Abstract
α-Methylacyl-CoA racemase (AMACR) is a peroxisomal and mitochondrial enzyme involved in the β-oxidation of branched fatty acids, shown to be elevated in prostate cancer by several recent studies. Sequence variants of AMACR have been linked to prostate cancer risk. Although mRNA transcript, protein, and sequence variants of AMACR have been studied in the context of prostate cancer, AMACR enzymatic activity has not been addressed. Here we present evidence that AMACR activity is consistently elevated in prostate cancer tissue specimens. This activity can be immunodepleted from prostate cancer tissue extracts. Furthermore, mock needle biopsy cores containing foci of prostate cancer exhibited increased AMACR enzymatic activity, correlating with both protein levels and histopathology. Taken together, our studies suggest that AMACR activity is increased in prostate cancer relative to benign epithelia and suggests that monitoring AMACR activity levels in prostate needle biopsies may have clinical applications.
High-throughput gene expression profiling of prostate cancers has lead to the discovery of many novel genes associated with cancers. α-Methylacyl-CoA racemase (AMACR) is one such gene, identified by our group and others, which is highly expressed in prostate carcinoma cells as compared with benign or normal prostate epithelial cells.1–6 AMACR is known to be responsible for the conversion of (R)-pristanoyl-CoA and (R)-C27-bile acyl-CoA to their (S)-stereoisomers, which can then be degraded via peroxisomal β-oxidation.7,8 Mutations in the gene encoding AMACR leading to AMACR enzyme deficiency have been linked to adult onset sensory motor neuropathy and pigmentary neuropathy.9 Other clinical conditions implicated with AMACR enzyme deficiency include atypical Refsum’s disease10 and neonatal cholestasis accompanied with fat-soluble vitamin malabsorption.11 Only recently, elevated levels of AMACR transcript and protein have been found to be associated with prostate cancer and the biological significance of this increase with respect to tumorigenesis is unclear. Epidemiological studies suggest red meat and dairy products, which are rich sources of branched chain fatty acids, are associated with increased risk of developing prostate cancer.12,13 Importantly, these branched chain fatty acids are substrates for AMACR.14 Chronically elevated levels of branched chain fatty acid substrates such as pristanic acid in Western diets, potentially may contribute to induction of AMACR. In addition, hydrogen peroxide generated during peroxisomal β-oxidation15 is a likely source of procarcinogenic oxidative damage.16,17 To assess the biochemical significance of increased AMACR levels in prostate cancer, we examined prostate cancer tissues for levels of AMACR enzymatic activity as compared to benign prostatic samples. Based on the formation of [3H] H2O from (R)-[2-3H]-pristanoyl-CoA, as described previously,7,18 we detected elevated levels of AMACR activity in extracts from frozen prostate cancer tissues. The increased AMACR activity associated with prostate cancer suggests a novel biochemical interaction between branched chain fatty acid metabolism and tumorigenesis. Although a mechanistic link between AMACR activity and tumorigenesis is a subject of further investigation, we explored measuring AMACR activity as a potential diagnostic tool. Needle biopsy core tissue extracts harvested from radical prostatectomy samples were examined for AMACR activity. Elevated levels of AMACR activity were specifically detected in needle biopsies containing foci of prostate cancer suggesting that tests designed to measure AMACR enzymatic activity may be useful in the real-time analysis of prostate needle biopsies in the clinic.
Materials and Methods
Procurement of Prostate Tissues and Mock Needle Biopsies
Prostate tissue samples were obtained from the radical prostatectomy performed at the University of Michigan Hospitals with prior approval from the Institutional Review Board. The prostate specimens were transported to the frozen section room located adjacent to the operating rooms and processed within 15 to 20 minutes after surgical resection. Using a previously established protocol, prostate specimens were inked and serially sliced from base to apex after removal of surgical margins.19 Approximately 50% of the tissue was saved for potential research purposes. This protocol has been previously evaluated in a formal study to ensure that partial sampling does not impair accurate staging and pathological evaluation.19 Tissue needle core samples were obtained with an 18-gauge needle biopsy device (Microvasive, Boston, MA) from the fresh sections saved for research by firing the biopsy device parallel to the plane of the section. Needle cores were primarily taken from bilateral peripheral zones to simulate ex vivo prostate biopsy procedure. An average of eight needle cores were taken per prostate. Needle cores were immediately snap-frozen in OCT gel media using isopentane. Hematoxylin and eosin-stained frozen sections of needle cores were evaluated for pathological correlation with AMACR enzyme activity.
Quantitative Real-Time Polymerase Chain Reaction (QRT-PCR)
QRT-PCR was performed using the Applied Biosystems Prism 7700 Sequence Detection System (Applied Biosystems, Foster City, CA). Briefly, 1 μg of total RNA isolated from benign, clinically localized prostate cancer and metastatic prostate samples was reverse-transcribed into first strand cDNA using Superscript II Reverse Transcriptase (Invitrogen, Carlsbad, CA) in the presence of GeneFilter Primer Poly dT and random hexamer primers (Invitrogen) and resuspended in 50 μl of DNase/RNase-free water (Life Technologies, Inc., Grand Island, NY). For each reaction, 0.5 μl of the cDNA product (containing the cDNA from 10 ng of original total RNA), 12.5 μl of 2× SYBR Green PCR Master Mix (Applied Biosystems), 1 μl containing 25 ng of both the forward and reverse primer, and 7.5 μl of DNase/RNase-free water was added to a final volume of 25 μl. Thermocylcling conditions, as suggested by the manufacturer, were 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. All reactions were performed in 96-well optical grade PCR plates (Applied Biosystems). To confirm the absence of nonspecific amplification and primer-dimer binding, a no-template control well was included and amplified products were separated on a 1.5% agarose gel. Threshold levels were set using the SDS v1.7 software (Applied Biosystems) and the quantity of DNA in each sample was calculated by interpolating its Ct value from a standard curve of Ct values obtained from serially diluted cDNA from one of the prostate cancer samples using Microsoft Excel (Microsoft, Redmond, WA). All standard curves had R2 values ≥0.99 over three orders of magnitude. The calculated quantity of AMACR from each sample was then divided by the average calculated quantity of the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) corresponding to each sample to give a relative expression of AMACR for each sample. Levels of another housekeeping gene hydroxymethylbilane synthase were also measured for each sample. Oligonucleotide primers were designed using Primer Select (DNASTAR, Madison, WI) to minimize primer-dimer formation and amplify cDNA products spanning at least one intron-exon junction to eliminate amplification of genomic DNA. Specificity of each primer set was confirmed by comparison to the known sequences in the National Center for Biotechnology Information BLAST database. Primer sequences are available on request.
Laser Capture Microdissection
Expression of AMACR was determined in benign prostate and prostate cancer epithelial cells isolated by laser capture microdissection. Laser capture microdissection was not required to measure AMACR activity in prostate needle biopsies. Briefly, a 6-μm slice was cut with a cryostat from the OCT-embedded tissue block and placed on a special manufactured membrane slide. The slide was then stained with Harris hematoxylin for 50 seconds, followed by two short water washes in distilled water. Eosin staining was done for 30 seconds. Excessive eosin was rinsed off with distilled water and slides were air-dried. The SL Microtest device (MMI, Manchester, NH) using the μCUT software was used for laser capture microdissection. Areas of interest were circled on the monitor under the supervision of a board certified pathologist (RBS), cut by the UV laser, and the dissectates were picked up using the adhesive surface of the lid of specially manufactured tubes (MMI). Three separate cancerous regions and one benign region from the same specimen were cut and placed in separate tubes. Total RNA was isolated from each sample using the Absolutely RNA Microprep Kit (Stratagene, La Jolla, CA) according to the manufacturer’s instructions, vacuum concentrated and reverse transcribed as above. QRT-PCR was performed in duplicate for AMACR and GAPDH expression in each sample and mean AMACR/GAPDH is reported.
Preparation of Recombinant HIS-AMACR Protein
HIS-AMACR plasmid was subcloned from AMACR-MBP fusion construct kindly provided by R.J. Wanders (University of Amsterdam, The Netherlands)9 into pET 28a(+) vector (Novagen, Madison, WI) as an N-terminal histidine-tagged fusion construct. Polyhistidine-tagged AMACR was purified from Escherichia coli by using standard nickel affinity chromatography as per the manufacturer’s instructions (Qiagen, Chatsworth, CA).
AMACR Activity Assay
Cells or tissues to be assayed were homogenized in 10 mmol/L sodium/potassium/phosphate buffer (Na/K/Pi), pH 6.8, 0.1% sodium azide, with a Polytron homogenizer, for 1 minute and the lysates were then sonicated (1 minute, microtip, 70% output). After centrifugation at 13,000 × g for 5 minutes to remove the cellular debris, the total protein content of the lysates was determined by the dye-binding method of Bradford20 using the Bio-Rad Protein Assay kit (Bio-Rad, Richmond, CA). Five μg of each lysate was used for the AMACR activity as previously described,7,18 with minor modifications. Briefly, the lysates were incubated with 100 μmol/L [2-3H]-pristanoyl-CoA (10,000 cpm), in 50 mmol/L Tris/HCl, pH 8.0, in 50 μl volume, at 37°C for 30 minutes. The reaction was stopped with 450 μl of 1% trichloroacetic acid and the whole reaction mixture was applied onto a reverse-phase silica gel (RP-18) column (Merck KGaA, Darmstadt, Germany). [3H]-H2O obtained, was quantified by liquid scintillation counting.
Immunoblot Analyses
Extracts prepared from frozen tissues or needle biopsy samples were separated on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The proteins were blotted onto nitrocellulose membranes (Amersham Biosciences, Arlington Heights, IL) and blocked for 4 hours in Tris-buffered saline containing 5% nonfat milk and 0.1% Tween-20 at room temperature. The membranes were then incubated with antibodies directed against human AMACR9,18 or, β-tubulin (NeoMarkers, Fremont, CA) at 4°C overnight. Membranes were washed with Tris-buffered saline containing 0.1% Tween-20 and incubated with horseradish peroxidase-conjugated secondary antibody (Amersham Biosciences) for 1 hour at room temperature. After washing the membrane with Tris-buffered saline containing 0.1% Tween-20, signals were visualized by the enhanced chemiluminescence system (Amersham Biosciences).
Immunodepletion
Tissues were lysed in 10 mmol/L sodium/potassium/phosphate buffer (Na/K/Pi), pH 6.8, 0.1% sodium azide and 5 μg each was incubated with either human AMACR antiserum18 or normal rabbit serum in a total volume of 400 μl of Na/K/Pi buffer and rotated end-over-end at 4°C for 60 minutes. Protein G-Sepharose beads (Amersham Biosciences) equilibrated in Na/K/Pi buffer were added and the reactions were incubated for a further 60 minutes. The lysates were separated from protein G-Sepharose antibody complex using a 301/2-gauge needle. Two more rounds of immunodepletions were performed. Immunodepleted lysates were assayed for AMACR activity as described in the previous section.
Results and Discussion
AMACR mRNA Transcript Is Overexpressed in Laser Capture Microdissected Prostate Cancer Epithelial Cells
AMACR is both highly specific and sensitive to prostate cancer, as reported earlier by DNA microarray analysis,2–5 and as noted here by quantitative RT-PCR of RNA isolated from grossly dissected prostate tissues (Figure 1A). To confirm that the increased AMACR mRNA transcript levels was because of increased expression in prostatic epithelia rather than stroma we performed laser capture microdissection and similar results were obtained (Figure 1B).
Figure 1.
AMACR mRNA transcript expression in prostatic cancer. A: Quantitative real-time PCR analysis of AMACR transcript levels in grossly dissected prostate tissues. The estimated quantity of AMACR divided by the average quantity of GAPDH was used to calculate the relative expression of AMACR (y axis) for each sample (x axis). An average of the three classes [benign, localized prostate cancer (PCA), and metastatic prostate cancer (MET)] is provided in the inset. B: Laser capture microscopic dissection of prostate epithelial cells followed by QRT-PCR analysis. Schematic of laser capture microdissection of prostate tissue (left) followed by quantitation of AMACR mRNA expression (right) as described in A.
AMACR Enzymatic Activity Is Elevated in Prostate Cancer
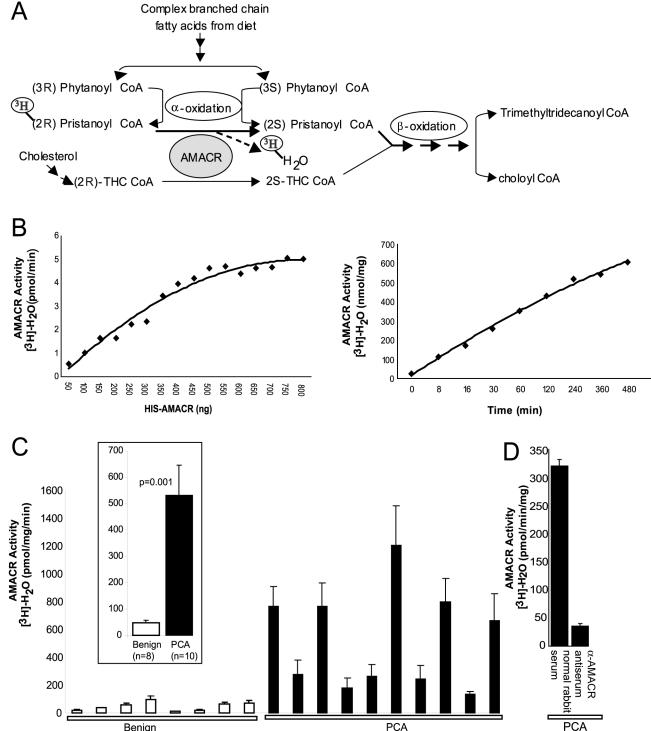
To assess if the increased levels of AMACR in prostate cancer tissues result in increased enzymatic activity, we made use of an AMACR activity assay reported earlier.7,18 This assay is based on the conversion of (R)-stereoisomer of [2-3H]-pristanoyl-CoA to the (S)-stereoisomer, facilitating its degradation by β-oxidation, resulting in release of [3H]-H2O (Figure 2A). The [3H]-H2O is recovered from the reaction mixture by passing through a reverse-phase silica gel column and quantified by liquid scintillation counting, thus providing a measure of AMACR activity. Before examining prostate cancer samples, we standardized the assay conditions and assessed the dynamic range of the assay using bacterially expressed recombinant HIS-AMACR protein incubated with the radiolabeled substrate. As seen in Figure 2B, the relationship between the enzyme concentration and the enzyme activity is linear throughout a range of concentrations of recombinant AMACR. This suggests that a dynamic range of detection of the enzyme activity is available for the AMACR assay. In addition, the magnitude of activity, as assessed throughout a period of time, is linear as well (Figure 2B, right). This is likely to be useful in increasing the sensitivity of the assay, involving low amounts of the protein.
Figure 2.
AMACR enzymatic activity in prostate cancer. A: Schematic representation of the role of AMACR in facilitating β-oxidation of branched chain fatty acids by converting (R)-stereoisomer of pristanoyl-CoA and THC-CoA to their respective (S)-stereoisomers. The schematic also depicts the rationale of AMACR activity assay involving breakdown of [3H]-(2R)-pristanoyl CoA leading to release of [3H]-H2O. B: Standardization of the AMACR activity assay using recombinant AMACR. Enzymatic activity of AMΑCR expressed as the amount of [3H]-H2O generated per unit time, on incubation of 100 μmol/L substrate, [2-3H] pristanoyl-CoA, with increasing amounts of purified HIS-AMACR protein, for 30 minutes (left). Enzymatic activity of AMACR expressed as the amount of [3H]-H2O generated per mg of enzyme, on incubation of 100 μmol/L substrate, [2-3H] pristanoyl-CoA, with 250 ng of purified HIS-AMACR protein, throughout a time course (right). C: AMACR enzymatic activity in extracts from frozen tissue samples of grossly dissected benign or clinically localized prostate cancers (PCA). Tissue extracts were incubated with 100 μmol/L substrate, [2-3H] pristanoyl-CoA, expressed as the amount of [3H]-H2O generated per unit time, per unit total protein concentration. The values shown are mean ± SEM of three independent experiments. The inset represents a consolidated plot of the values for benign and PCA samples. D: Specific immunodepletion of AMACR enzymatic activity from prostate cancer extracts. The values shown are mean ± SEM of three independent assays.
With the assay parameters in place and standardized, we next examined AMACR activity profile in a panel of benign and localized prostate cancer tissue extracts. The activity profile, as shown in Figure 2C, clearly demonstrates increased levels of activity in the prostate cancer samples as compared to the benign tissue. To confirm that these observations are specifically a result of AMACR activity, a prostate cancer tissue lysate was assayed for activity after being immunodepleted with either α-AMACR antiserum or normal rabbit serum. AMACR activity was attenuated on immunodepletion with α-AMACR antiserum (Figure 2D), but not with normal rabbit serum, confirming that the assay is indeed specific to AMACR activity.
AMACR Activity Is Elevated in Prostate Needle Biopsies Containing Foci of Prostate Cancer
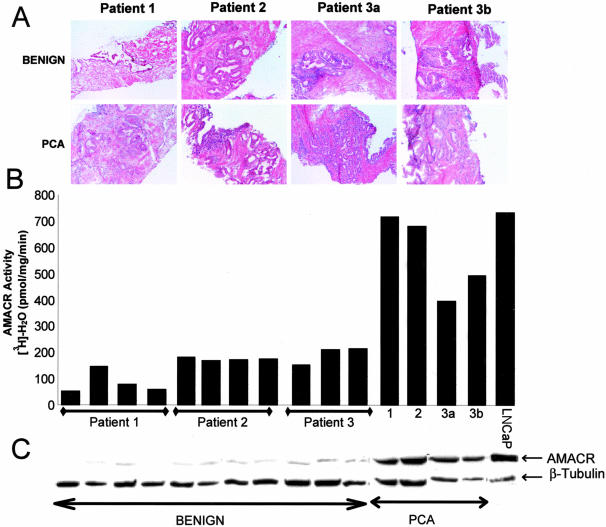
Needle biopsy cores obtained from three prostatectomies, that were either positive or negative for prostate cancer were used for histopathological assessment and AMACR assays as shown in Figure 3. Based on histopathology, a total of four needle cores clearly demonstrated prostate cancer and 14 demonstrated only benign tissue (representative data shown) (Figure 3A). Extracts derived from the needle biopsies were subjected to the AMACR activity assay. Similar to the LNCaP prostate cancer cell line, prostate needle biopsies that contained foci of prostate cancer had high AMACR activity levels (Figure 3B). Much lower AMACR activity levels were seen in prostate needle biopsies containing only benign or high-grade prostatic intraepithelial cells (data not shown) and AMACR protein levels correlated well with AMACR activity levels in matched extracts (Figure 3C). To further validate our results we analyzed 50 ex vivo needle biopsies for AMACR activity levels. The prostate needle biopsies that contained foci of cancer exhibited on average 6.6-fold more AMACR activity than benign needle biopsies. Importantly, 12 of 13 prostate needle biopsies that contained foci of cancer had high AMACR activity whereas only 4 of 37 benign needle biopsies had high AMACR activity. Thus, in this testing cohort of prostate needle biopsies, the AMACR activity test exhibited 92.3% sensitivity and 89.2% specificity.
Figure 3.
Correlation of histopathology, AMACR enzymatic activity and AMACR protein levels in prostate needle biopsies. A: H&E-stained frozen sections of ex vivo prostate needle biopsies performed on three patients. Both benign and prostate cancer biopsies were used from each patient. B: AMACR enzymatic activity of matched tissue extracts from biopsies shown in A. See Figure 2 for assay details. LnCaP is a prostate cancer cell line used as a positive control. Patient 3a and 3b represent tissue from the same patient but different areas of the prostate. C: Matched immunoblot analysis of prostate needle biopsy extracts measuring AMACR protein levels. Blots were probed with α-AMACR antiserum. Human β-tubulin is used as loading control.
The advent of DNA microarrays and tissue microarrays has led to the identification of numerous prostate cancer-associated genes and proteins.1,21,22 Using the above approach our group and others identified and validated Hepsin,1,21–23 AMACR,2,3,5 and EZH2,24 as potential biomarkers for prostate cancer. Among these, AMACR has been shown to be potentially useful in assessing prostate cancer in the context of needle biopsies.2–5 Several studies have demonstrated the remarkable specificity of AMACR protein in prostate cancer epithelia relative to benign epithelia. In this study, we demonstrated that AMACR enzymatic activity is up-regulated in prostate cancer. Future studies will address the role of AMACR activity and the β-oxidation of branched fatty acids in prostate carcinogenesis. This study also suggests that further epidemiological studies measuring levels of AMACR substrates such as diastereoisomers of pristanic acid, phytol, phytanic acid, and the bile acid intermediates di- and trihydroxycholestanoic acid are warranted.
In addition to demonstrating AMACR activity in prostate cancer, we demonstrated that AMACR activity could be measured in prostate needle biopsies. Elevated levels of AMACR activity were specifically detected in needle biopsies containing foci of prostate cancer suggesting that an assay designed to measure AMACR enzymatic activity may be useful as a rapid diagnostic test for prostate cancer at the time of a prostate needle biopsy. Although the radioactivity-based AMACR assay used in this study is not conducive to clinical implementation, a colorimetric assay25 could be developed. Such an assay could instantaneously measure AMACR activity in prostate needle biopsies so that fewer needle cores may be necessary for the detection of prostate cancer.
Acknowledgments
We thank Dr. Atreya Dash for technical help with ex vivo needle biopsy; Daniel Rhodes for input on statistical analysis; and Dr. R. J. Wanders, University of Amsterdam, The Netherlands, for providing us the AMACR-MBP clone.
Footnotes
Address reprint requests to Arul M. Chinnaiyan, M.D., Ph.D., Department of Pathology, University of Michigan Medical School, 1301 Catherine Rd., MSI Rm. 4237, Ann Arbor, MI 48109-0602. E-mail: arul@umich.edu.
Supported in part by the American Cancer Society (RSG-02-179-MGO to A.M.C. and M.A.R.), the National Institutes of Health (Prostate SPORE P50CA69568 to A.M.C., M.A.R., R.B.S., J.T.W.; R01AG21404 to A.M.C. and M.A.R.), R01 CA8241901-A1 (to M.G.S), and the V Foundation (to A.M.C.).
A.M.C. is a Pew Biomedical Scholar and S.A.T. is a Fellow of the Medical Scientist Training Program.
References
- Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K, Pienta KJ, Rubin MA, Chinnaiyan AM. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412:822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Woda BA, Rock KL, Xu Y, Savas L, Khan A, Pihan G, Cai F, Babcook JS, Rathanaswami P, Reed SG, Xu J, Fanger GR. P504S: a new molecular marker for the detection of prostate carcinoma. Am J Surg Pathol. 2001;25:1397–1404. doi: 10.1097/00000478-200111000-00007. [DOI] [PubMed] [Google Scholar]
- Luo J, Zha S, Gage WR, Dunn TA, Hicks JL, Bennett CJ, Ewing CM, Platz EA, Ferdinandusse S, Wanders RJ, Trent JM, Isaacs WB, De Marzo AM. Alpha-methylacyl-CoA racemase: a new molecular marker for prostate cancer. Cancer Res. 2002;62:2220–2226. [PubMed] [Google Scholar]
- Kuefer R, Varambally S, Zhou M, Lucas PC, Loeffler M, Wolter H, Mattfeldt T, Hautmann RE, Gschwend JE, Barrette TR, Dunn RL, Chinnaiyan AM, Rubin MA. Alpha-methylacyl-CoA racemase: expression levels of this novel cancer biomarker depend on tumor differentiation. Am J Pathol. 2002;161:841–848. doi: 10.1016/s0002-9440(10)64244-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin MA, Zhou M, Dhanasekaran SM, Varambally S, Barrette TR, Sanda MG, Pienta KJ, Ghosh D, Chinnaiyan AM. Alpha-methylacyl coenzyme A racemase as a tissue biomarker for prostate cancer. JAMA. 2002;287:1662–1670. doi: 10.1001/jama.287.13.1662. [DOI] [PubMed] [Google Scholar]
- Yang XJ, Wu CL, Woda BA, Dresser K, Tretiakova M, Fanger GR, Jiang Z. Expression of alpha-methylacyl-CoA racemase (P504S) in atypical adenomatous hyperplasia of the prostate. Am J Surg Pathol. 2002;26:921–925. doi: 10.1097/00000478-200207000-00011. [DOI] [PubMed] [Google Scholar]
- Schmitz W, Fingerhut R, Conzelmann E. Purification and properties of an alpha-methylacyl-CoA racemase from rat liver. Eur J Biochem. 1994;222:313–323. doi: 10.1111/j.1432-1033.1994.tb18870.x. [DOI] [PubMed] [Google Scholar]
- Cuebas DA, Phillips C, Schmitz W, Conzelmann E, Novikov DK. The role of alpha-methylacyl-CoA racemase in bile acid synthesis. Biochem J. 2002;363:801–807. doi: 10.1042/0264-6021:3630801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferdinandusse S, Denis S, Clayton PT, Graham A, Rees JE, Allen JT, McLean BN, Brown AY, Vreken P, Waterham HR, Wanders RJ. Mutations in the gene encoding peroxisomal alpha-methylacyl-CoA racemase cause adult-onset sensory motor neuropathy. Nat Genet. 2000;24:188–191. doi: 10.1038/72861. [DOI] [PubMed] [Google Scholar]
- Wierzbicki AS, Lloyd MD, Schofield CJ, Feher MD, Gibberd FB. Refsum’s disease: a peroxisomal disorder affecting phytanic acid alpha-oxidation. J Neurochem. 2002;80:727–735. doi: 10.1046/j.0022-3042.2002.00766.x. [DOI] [PubMed] [Google Scholar]
- Setchell KD, Heubi JE, Bove KE, O’Connell NC, Brewsaugh T, Steinberg SJ, Moser A, Squires RH., Jr Liver disease caused by failure to racemize trihydroxycholestanoic acid: gene mutation and effect of bile acid therapy. Gastroenterology. 2003;124:217–232. doi: 10.1053/gast.2003.50017. [DOI] [PubMed] [Google Scholar]
- Giovannucci E, Rimm EB, Colditz GA, Stampfer MJ, Ascherio A, Chute CC, Willett WC. A prospective study of dietary fat and risk of prostate cancer. J Natl Cancer Inst. 1993;85:1571–1579. doi: 10.1093/jnci/85.19.1571. [DOI] [PubMed] [Google Scholar]
- Kushi L, Giovannucci E. Dietary fat and cancer. Am J Med. 2002;113(Suppl 9B):63S–70S. doi: 10.1016/s0002-9343(01)00994-9. [DOI] [PubMed] [Google Scholar]
- Wanders RJ, Jacobs C, Skjeldal OH. London: McGraw Hill,; The Metabolic and Molecular Bases of Inherited Disease. 2001:pp 3303–3321. [Google Scholar]
- Wanders RJ, Vreken P, Ferdinandusse S, Jansen GA, Waterham HR, van Roermund CW, Van Grunsven EG. Peroxisomal fatty acid alpha- and beta-oxidation in humans: enzymology, peroxisomal metabolite transporters and peroxisomal diseases. Biochem Soc Trans. 2001;29:250–267. doi: 10.1042/0300-5127:0290250. [DOI] [PubMed] [Google Scholar]
- Tamatani T, Hattori K, Nakashiro K, Hayashi Y, Wu S, Klumpp D, Reddy JK, Oyasu R. Neoplastic conversion of human urothelial cells in vitro by overexpression of H2O2-generating peroxisomal fatty acyl CoA oxidase. Int J Oncol. 1999;15:743–749. doi: 10.3892/ijo.15.4.743. [DOI] [PubMed] [Google Scholar]
- Ockner RK, Kaikaus RM, Bass NM. Fatty-acid metabolism and the pathogenesis of hepatocellular carcinoma: review and hypothesis. Hepatology. 1993;18:669–676. [PubMed] [Google Scholar]
- Schmitz W, Albers C, Fingerhut R, Conzelmann E. Purification and characterization of an alpha-methylacyl-CoA racemase from human liver. Eur J Biochem. 1995;231:815–822. doi: 10.1111/j.1432-1033.1995.tb20766.x. [DOI] [PubMed] [Google Scholar]
- Hollenbeck BK, Bassily N, Wei JT, Montie JE, Hayasaka S, Taylor JM, Rubin MA. Whole mounted radical prostatectomy specimens do not increase detection of adverse pathological features. J Urol. 2000;164:1583–1586. [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Welsh JB, Sapinoso LM, Su AI, Kern SG, Wang-Rodriguez J, Moskaluk CA, Frierson HF, Jr, Hampton GM. Analysis of gene expression identifies candidate markers and pharmacological targets in prostate cancer. Cancer Res. 2001;61:5974–5978. [PubMed] [Google Scholar]
- Luo J, Duggan DJ, Chen Y, Sauvageot J, Ewing CM, Bittner ML, Trent JM, Isaacs WB. Human prostate cancer and benign prostatic hyperplasia: molecular dissection by gene expression profiling. Cancer Res. 2001;61:4683–4688. [PubMed] [Google Scholar]
- Magee JA, Araki T, Patil S, Ehrig T, True L, Humphrey PA, Catalona WJ, Watson MA, Milbrandt J. Expression profiling reveals hepsin overexpression in prostate cancer. Cancer Res. 2001;61:5692–5696. [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Van Veldhoven PP, Croes K, Casteels M, Mannaerts GP. 2-Methylacyl racemase: a coupled assay based on the use of pristanoyl-CoA oxidase/peroxidase and reinvestigation of its subcellular distribution in rat and human liver. Biochim Biophys Acta. 1997;1347:62–68. doi: 10.1016/s0005-2760(97)00053-2. [DOI] [PubMed] [Google Scholar]