Abstract
Despite years of discovery and attempts at validation, few molecular biomarkers achieve acceptance in the clinical setting. Tissue-based markers evaluated by immunohistochemistry suffer from a high degree of inter- and intraobserver variability. One recent advance in this field that promises to automate this process is the development of AQUA, a molecular-based method of quantitative assessment of protein expression. This system integrates a set of algorithms that allows for the rapid, automated, continuous, and quantitative analysis of tissue samples, including the separation of tumor from stromal elements and the subcellular localization of signals. This study uses the AQUA system to assess a recently described prostate cancer biomarker, α-methylacyl-CoA-racemase (AMACR), and to determine the effectiveness of the quantitative measurement of this marker as a means for making the diagnosis of prostate cancer. Using a prostate cancer progression tissue microarray containing a wide range of prostate tissues, AQUA was directly compared to standard immunohistochemical evaluation for AMACR protein expression using the p504s monoclonal antibody. Both methods produced similar results showing AMACR protein expression to be strongest in the clinically localized prostate cancer, followed by the metastatic tumor samples. Benign prostate tissue was categorized as negative for most tissue samples by immunohistochemistry. However, AMACR was detectable using the AQUA system at low levels using the standard 1:25 dilution but also at 1:250 dilution, which is not detectable by light microscopy. The AQUA system was also able to discriminate foamy gland prostate cancers, which are known to have a lower AMACR expression than typical acinar prostate cancers, from benign prostate tissue samples. Finally, a receiver-operating-characteristic curve was plotted to determine the specificity of the AMACR AQUA Z-score (normalized AQUA score) to predict that a given tissue microarray sample contains cancer. The area under the curve was calculated at 0.90 (P < 0.00001; 95% CI, 0.84 to 0.95). At an AMACR AQUA Z-score score of −0.3, 91% of the 70 samples classified as prostate cancer were correctly categorized without the intervention of a pathologist reviewing the tissue microarray slide. In conclusion, the AQUA system provides a continuous measurement of AMACR on a wide range of prostate tissue samples. In the future, the AMACR AQUA Z-score may be useful in the automated screening and evaluation of prostate tissue biomarkers.
Despite years of discovery and attempts at validation, few molecular biomarkers have achieved acceptance in the clinical setting. Often exciting initial reports fail confirmation when tested in other laboratories. Many factors are involved in the standardization and adaptation of putative biomarkers. Even Federal Drug Administration-approved tests, such as the Herceptin test, vary so greatly as to question the reproducibility of the test outside of high-volume reference laboratories.1,2 Tissue-based biomarkers evaluated by immunohistochemistry can be quantified. However, one important limitation with standard immunohistochemistry is that low antibody concentrations lack sensitivity at the low end of protein expression and high antibody concentrations fail to distinguish protein expression at the mid to high levels of expression because of saturation, higher background, and nonspecific staining. These inherent shortcomings of standard immunohistochemistry relegate evaluation to subjective nominal categories, which may demonstrate poor inter- and intraobserver agreement. One general concern is that excellent biomarkers may and have been dismissed prematurely because of limitation in tissue-based evaluations. The cell-cycle inhibitor, p27(kip1), is an example of a promising prostate cancer biomarker that has never gained a foothold in the clinical setting.3–5
This dilemma becomes more urgent with the advent of high-throughput technologies that identify hundreds of dysregulated genes but offer no clear strategies to advance our understanding of these genes as tissue markers. The analysis is limited by standard pathology evaluation, creating a biomarker bottleneck on the microscope stage of academic pathologists. Our group and others have extensively used tissue microarrays (TMAs) to screen large numbers of gene products with the goal of characterizing novel cancer biomarkers. Yet the rate-limiting factor remains a pathologist’s ability to review, interpret, and subjectively qualify the staining intensity of the tissue samples using standard immunohistochemistry.
One recent advance in this field that promises to automate this process is the development of the AQUA system.6,7 The AQUA system integrates a set of algorithms that allows for the rapid, automated, continuous, and quantitative analysis of TMAs, including the separation of tumor from stromal elements and the subcellular localization of signals. Proof of principle experiments validated this approach using estrogen receptor and Her2-neu analysis in breast carcinoma and showed that this technology was able to discover relationships between expression and outcome that were impossible for conventional pathologist-based immunohistochemical methods.6,7 Automated analysis and subcellular localization of β-catenin in colon cancer identified two novel, prognostically significant tumor subsets, not detected by traditional pathologist-based scoring.6 From the perspective of screening novel biomarkers the AQUA system promises to make the process more objective and quantitative.
In the field of prostate cancer biomarker development, we believed that the AQUA system could be used to help screen for new prognostic and predictive biomarkers. Therefore the goal of the current study was to characterize one recently identified prostate cancer biomarker, α-methylacyl-Co-A racemase (AMACR). As previously described by several groups, AMACR is expressed in prostate cancer and high-grade prostatic intraepithelial neoplasia (PIN).8–17 However, this sensitive prostate cancer biomarker is variably expressed in some clinically localized prostate cancers and metastatic tumors.10,13 Therefore one of the challenges in using AMACR in the detection of prostate cancer is the variability of expression. The current study examines AMACR expression using the AQUA system on a prostate cancer progression TMA with the goal of determining AMACR protein expression throughout a continuous range in a wide variety of prostate samples.
Materials and Methods
Case Selection
To test the feasibility of using the AQUA system in combination with AMACR to quantify prostate cancer biomarkers, we developed a prostate cancer progression TMA. This TMA is composed of benign prostate tissue, localized prostate cancer, and hormone naïve and hormone-refractory metastatic prostate cancer (Figure 1A). These cases came from well-fixed radical prostatectomy specimens from the University of Michigan (Ann Arbor, MI), the University Hospital Ulm (Ulm, Germany), and the rapid autopsy program from the University of Michigan Specialized Program of Research Excellence in Prostate Cancer.18 All samples were collected with prior institutional review board approval at each respective institution. A second focused TMA was created to test foamy gland tumors, which have been previously reported to have a significantly lower expression of AMACR when compared to clinically localized prostate cancers.19 This TMA was composed of classic acinar prostate cancers and areas demonstrating foamy gland features from the same cases. Benign tissue samples were also placed in the TMA to serve as a negative control.
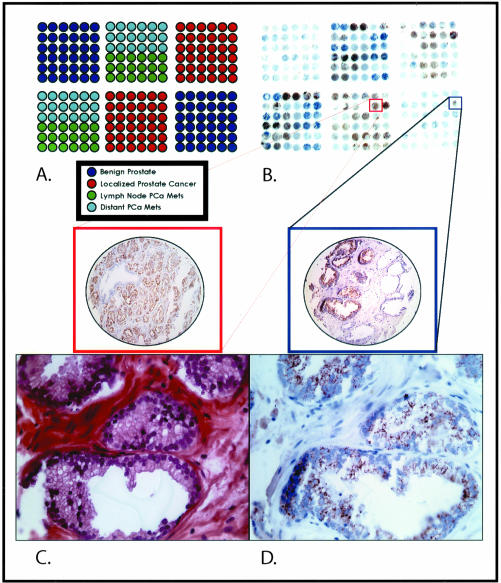
Figure 1.
A–D: Prostate cancer progression chip to measure AMACR expression in benign and neoplastic prostate tissue samples. A prostate cancer progression tissue microarray was designed composed of benign prostate sectors (dark blue), clinically localized prostate cancer sectors (red), regional lymph node metastatic prostate tumors (green), and distant hormone-refractory metastatic prostate tumors (light blue). A: AMACR expression can be seen at the macroscopic level with strong brown expression seen in the prostate cancer sectors. Rare focal expression can be appreciated in the benign section (blue square and TMA sample) and at a similar intensity as seen in an example of clinically localized prostate cancer (red square and TMA sample). C: This sample demonstrates high-grade PIN, a precursor lesion to prostate cancer. D: This area of high-grade PIN demonstrates moderate levels of AMACR expression. Original magnifications: ×200 (A, B); ×630 (C, D).
Immunohistochemistry for AMACR
Pretreatment conditions and incubations have already been worked out for AMACR immunostaining using the commercially available monoclonal antibody directed against AMACR (p504s; Zeta Corp., Sierra Madre, CA). The TMA was soaked in xylene overnight to remove adhesive tape used for construction of the TMA. Pretreatment included placing the slide in a 6.0- pH citrate buffer and microwaving for 30 minutes. Primary p504s antibody was incubated for 40 minutes at room temperature. Secondary anti-mouse antibodies were applied for 30 minutes and the enzymatic reaction was completed using a streptavidin biotin detection kit (DAKO Developing System; DAKO, Carpinteria, CA). The slides were developed using diaminobenzidine for 5 minutes. Optimal primary antibody concentration was determined by serial dilutions, optimizing for maximal signal without background immunostaining. After extensive testing, a concentration of 1:25 dilution was determined to be optimal under these conditions.
Scoring of AMACR Protein Expression
AMACR protein expression was determined using a validated scoring method,10,13,20–22 in which staining was evaluated for intensity. Benign epithelial glands and prostate cancer cells were scored for AMACR staining intensity on a four-tiered system ranging from negative to strong expression.
AQUA System Analysis of AMACR
AMACR expression was determined using the AQUA system by first localizing epithelial cells using a fluorescently tagged anti-cytokeratin antibody cocktail (AE1/AE3, DAKO). 4,6-Diamidino-2-phenylindole (DAPI) was added to visualize nuclei. AMACR was visualized with a fluorescent chromogen (Cy-5-tyramide; NEN Life Science Products, Boston, MA) which, like diaminobenzidine, is activated by horseradish peroxidase and results in the deposition of numerous covalently associated Cy-5 dyes immediately adjacent to the horseradish peroxidase-conjugated secondary antibody. As previously described, Cy-5 (red) was used because its emission peak is well outside the green-orange spectrum of tissue autofluorescence.6,7 Using this approach, classical compartments can be identified on the basis of molecular co-localization. The cytokeratin compartment is equivalent to all epithelial cells in the 0.6-mm diameter TMA spot. DAPI is the area defined as the cell nucleus. The AMACR-positive compartment is consistent with prostate cancer or high-grade PIN. This approach is presented in schematic form in Figure 2. A complete and detailed description of the staining process and the exact methods of the system used for analysis (AQUA) are available in the article and supplemental material from Camp and colleagues.6
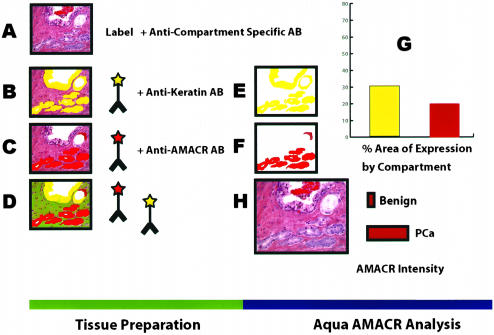
Figure 2.
A–H: Schematic view for using AQUA to measure AMACR in prostate samples. A: Prostate cancers are heterogeneous with a mixture of benign and infiltrating cancer cells. B: Using a single antibody against keratin, the AQUA algorithm can measure the intensity and area of keratin-positive cells using fluorescent dyes. This area is shown in yellow. C: A second antibody against AMACR (p504s) is used to target neoplastic prostate glands. This area is shown in red. D: The fluorescent dyes allow for the simultaneous evaluation of up to four antibodies. In this example AMACR and keratin are shown. The area for each marker is measured by the system (E and F) and can be considered a virtual compartment representing a keratin-positive area and AMACR-positive area (G). Dual expression of keratin and AMACR define a neoplastic compartment (ie, prostate cancer or prostatic intraepithelial neoplasia). Keratin-positive and AMACR-negative areas are consistent with benign epithelial compartment. Negative staining for both AMACR and keratin are consistent with a stromal compartment. Using these definitions, the intensities of different compartments can be measured using arbitrary units (H).
Automated Image Acquisition and Analysis
Automated image acquisition and analysis using AQUA has been previously described.6,7 In brief, multiple, monochromatic, high-resolution (1024 × 1024 pixels, 0.5-μm resolution) images were obtained of each TMA spot using an Olympus AX-51 epifluorescence microscope (Olympus, Melville, NY) set up with a series of user modifications including an automated Prior microscope stage and both video and digital image (Roper Cool Snap) acquisition driven by custom program and macro-based interfaces with IPLabs (Scanalytics Inc., Fairfax, VA) software, through a 10× Plan Apo objective. As described above, areas of epithelial cells were distinguished from stromal elements by creating a mask from the cytokeratin signal (Figure 2). To obtain a final normalized score for AMACR, theAMACR signal intensity from pixels within the cytokeratin mask were measured on a scale of 0 to 255, and expressed as signal intensity divided by the cytokeratin pixel area. The resultant score has three significant figures and is directly proportional to the number of molecules per unit area, however that conversion is not yet possible for AMACR.
RESA/PLACE Algorithmic Analysis of Images
This algorithm has been previously described.6,7 First, a tumor-specific mask is generated by thresholding the image of a marker that differentiates tumor from surrounding stroma and/or leukocytes. This creates a binary mask (each pixel is either on or off). In this study we used cytokeratin and AMACR to create tumor masks. Because formalin-fixed tissues can exhibit autofluorescence, analysis may give multiple background peaks. The RESA/PLACE algorithms determine which of these peaks is predominant and sets a binary mask threshold at a slightly higher intensity level. This provides an adaptive (unique to each TMA sample) thresholding system that ensures that only the target signal from the tumor and not the surrounding elements is analyzed. Thresholding levels were verified by spot-checking a few images and then automated for the remaining images. This binary mask can be modified using standard image manipulations. In most cases this involves filling holes of a particular size (eg, less than 500 pixels, to fill in tumor nuclei that do not stain for either cytokeratin or AMACR) and removing extraneous single pixels. Once set, these image manipulations are performed automatically on all images. All subsequent image manipulations involve only image information from the masked area.
Next, two images (one in-focus, one slightly deeper) are taken of the compartment-specific tags and the target marker. A percentage of the out-of-focus image is subtracted from the in-focus image, based on a pixel-by-pixel analysis of the two images. This percentage is determined according to the ratio of the highest/lowest intensity pixels in the in-focus image—representing the signal to noise ratio of the image. By using an exponential scale, this allows RESA to subtract low-intensity pixels in images with a low signal/noise ratio less heavily than low-intensity pixels from images with a high signal/noise ratio. The overall degree of subtraction is based on a user-defined percentage for each subcellular compartment. For most applications this is empirically set to 40% of the total signal, and remains constant for images from an entire microarray. RESA thus eliminates all out-of-focus information. The algorithm has the added benefit of enhancing the interface between areas of higher intensity staining and adjacent areas of lower intensity staining, allowing more accurate assignment of pixels of adjacent compartments. In contrast to the compartment-specific tags, the RESA subtraction of the target signal is uniform and not based on overall intensity of the image intensity. This ensures that the same amount of subtraction occurs with the target signal from all specimens.
Finally, the PLACE algorithm assigns each pixel in the image to a specific subcellular compartment. Pixels that cannot be accurately assigned to a compartment to within a user-defined degree of confidence (usually 95%) are discarded. This is accomplished by iteratively determining the ratio of signal from two compartment-specific markers that minimizes the spillover of marker from one compartment into another. Pixels in which the nuclear and membrane pixel intensities are too similar to be accurately assigned are negated (usually comprising <8% of the total pixels). A third compartment (the cytoplasm) can be defined by exclusion (nonmembrane, nonnuclear). Once each pixel is assigned to a subcellular compartment (or excluded as described above), the signal in each location is added up. The data are saved and can subsequently be expressed either as a percentage of total signal or as the average signal intensity per compartment area. The score is expressed on a scale of 1 to 1000 because the total intensity detectable in a pixel ranges from 1 to 255 creating three significant figures. These algorithms are described in a recently submitted patent of this technology owned by Yale University. In this study, for AMACR only cytoplasmic-localized signal was used over a cytokeratin-positive compartment.
Statistical Analysis
AMACR protein expression was evaluated between groups using analysis of variance. To account for multiple associations between tissue categories, which might tend to favor a significant association because of chance, posthoc analysis according to the Scheffé method was performed as previously used.13 The AMACR protein expression data were presented graphically using error bars with 95% confidence intervals. For the analysis of the foamy gland tumors, the data needed to be normalized to compare AQUA scores from two separate experiments. A Z-score was calculated to compare the arbitrary AQUA units from one study to the next. This approach has been previously validated using cDNA expression array data sets.23 The Z-score was defined as follows: [(AQUA score) − (mean score of all AQUA scores on TMA X)]/standard deviation, where X is a given TMA experiment. Using this approach it is possible to compare results from different experiments. After normalization, AMACR expression was compared between foamy gland tumors and other tissue types. A receiver-operating-characteristic (ROC) curve was then created to plot the sensitivity versus 1 minus the specificity of the AQUA Z-score to predict that a given TMA sample contains cancer. The area under the curve was calculated using a nonparametric method. An area under the curve of 1.0 indicates a test with perfect discrimination between TMA samples with and those without prostate cancer. An area under the curve of 0.5 indicates a test with no discriminatory power. All statistical analyses were performed using a commercially available software package (SPSS 11.1; SPSS Inc., Chicago, IL).
Results
AMACR Expression Determined by the Study Pathologist Using Immunohistochemistry
The expression of AMACR (p504s) was determined using a prostate cancer progression TMA (Figure 1). Expression was scored as negative (score = 1), weak (score = 2), moderate (score = 3), and strong (score = 4). The results for each tissue type are presented as error bars of AMACR (p504s) protein expression with 95% confidence intervals (Figure 3D). AMACR demonstrated moderate to strong cytoplasmic protein expression by standard immunohistochemistry in clinically localized prostate cancer with a mean expression score of 3.14 (SE, 0.1; 95% CI, 2.91 to 3.36). By using the prostate progression TMA, more variable expression was seen in the metastatic prostate cancers (Table 1). AMACR protein expression using standard immunohistochemistry demonstrated a significant difference in staining intensity between localized prostate cancer and benign prostate tissue with mean staining intensities of 3.14 and 1.3, respectively (mean difference, 1.84; analysis of variance posthoc Scheffé analysis, P < 0.00001). Expression of AMACR was significantly higher for both hormone-sensitive and hormone-refractory prostate cancer, but the mean expression was lower than for clinically localized prostate cancer. These results are consistent with previous observations.10,13 No differences were detected in AMACR protein expression based on tumor Gleason score, consistent with previous observations.12–14,16
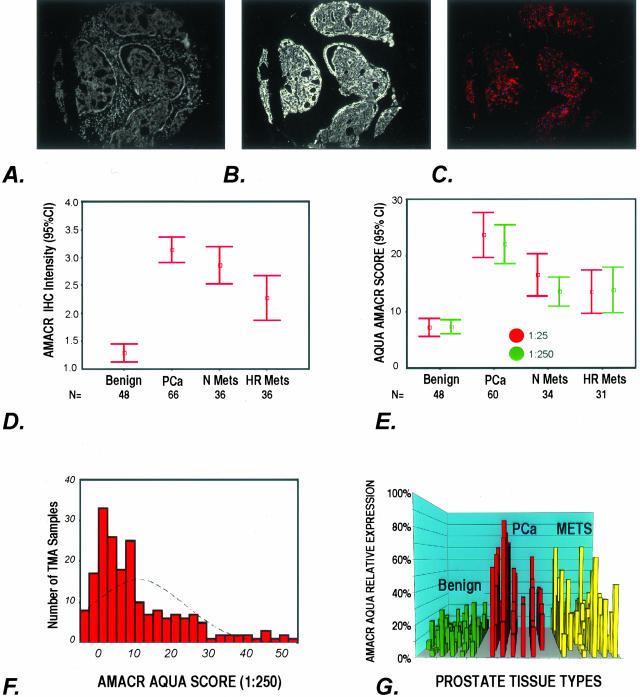
Figure 3.
A–G: AMACR expression as determined by AQUA and standard pathology review. A to C demonstrate how AQUA identifies prostate cancer from benign tissue in a tissue microarray sample (0.6 mm diameter). A: Using DAPI staining, all nucleated cells in both the stroma and epithelial compartments are identified. The cytokeratin-positive compartment identifies all epithelial cells (B) and Cy-5 expression (red) demonstrates the area of AMACR expression (C). The staining area and intensity of each compartment can be calculated. The AMACR-positive area, which overlaps with the cytokeratin mask is equivalent to the prostate cancer compartment. D: AMACR (p504s) protein expression by standard pathology review using immunohistochemistry. The expression of AMACR (p504s) was determined using the prostate cancer progression tissue microarray. Expression was scored as negative (score = 1), weak (score = 2), moderate (score = 3), and strong (score = 4). The results for each tissue type are presented as error bars of AMACR (p504s) protein expression with 95% confidence intervals. E: AQUA evaluation of AMACR expression using the prostate cancer progression tissue microarray at two antibody concentrations. AQUA measured similar AMACR protein expression levels for the different tissue categories [ie, benign, localized prostate cancer (PCa), metastatic prostate cancer to regional lymph nodes (Mets), and distant hormone-refractory metastatic prostate cancer (HR Mets)] at standard antibody concentrations (1:25) and at a concentration that would not be visible using standard immunohistochemistry (1:250). The intensity is corrected for area for each tissue microarray spot and measured using arbitrary units. The errors bars demonstrate that within 95% confidence intervals, benign prostate tissue does not score greater than 10 units. F: AMACR protein expression histogram for AQUA analysis. Using the arbitrary unit, this histogram demonstrates a relatively normal distribution of AMACR protein expression using samples from the prostate progression chip. This demonstrates the ability of the AQUA system to give a continuous readout of protein expression. G: Variable AMACR protein expression is appreciated by presenting the individual staining intensities for each sample based on tissue category. Although AMACR protein expression for the clinically localized prostate tumors is significantly higher than the population of benign samples, AMACR expression is still observed in benign tissues and can also be found to be low in other categories.
Table 1.
AMACR (p504s) Protein Expression by Standard Immunohistochemical Analysis by Pathologist
| Tissue type | n | Mean | SE | 95% CI
|
|
|---|---|---|---|---|---|
| Lower bound | Upper bound | ||||
| Benign | 48 | 1.29 | 0.079 | 1.13 | 1.45 |
| Clinically localized Pca | 66 | 3.14 | 0.114 | 2.91 | 3.36 |
| Hormone-naïve mets | 36 | 2.86 | 0.165 | 2.53 | 3.2 |
| Hormone-refractory mets | 36 | 2.28 | 0.198 | 1.88 | 2.68 |
Mets, metastatic tumor; CI, confidence intervals.
AQUA Evaluation of AMACR Expression Using the Prostate Cancer Progression Chip
AQUA measured similar AMACR protein expression levels for the different tissue categories (ie, benign, localized prostate cancer, metastatic prostate cancer to regional lymph nodes, and distant hormone-refractory metastatic prostate cancer) at standard antibody concentrations (1:25) and at a lower concentration that would not be detected using standard immunohistochemistry (1:250). The results are presented in Figure 3E and Table 2. The intensity of AMACR protein expression is corrected for the area for each TMA spot and measured using arbitrary units. The errors bars demonstrate that benign prostate tissue does not score greater than 10 arbitrary units. A second finding is that AMACR at both concentrations demonstrates a similar range of staining intensities for each tissue type (Figure 3E). The results also demonstrate that AMACR expression is quite variable in the hormone-refractory metastatic prostate cancer cases, consistent with our previous observations using other methods.10,13 Significant differences were seen between benign prostate tissue (mean AQUA score, 7.3) and clinically localized prostate cancer (mean AQUA score, 21.6) with a mean difference of 14.3 (95% CI, 8.5 to 20.2; P < 0.00001). Significant differences were seen between localized prostate cancer and hormone-naïve prostate cancer (mean difference, 7.9; 95%CI, 1.5 to 14.3; P = 0.006) and between localized prostate cancer and hormone-refractory prostate cancer (mean difference, 8.1; 95% CI, 1.7 to 14.6; P = 0.005). There is no significant difference in AMACR expression between the hormone-naïve and -refractory tumors (mean difference, 0.2).
Table 2.
AMACR (p504s) Protein Expression by AQUA Analysis (1:25 Dilution)
| Tissue Type | n | Mean | SE | 95% CI
|
|
|---|---|---|---|---|---|
| Lower bound | Upper bound | ||||
| Benign | 48 | 7.3 | 0.62 | 6.1 | 8.6 |
| Clinically localized Pca | 63 | 21.6 | 1.69 | 18.3 | 25 |
| Hormone-naïve Pca | 36 | 13.7 | 1.25 | 11.2 | 16.3 |
| Hormone-refractory Pca | 35 | 13.5 | 1.8 | 9.8 | 17.2 |
Comparison of AMACR Expression Using AQUA versus Standard Pathology Review
As schematically demonstrated in Figure 2, AQUA can identify prostate cancer from benign tissue in a TMA sample (0.6 mm diameter) (Figure 3; A to C). Using DAPI staining, all nucleated cells in both the stroma and epithelial compartments are identified (Figure 3A). The cytokeratin-positive compartment identifies all epithelial cells (Figure 3B) and Cy-5 expression (red) demonstrates the area of AMACR expression (Figure 3C). The staining area and intensity of each compartment can be calculated. The AMACR-positive area, which overlaps with the cytokeratin mask, is equivalent to the prostate cancer compartment. AMACR (p504s) protein expression as determined by standard pathology review using immunohistochemistry is presented in Figure 3D. The expression of AMACR (p504s) was determined using the prostate cancer progression TMA. Expression was scored as negative (score = 1), weak (score = 2), moderate (score = 3), and strong (score = 4). The results for each tissue type is presented as error bars of AMACR (p504s) protein expression with 95% confidence intervals. AQUA evaluation of AMACR expression using the prostate cancer progression TMA at two antibody concentrations is presented in Figure 3E. AQUA measured similar AMACR protein expression levels for the different tissue categories [ie, benign, localized prostate cancer (PCa), metastatic prostate cancer to regional lymph nodes (Mets), and distant hormone-refractory metastatic prostate cancer (HR Mets)] at standard antibody concentrations (1:25) and at a concentration that would not be visible using standard immunohistochemistry (1:250). The intensity is corrected for area for each TMA spot and measured using arbitrary units. The error bars demonstrate that within 95% confidence intervals, benign prostate tissue does not score greater than 10 units. AMACR protein expression as determined by AQUA is presented as a histogram in Figure 3F. Using the arbitrary unit, this histogram demonstrates a relatively normal distribution of AMACR protein expression using samples from the prostate progression TMA. This demonstrates the ability of the AQUA system to give a continuous read-out of protein expression. Variable AMACR protein expression is appreciated by presenting the individual staining intensities for each sample based on tissue category (Figure 3G). Although AMACR protein expression for the clinically localized prostate tumors is significantly higher than the population of benign samples, AMACR expression is still observed in benign tissues and can also be found to be low in other categories.
Although AMACR protein expression for the clinically localized prostate tumors is significantly higher than the population of benign samples, AMACR expression is still observed in benign tissues and can also be found to be low in other tissue categories. For example, as has been previously reported, AMACR expression is decreased in a subset of metastatic samples.10,13 Variability within the clinically localized prostate cancers demonstrates that some cases have weak AMACR expression.
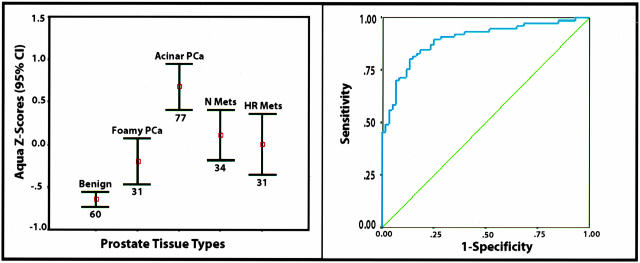
Recent work by Zhou and colleagues19 report a decrease in AMACR expression in a subtype of acinar prostate cancers referred to as foamy gland cancers. To confirm that the variability seen on the prostate progression TMA was associated with previously described variability, we created a focused TMA composed of prostate cancers with foamy gland features. Both the standard acinar tumors and the foamy gland subcomponent were represented on the TMA. As presented in Figure 4A, after normalization the AQUA Z-score for AMACR (p504s) expression in foamy gland prostate cancer is weaker compared to the more typical prostatic adenocarcinoma. However, the expression is significantly higher than benign samples as demonstrated by the nonoverlapping error bars with 95% confidence intervals (Figure 4A).
Figure 4.
Evaluation of normalized AMACR-AQUA scores and ability to predict the presence of prostate cancer. The AQUA results were normalized creating an AMACR AQUA Z-score (left). Expression results demonstrate measurable levels of AMACR in the foamy gland tumor population that are statistically significantly higher than the benign prostate tissue population as demonstrated by nonoverlapping error bars with 95% confidence intervals. A ROC plot demonstrates the sensitivity and specificity of using an AMACR AQUA Z-score to diagnose prostate cancer in an unsupervised manner (ie, without previous review by a pathologist) (right). The area under the ROC curve is 0.90 (P < 0.00001; 95% CI, 0.84 to 0.95). An area under the curve of 1.00 represents perfect discrimination of the test to predict prostate cancer and 0.50 no discriminatory power.
Finally, ROC curves were plotted to evaluate the specificity and sensitivity of using AMACR as measured by the AQUA system to detect prostate cancer in an unsupervised manner (ie, without a pathologist’s review). Including all localized prostate cancer cases (ie, acinar and foamy gland), the area under the curve was 0.844 (P < 0.00001; 95% CI, 0.79 to 0.90). By excluding the foamy gland tumors and evaluating only classic acinar tumors from cases with a range of Gleason scores from 6 to 9, the area under the curve was 0.90 (P < 0.00001; 95% CI, 0.84 to 0.95). The ROC plot is presented in Figure 4B. Therefore an AQUA Z-score of −0.30 is associated with 81% sensitivity and 87% specificity; an AQUA Z-score of −0.50 is associated with 88% sensitivity and 75% specificity; and an AQUA Z-score of −0.70 has a sensitivity of 94% and specificity of 53%. For an AQUA Z-score of −0.30, 91% of the 70 samples classified as prostate cancer are correctly classified without the intervention of a pathologist reviewing the TMA slide.
Discussion
AMACR protein expression is developing as an important prostate cancer biomarker. Since its identification by several groups,12,13,17 AMACR is gaining acceptance as a tool in the clinical diagnosis of prostate cancer biopsies.8,12–15 Although to date there is no evidence that AMACR plays a causative role in the development of prostate cancer, it is emerging as a useful marker of prostate cancer. For example in work performed by our group, a measurable AMACR humoral response is detected in men with prostate cancer (unpublished observations). This suggests that AMACR may become a clinically useful serum test. We have also recently observed that AMACR enzymatic activity can be detected in fresh biopsy samples, suggesting a means of screening for positive biopsies in the urologist’s office during ultrasound-guided needle biopsy to detect cancer (unpublished observations). Several groups recognized that AMACR is expressed in the precursor lesion to prostate cancer, high-grade PIN.12,13,16 However, we have observed that even in areas suggestive of low-grade PIN and histologically benign prostate tissue, AMACR expression may be observed. This potential harbinger effect may prove to be a useful way to measure the earliest alterations associated with prostate cancer development. How would we be able to assess these changes? Currently, standard pathology is limited to morphological alterations such as PIN. However, the incorporation of biomarkers into clinical trials as measures of endpoints may prove to be more reliable as rare morphological events such as PIN are often missed in needle biopsies. The AQUA system might represent a useful way of measuring these events that cannot be distinguished by the trained pathologist. As demonstrated in this study, AMACR could be detected at significantly lower levels than standard light microscopy. AMACR protein expression is detectable at 1:25 dilutions using standard immunohistochemistry. Concentrations of 1:50 are weak and give inconsistent protein expression in prostate cancer samples. No detectable signal was observed using concentrations of 1:100 and higher dilutions. The AQUA system was able to detect AMACR at 1:250 concentrations. Moreover, AMACR could be measured in a continuous manner. We are currently trying to quantify this wide range of expression to be able to determine expression based on the number of protein molecules. Work that is in progress is measuring HER2 levels in breast cancer using AQUA on a series of cell lines that have widely variable levels of expression, from a few hundred molecules to more than a million per cell. Preliminary evidence suggests we cover ∼2 logs but can cover 4 logs with two exposure times. This is considerably better that standard interpretation of immunohistochemistry stains, which are linear although ∼1 log of expression and tend to use nominal categories such as negative, weak, moderate, and strong to describe the expression of tissue biomarkers.
We also analyzed an intriguing subtype of localized prostate cancers, the foamy gland variant that has been reported to be associated with more aggressive prostate cancer despite its bland light microscopic appearance.24,25 Although not the major focus of this study, we wanted to determine whether the foamy gland components, which by standard AMACR immunohistochemistry have the lowest expression of any of the localized prostate cancer studies thus to date, could be measured by the AQUA system. The expression of AMACR in the foamy gland variant was significantly higher than benign prostate tissue but lower than clinically localized prostate cancer. Interestingly, AMACR expression has been previously shown to be up-regulated very early in the neoplastic process with expression seen in high-grade PIN.12,13,16
This study also points to the potential use of AMACR as a marker to distinguish benign prostate tissue from neoplastic tissues using the AQUA system for biomarker screening. This would represent a significant improvement in the throughput of prostate cancer biomarker evaluation. One concern is that because of the variable expression of AMACR in prostate tumors as seen with the foamy gland variant and the metastatic tumors, AMACR would not have sufficient discriminatory ability. However, as demonstrated in the ROC plot, the ability to use AMACR AQUA Z-scores to discriminate prostate cancers from benign tissue is highly feasible with an area under of the curve of 0.90. Therefore by determining an ideal threshold, the AMACR expression can be used to screen other prostate cancer biomarkers in an unsupervised manner. For example, if one wanted to determine the range of p27 expression in a large number of prostate cancer samples, the expression of AMACR in the same TMA samples would provide sufficient information to distinguish benign and tumors samples. Performing this process on a large number of biomarkers would represent an excellent strategy to perform early phase analysis of prostate tissue biomarkers. In this preliminary study, we determined that by using an AMACR AQUA Z-score of −0.3, one could select 91 prostate cancers from 100 samples. Although this would clearly not be acceptable in a clinical practice, this would be suitable for automated screening of multiple prostate cancer biomarkers. Another potential limitation is that both PIN and prostate cancer express AMACR, however for the purpose of screening biomarkers in a high-throughput manner, high-grade PIN would be a relatively rare event that should not pose a large problem in interpreting the data provided that enough samples were used for the analysis. Alternatively, use of a basal cell marker such a p63 in combination with AMACR as previously described by Luo and colleagues,12 might further improve the discriminatory ability of using the AQUA system to automate prostate cancer biomarker analysis.
Although this work concentrated on prostate cancer, AMACR is known to be expressed in other tissues including colon cancer.10,11 Therefore, this type of analysis using AMACR is not limited to prostate cancer. Other strategies may be useful for other tumor types with the goal of automating the evaluation of tissue-based biomarkers.
In summary, we demonstrate that the AQUA system can perform continuous measurements of AMACR in a large range of prostate tissue samples. Expression was found at low antibody concentrations because of the use of more sensitive and quantitative detection methods. Continuous measurements confirmed previous observations that some types of prostate tumors have a decrease in AMACR expression including foamy gland tumors and metastatic prostate tumors. This study also suggests that AMACR may also be a useful means of distinguishing prostate cancer from benign samples in an automated manner for use in the unsupervised evaluation of prostate cancer biomarkers.
Acknowledgments
We thank Celina Kleer for her thoughtful review and comments; Lela Schumacher, Martina Storz-Schweizer, and Tatiana Zolotarev for technical assistance in the construction of tissue microarrays and immunohistochemistry; and our clinical colleagues at the University of Michigan (James Montie, Kenneth Pienta, and Martin Sanda) and the University of Ulm (Richard Hautmann, Juergen Gshwend, and Peter Moeller) for their continued important support.
Footnotes
Address reprint requests to Mark A. Rubin, M.D, Department of Pathology (Amory 3-195), Brigham and Women’s Hospital/Harvard Medical School, 75 Francis St., Boston, MA 02115. E-mail: marubin@partners.org.
Supported by the Specialized Program of Research Excellence for Prostate Cancer from the National Cancer Institute (grants P50CA90381 to M.A.R., P50CA69568 to M.A.R. and A.M.C., R01AG21404 to M.A.R., and NCI R21 CA100825 to D.L.R.).
References
- Dowsett M, Bartlett J, Ellis IO, Salter J, Hills M, Mallon E, Watters AD, Cooke T, Paish C, Wencyk PM, Pinder SE. Correlation between immunohistochemistry (HercepTest) and fluorescence in situ hybridization (FISH) for HER-2 in 426 breast carcinomas from 37 centres. J Pathol. 2003;199:418–423. doi: 10.1002/path.1313. [DOI] [PubMed] [Google Scholar]
- Paik S, Bryant J, Tan-Chiu E, Romond E, Hiller W, Park K, Brown A, Yothers G, Anderson S, Smith R, Wickerham DL, Wolmark N. Real-world performance of HER2 testing—National Surgical Adjuvant Breast and Bowel Project experience. J Natl Cancer Inst. 2002;94:852–854. doi: 10.1093/jnci/94.11.852. [DOI] [PubMed] [Google Scholar]
- Fernandez PL, Arce Y, Farre X, Martinez A, Nadal A, Rey MJ, Peiro N, Campo E, Cardesa A. Expression of p27/Kip1 is down-regulated in human prostate carcinoma progression. J Pathol. 1999;187:563–566. doi: 10.1002/(SICI)1096-9896(199904)187:5<563::AID-PATH292>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Yang RM, Naitoh J, Murphy M, Wang HJ, Phillipson J, deKernion JB, Loda M, Reiter RE. Low p27 expression predicts poor disease-free survival in patients with prostate cancer. J Urol. 1998;159:941–945. [PubMed] [Google Scholar]
- Guo Y, Sklar GN, Borkowski A, Kyprianou N. Loss of the cyclin-dependent kinase inhibitor p27 (Kip1) protein in human prostate cancer correlates with tumor grade. Clin Cancer Res. 1997;3:2269–2274. [PubMed] [Google Scholar]
- Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002;8:1323–1327. doi: 10.1038/nm791. [DOI] [PubMed] [Google Scholar]
- Camp RL, Dolled-Filhart M, King BL, Rimm DL. Quantitative analysis of breast cancer tissue microarrays shows that both high and normal levels of HER2 expression are associated with poor outcome. Cancer Res. 2003;63:1445–1448. [PubMed] [Google Scholar]
- Leav I, McNeal JE, Ho SM, Jiang Z. Alpha-methylacyl-CoA racemase (P504S) expression in evolving carcinomas within benign prostatic hyperplasia and in cancers of the transition zone. Hum Pathol. 2003;34:228–233. doi: 10.1053/hupa.2003.42. [DOI] [PubMed] [Google Scholar]
- Zheng SL, Chang BL, Faith DA, Johnson JR, Isaacs SD, Hawkins GA, Turner A, Wiley KE, Bleecker ER, Walsh PC, Meyers DA, Isaacs WB, Xu J. Sequence variants of alpha-methylacyl-CoA racemase are associated with prostate cancer risk. Cancer Res. 2002;62:6485–6488. [PubMed] [Google Scholar]
- Kuefer R, Varambally S, Zhou M, Lucas PC, Loeffler M, Wolter H, Mattfeldt T, Hautmann RE, Gschwend JE, Barrette TR, Dunn RL, Chinnaiyan AM, Rubin MA. Alpha-methylacyl-CoA racemase: expression levels of this novel cancer biomarker depend on tumor differentiation. Am J Pathol. 2002;161:841–848. doi: 10.1016/s0002-9440(10)64244-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Chinnaiyan AM, Kleer CG, Lucas PC, Rubin MA. Alpha-methylacyl-CoA racemase: a novel tumor marker over-expressed in several human cancers and their precursor lesions. Am J Surg Pathol. 2002;26:926–931. doi: 10.1097/00000478-200207000-00012. [DOI] [PubMed] [Google Scholar]
- Luo J, Zha S, Gage WR, Dunn TA, Hicks JL, Bennett CJ, Ewing CM, Platz EA, Ferdinandusse S, Wanders RJ, Trent JM, Isaacs WB, De Marzo AM. Alpha-methylacyl-CoA racemase: a new molecular marker for prostate cancer. Cancer Res. 2002;62:2220–2226. [PubMed] [Google Scholar]
- Rubin MA, Zhou M, Dhanasekaran SM, Varambally S, Barrette TR, Sanda MG, Pienta KJ, Ghosh D, Chinnaiyan AM. Alpha-methylacyl coenzyme A racemase as a tissue biomarker for prostate cancer. JAMA. 2002;287:1662–1670. doi: 10.1001/jama.287.13.1662. [DOI] [PubMed] [Google Scholar]
- Beach R, Gown AM, De Peralta-Venturina MN, Folpe AL, Yaziji H, Salles PG, Grignon DJ, Fanger GR, Amin MB. P504S immunohistochemical detection in 405 prostatic specimens including 376 18-gauge needle biopsies. Am J Surg Pathol. 2002;26:1588–1596. doi: 10.1097/00000478-200212000-00006. [DOI] [PubMed] [Google Scholar]
- Yang XJ, Wu CL, Woda BA, Dresser K, Tretiakova M, Fanger GR, Jiang Z. Expression of alpha-methylacyl-CoA racemase (P504S) in atypical adenomatous hyperplasia of the prostate. Am J Surg Pathol. 2002;26:921–925. doi: 10.1097/00000478-200207000-00011. [DOI] [PubMed] [Google Scholar]
- Jiang Z, Woda BA, Rock KL, Xu Y, Savas L, Khan A, Pihan G, Cai F, Babcook JS, Rathanaswami P, Reed SG, Xu J, Fanger GR. P504S: a new molecular marker for the detection of prostate carcinoma. Am J Surg Pathol. 2001;25:1397–1404. doi: 10.1097/00000478-200111000-00007. [DOI] [PubMed] [Google Scholar]
- Xu J, Stolk JA, Zhang X, Silva SJ, Houghton RL, Matsumura M, Vedvick TS, Leslie KB, Badaro R, Reed SG. Identification of differentially expressed genes in human prostate cancer using subtraction and microarray. Cancer Res. 2000;60:1677–1682. [PubMed] [Google Scholar]
- Rubin MA, Putzi M, Mucci N, Smith DC, Wojno K, Korenchuk S, Pienta KJ. Rapid (“warm”) autopsy study for procurement of metastatic prostate cancer. Clin Cancer Res. 2000;6:1038–1045. [PubMed] [Google Scholar]
- Zhou M, Jiang Z, Epstein JI. Expression and diagnostic utility of alpha-methylacyl-CoA-racemase (P504S) in foamy gland and pseudohyperplastic prostate cancer. Am J Surg Pathol. 2003;27:772–778. doi: 10.1097/00000478-200306000-00007. [DOI] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;10:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Dhanasekaran SM, Barrette TR, Ghosh D, Shah R, Varambally S, Kurachi K, Pienta KJ, Rubin MA, Chinnaiyan AM. Delineation of prognostic biomarkers in prostate cancer. Nature. 2001;412:822–826. doi: 10.1038/35090585. [DOI] [PubMed] [Google Scholar]
- Rubin MA, Mucci NR, Figurski J, Fecko A, Pienta KJ, Day ML. E-cadherin expression in prostate cancer: a broad survey using high-density tissue microarray technology. Hum Pathol. 2001;32:690–697. doi: 10.1053/hupa.2001.25902. [DOI] [PubMed] [Google Scholar]
- Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using z score transformation. J Mol Diagn. 2003;5:73–81. doi: 10.1016/S1525-1578(10)60455-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TT, Sengupta E, Yang XJ. Prostatic foamy gland carcinoma with aggressive behavior: clinicopathologic, immunohistochemical, and ultrastructural analysis. Am J Surg Pathol. 2001;25:618–623. doi: 10.1097/00000478-200105000-00008. [DOI] [PubMed] [Google Scholar]
- Nelson RS, Epstein JI. Prostatic carcinoma with abundant xanthomatous cytoplasm. Foamy gland carcinoma. Am J Surg Pathol. 1996;20:419–426. doi: 10.1097/00000478-199604000-00004. [DOI] [PubMed] [Google Scholar]