA, RhoB, and RhoC in both A2B5+ and O4+ cells. A new Rho family member called RhoN was upregulated in O4+ cells, as well as a RhoN effector called rapostlin. RhoN and rapostlin have been shown to induce neurite branching in PC12 cells (Fujita et al., 2002), which suggests that this pathway may be required for the extensive process branching that takes place during oligodendrocyte differentiation. Several transcripts important for actin stabilization were also identified as upregulated in O4+ cells including Tmod2 and Gelsolin. The microarrays detected the presence of several subunits of the APC2/3 actin polymerization complex, as well as the upregulation of Arpc1a in O4+ cells. The detection of transcripts encoding proteins involved with different aspects of actin dynamics including actin polymerization and stabilization suggests that multiple pathways are working simultaneously to achieve the changes in morphology required for process growth and myelination.
The number of cell adhesion and cytoskeleton transcripts upregulated in O4+ cells likely reflects the rapid changes taking place as oligodendrocytes begin the myelination process. We identified a number of candidate genes that encoded putative membrane proteins for additional study, one of which we describe here. Prmp is a novel gene that contains a single predicted transmembrane domain. The extracellular domain of Prmp contains a short region of homology to E-cadherin with 80% identity. The cytoplasmic domain of Prmp has a proline-rich region that contains a RhoA effector conserved domain. RhoA is a small GTPase that is important for focal adhesions and stress fiber formation (Wennerberg and Der, 2004). Proteins with a RhoA effector domain include the formin family of proteins that contain an FH2 domain, which polymerizes actin and a proline-rich FH1 domain that can bind a number of proteins involved with actin regulation including profilins (Higgs, 2005). Prmp is unusual because it does not contain an actin polymerization domain, but the homology to E-cadherin and the RhoA effector domain both suggested a potential cell adhesion function for Prmp.
Immunocytochemistry indicated that the transfected Prmp protein appears to be localized to cell–cell junctions consistent with a possible cell adhesion function. To begin to investigate Prmp function, Prmp and a Prmp deletion mutant lacking the cytoplasmic domain (Prmp-ct) were transfected into cell lines. Membrane spreading and process extension in both COS7 and CG4 cells were dramatically inhibited in the Prmp-ct transfected cells. Cells transfected with Prmp-ct were also less adherent than control or Prmp-wt transfected cells. These data are consistent with a model in which Prmp functions to link the extracellular matrix to the actin cytoskeleton. This hypothesis is further supported by the detergent-insoluble properties of Prmp, which may be caused by a close association of Prmp with the detergent-insoluble cytoskeleton. The absence of any obvious effect in the Prmp full-length transfections and the absence of an FH1 domain that is required for actin polymerization, suggests that Prmp may function to stabilize plasma membrane structures rather than promoting actin polymerization and membrane growth. However, additional studies of the endogenous Prmp protein in primary cells will be required to fully elucidate Prmp function.
In summary, the combination of FACS and microarrays has identified many of the transcripts that are differentially regulated as oligodendrocytes differentiate from A2B5+ progenitors into O4+ oligodendrocytes. The gene lists generated in this study may provide a useful reference for the continued study of the proteins important for myelination. In addition, these lists may be useful for the identification of disease genes relevant to myelin disorders. In this study, we have focused on events occurring at the plasma membrane of oligodendrocytes, and have identified Prmp as a novel oligodendrocyte cell adhesion protein. The future challenge remains to identify and characterize additional novel proteins, and to unravel how the individual proteins of the myelination machinery interact during myelination.
Footnotes
This work was supported by the National Institute of Neurological Disorders and Stroke (NINDS) Division of Intramural Research. We thank Abdel Elkahloun (National Human Genome Research Institute, Bethesda, MD) and his staff for carrying out the microarray hybridization, washing, and scanning; Elena Romm (NINDS, Bethesda, MD) for the antibody labeling; Catherine Campbell (NINDS) for assistance with the analysis of the microarray data; Gabor Lovas (Semmelweis University, Budapest, Hungary) for demonstrating the oligodendrocyte purification protocol; Steve Pfeiffer for helpful discussions; Regina Armstrong (Uniformed Services University of the Health Sciences, Bethesda, MD), Chang-Yu Wang (United States Patent and Trademark Office, Alexandria, VA), Naser Muja (NINDS), Sara Szuchet (University of Chicago, Chicago, IL), and the Fellow Editorial Board (National Cancer Institute, Frederick, MD) for critical reading of this manuscript. We acknowledge Aurea Pimenta (Vanderbilt University, Nashville, TN) for providing the anti-Lsamp antibody and Toshiyuki Araki (Washington University, St. Louis, MO) for providing the anti-Ninjurin2.
References
- Abney ER, Williams BP, Raff MC. Tracing the development of oligodendrocytes from precursor cells using monoclonal antibodies, fluorescence-activated cell sorting, and cell culture. Dev Biol. 1983;100:66–171. doi: 10.1016/0012-1606(83)90207-5. [DOI] [PubMed] [Google Scholar]
- Araki T, Milbrandt J. Ninjurin2, a novel homophilic adhesion molecule, is expressed in mature sensory and enteric neurons and promotes neurite outgrowth. J Neurosci. 2000;20:187–195. doi: 10.1523/JNEUROSCI.20-01-00187.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong RC. Isolation and characterization of immature oligodendrocyte lineage cells. Methods. 1998;16:282–292. doi: 10.1006/meth.1998.0685. [DOI] [PubMed] [Google Scholar]
- Asou H, Hamada K, Miyazaki T, Sakota T, Hayashi K, Takeda Y, Marret S, Delpech B, Itoh K, Uyemura K. CNS myelinogenesis in vitro: time course and pattern of rat oligodendrocyte development. J Neurosci Res. 1995;40:519–534. doi: 10.1002/jnr.490400411. [DOI] [PubMed] [Google Scholar]
- Barrett T, Suzek TO, Troup DB, Wilhite SE, Ngau WC, Ledoux P, Rudnev D, Lash AE, Fujibuchi W, Edgar R. NCBI GEO: mining millions of expression profiles—database and tools. Nucleic Acids Res. 2005;33:D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourdon JC, Renzing J, Robertson PL, Fernandes KN, Lane DP. Scotin, a novel p53-inducible proapoptotic protein located in the ER and the nuclear membrane. J Cell Biol. 2002;158:235–246. doi: 10.1083/jcb.200203006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttery PC, ffrench-Constant C. Laminin-2/integrin interactions enhance myelin membrane formation by oligodendrocytes. Mol Cell Neurosci. 1999;14:199–212. doi: 10.1006/mcne.1999.0781. [DOI] [PubMed] [Google Scholar]
- Charles P, Tait S, Faivre-Sarrailh C, Barbin G, Gunn-Moore F, Denisenko-Nehrbass N, Guennoc AM, Girault JA, Brophy PJ, Lubetzki C. Neurofascin is a glial receptor for the paranodin/Caspr-contactin axonal complex at the axoglial junction. Curr Biol. 2002;12:217–220. doi: 10.1016/s0960-9822(01)00680-7. [DOI] [PubMed] [Google Scholar]
- Cohen RI, Rottkamp DM, Maric D, Barker JL, Hudson LD. A role for semaphorins and neuropilins in oligodendrocyte guidance. J Neurochem. 2003;85:1262–1278. doi: 10.1046/j.1471-4159.2003.01722.x. [DOI] [PubMed] [Google Scholar]
- Eisenbarth GS, Walsh FS, Nirenberg M. Monoclonal antibody to a plasma membrane antigen of neurons. Proc Natl Acad Sci USA. 1979;76:4913–4917. doi: 10.1073/pnas.76.10.4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MA, Afshari FS, Alexander JK, Colello RJ, Fuss B. Growth conelike sensorimotor structures are characteristic features of postmigratory, premyelinating oligodendrocytes. Glia. 2006;53:563–566. doi: 10.1002/glia.20293. [DOI] [PubMed] [Google Scholar]
- Fujita H, Katoh H, Ishikawa Y, Mori K, Negishi M. Rapostlin is a novel effector of Rnd2 GTPase inducing neurite branching. J Biol Chem. 2002;277:45428–45434. doi: 10.1074/jbc.M208090200. [DOI] [PubMed] [Google Scholar]
- Gard AL, Pfeiffer SE. Two proliferative stages of the oligodendrocyte lineage (A2B5+O4− and O4+GalC−) under different mitogenic control. Neuron. 1990;5:615–625. doi: 10.1016/0896-6273(90)90216-3. [DOI] [PubMed] [Google Scholar]
- Higgs HN. Formin proteins: a domain-based approach. Trends Biochem Sci. 2005;30:342–353. doi: 10.1016/j.tibs.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Hu JG, Fu SL, Zhang KH, Li Y, Yin L, Lu PH, Xu XM. Differential gene expression in neural stem cells and oligodendrocyte precursor cells: a cDNA microarray analysis. J Neurosci Res. 2004;78:637–646. doi: 10.1002/jnr.20317. [DOI] [PubMed] [Google Scholar]
- Jurevics H, Morell P. Cholesterol for synthesis of myelin is made locally, not imported into brain. J Neurochem. 1995;64:895–901. doi: 10.1046/j.1471-4159.1995.64020895.x. [DOI] [PubMed] [Google Scholar]
- Kachar B, Behar T, Dubois-Dalcq M. Cell shape and motility of oligodendrocytes cultured without neurons. Cell Tissue Res. 1986;244:27–38. doi: 10.1007/BF00218378. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34:D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessaris N, Fogarty M, Iannarelli P, Grist M, Wegner M, Richardson WD. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat Neurosci. 2006;9:173–179. doi: 10.1038/nn1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khyrul WA, LaLonde DP, Brown MC, Levinson H, Turner CE. The integrin-linked kinase regulates cell morphology and motility in a Rho-associated kinase-dependent manner. J Biol Chem. 2004;279:54131–54139. doi: 10.1074/jbc.M410051200. [DOI] [PubMed] [Google Scholar]
- Kim HJ, DiBernardo AB, Sloane JA, Rasband MN, Solomon D, Kosaras B, Kwak SP, Vartanian TK. WAVE1 is required for oligodendrocyte morphogenesis and normal CNS myelination. J Neurosci. 2006;26:5849–5859. doi: 10.1523/JNEUROSCI.4921-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Leblanc SE, Srinivasan R, Ferri C, Mager GM, Gillian-Daniel AL, Wrabetz L, Svaren J. Regulation of cholesterol/lipid biosynthetic genes by Egr2/Krox20 during peripheral nerve myelination. J Neurochem. 2005;93:737–748. doi: 10.1111/j.1471-4159.2005.03056.x. [DOI] [PubMed] [Google Scholar]
- Levitt P. A monoclonal antibody to limbic system neurons. Science. 1984;223:299–301. doi: 10.1126/science.6199842. [DOI] [PubMed] [Google Scholar]
- Liang X, Draghi NA, Resh MD. Signaling from integrins to Fyn to Rho family GTPases regulates morphologic differentiation of oligodendrocytes. J Neurosci. 2004;24:7140–7149. doi: 10.1523/JNEUROSCI.5319-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maric D, Barker JL. Neural stem cells redefined: a FACS perspective. Mol Neurobiol. 2004;30:49–76. doi: 10.1385/MN:30:1:049. [DOI] [PubMed] [Google Scholar]
- Maric D, Maric I, Chang YH, Barker JL. Prospective cell sorting of embryonic rat neural stem cells and neuronal and glial progenitors reveals selective effects of basic fibroblast growth factor and epidermal growth factor on self-renewal and differentiation. J Neurosci. 2003;23:240–251. doi: 10.1523/JNEUROSCI.23-01-00240.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan R, Le N, Mahoney H, Araki T, Milbrandt J. Deciphering peripheral nerve myelination by using Schwann cell expression profiling. Proc Natl Acad Sci USA. 2002;99:8998–9003. doi: 10.1073/pnas.132080999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterhout DJ, Wolven A, Wolf RM, Resh MD, Chao MV. Morphological differentiation of oligodendrocytes requires activation of Fyn tyrosine kinase. J Cell Biol. 1999;145:1209–1218. doi: 10.1083/jcb.145.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saher G, Brugger B, Lappe-Siefke C, Mobius W, Tozawa R, Wehr MC, Wieland F, Ishibashi S, Nave KA. High cholesterol level is essential for myelin membrane growth. Nat Neurosci. 2005;8:468–475. doi: 10.1038/nn1426. [DOI] [PubMed] [Google Scholar]
- Scarlato M, Beesley J, Pleasure D. Analysis of oligodendroglial differentiation using cDNA arrays. J Neurosci Res. 2000;59:430–435. doi: 10.1002/(SICI)1097-4547(20000201)59:3<430::AID-JNR18>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Sherman DL, Brophy PJ. Mechanisms of axon ensheathment and myelin growth. Nat Rev Neurosci. 2005;6:683–690. doi: 10.1038/nrn1743. [DOI] [PubMed] [Google Scholar]
- Simpson PB, Armstrong RC. Intracellular signals and cytoskeletal elements involved in oligodendrocyte progenitor migration. Glia. 1999;26:22–35. [PubMed] [Google Scholar]
- Sommer I, Schachner M. Monoclonal antibodies (O1 to O4) to oligodendrocyte cell surfaces: an immunocytological study in the central nervous system. Dev Biol. 1981;83:311–327. doi: 10.1016/0012-1606(81)90477-2. [DOI] [PubMed] [Google Scholar]
- Song J, Goetz BD, Baas PW, Duncan ID. Cytoskeletal reorganization during the formation of oligodendrocyte processes and branches. Mol Cell Neurosci. 2001;17:624–636. doi: 10.1006/mcne.2001.0974. [DOI] [PubMed] [Google Scholar]
- Taylor CM, Marta CB, Claycomb RJ, Han DK, Rasband MN, Coetzee T, Pfeiffer SE. Proteomic mapping provides powerful insights into functional myelin biology. Proc Natl Acad Sci USA. 2004;101:4643–4648. doi: 10.1073/pnas.0400922101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheijen MH, Chrast R, Burrola P, Lemke G. Local regulation of fat metabolism in peripheral nerves. Genes Dev. 2003;17:2450–2464. doi: 10.1101/gad.1116203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wennerberg K, Der CJ. Rho-family GTPases: it's not only Rac and Rho (and I like it) J Cell Sci. 2004;117:1301–1312. doi: 10.1242/jcs.01118. [DOI] [PubMed] [Google Scholar]
- Wolf RM, Draghi N, Liang X, Dai C, Uhrbom L, Eklof C, Westermark B, Holland EC, Resh MD. p190RhoGAP can act to inhibit PDGF-induced gliomas in mice: a putative tumor suppressor encoded on human chromosome 19q13.3. Genes Dev. 2003;17:476–487. doi: 10.1101/gad.1040003. [DOI] [PMC free article] [PubMed] [Google Scholar]