Abstract
Human Polycomb-group (PcG) genes play a crucial role in the regulation of embryonic development and regulation of the cell cycle and hematopoiesis. PcG genes encode proteins that form two distinct PcG complexes, involved in maintenance of cell identity and gene silencing patterns. We recently showed that expression of the BMI-1 and EZH2 PcG genes is separated during normal B-cell development in germinal centers, whereas Hodgkin/Reed-Sternberg (H/RS) cells co-express BMI-1 and EZH2. In the current study, we used immunohistochemistry and immunofluorescence to determine whether the binding partners of these PcG proteins are also present in H/RS cells and H/RS-derived cell lines. PcG expression profiles were analyzed in combination with expression of the cell cycle inhibitor p16INK4a, because experimental model systems indicate that p16 is a downstream target of Bmi-1. We found that H/RS cells and HL-derived cell lines co-express all core proteins of the two known PcG complexes, including BMI-1, MEL-18, RING1, HPH1, HPC1, and -2, EED, EZH2, YY1, and the HPC2 binding partner, CtBP. Expression of HPC1 has not been found in normal mature B cells and other malignant lymphomas of B-cell origin, suggesting that the PcG expression profile of H/RS is unique. In contrast to Bmi-1 transgenic mice where p16INK4a is down-regulated, 27 of 52 BMI-1POS cases of HL revealed strong nuclear expression of p16INK4a. We propose that abnormal expression of BMI-1 and its binding partners in H/RS cells contributes to development of HL. However, abnormal expression of BMI-1 in HL is not necessarily associated with down-regulation of p16INK4a.
Hodgkin Lymphoma (HL) is a malignant proliferation of mature lymphocytes, characterized by the presence of low numbers of neoplastic cells surrounded by high numbers of reactive T and B lymphocytes, histiocytes, and eosinophils.1 The neoplastic cells in HL are the Reed-Sternberg cells (RS cells) and their mononuclear variants, the Hodgkin cells (H cells). The majority of classical H/RS cells are thought to be clonal expansions originating from germinal center B cells.2–4 The exact mechanism responsible for malignant transformation and subsequent development of HD is unclear. Various mechanisms might be involved, including aberrant expression of tumor necrosis factor family members,5 infection with Epstein-Barr virus,6 and deregulation of the apoptosis pathway.7,8 We recently proposed that altered expression of Polycomb-group (PcG) genes could also contribute to malignant transformation.9
PcG genes play a crucial role in embryonic development and have been associated with regulation of hematopoiesis and the cell cycle.10–17 PcG proteins form two evolutionary conserved multimeric complexes, that bind to DNA and maintain cell identity by gene suppression.18,19 The PRC1/HPC-HPH “maintenance” complex is the mammalian counterpart of the Drosophila Polycomb repressive complex 1 (PRC1) and contains the BMI-1, RING1, HPH, and HPC PcG proteins, and probably MEL-18.20–27 The PRC2/EED-EZH or Esc-Ez “initiation” complex, is much smaller and is composed of the EED, EZH, SU(Z)12, and YY1 PcG proteins.28–33 PcG complexes can contain various transcription factors that may contribute to DNA binding.34,35 In addition, the presence of histone-methyltransferases and histone deacetylases strongly suggests that PcG complexes silence genes by histone modification.36–39
The essential role of PcG genes in maintenance of cellular identity and normal cellular development is reflected by the relationship between abnormal PcG gene expression and malignant transformation. The best-known example is the Bmi-1 transgenic mouse, where Bmi-1 up-regulation induces down-regulation of p16INK4a and p19ARF, increased lymphoid proliferation, and development of lymphomas.40–42 Various BMI-1 binding partners have also been associated with malignant transformation, but appear to do so by mechanisms that do not involve the p16 pathway.22,43 In addition, overexpression of the Eed PcG gene results in diminished lymphoproliferation, suggesting that the two PcG complexes have opposing effects.44 We recently discovered that predominance of PcG proteins associated with the PRC1/HPC-HPH complex is associated with resting cells in the germinal center, whereas predominance of PRC2/EED-EZH PcG complex proteins is related to proliferation.45,46 The opposing effects on lymphoproliferation in Bmi-1- and Eed-transgenic mice, and the PcG expression profiles in human germinal centers, suggest that a balance between the two PcG complexes is essential for proper regulation of lymphoproliferation.
Despite the correlation between malignant transformation and altered expression of PcG genes observed in experimental models, the role of human PcG genes in oncogenesis remains relatively unexplored. Previous data of our group showed that H/RS cells and neoplastic cells in high-grade large B-cell lymphomas and mantle-cell lymphomas are associated with nuclear co-expression of BMI-1 and EZH2.9,47,48 Co-expression of these two PcG genes is generally not observed in the normal counterparts of these cells,45,46 suggesting that aberrant PcG expression contributes to development of malignant lymphomas in man. It is unclear, however, whether BMI-1 expression in EZH2POS malignant lymphomas has functional consequences for development of these tumors. One condition that should be met is that binding partners of BMI-1 and EZH2 are present in the tumor cells. In the current study, we investigated whether H/RS cells and HL-derived cell lines express other components of the PRC1/HPC-HPH- and PRC2/EED-EZH PcG complexes, and compared these patterns with the PcG expression profile of normal B cells. In addition, PcG expression was correlated with p16INK4a expression.
Materials and Methods
Cell Lines
The characteristics of the HL-derived cell lines L428 and L1236 were described in detail previously.49 Cell lines were maintained in RPMI 1640 (BioWhittaker, East Rutherford, NJ) medium supplemented with 25 mmol/L HEPES, 2 mmol/L L-glutamine, 10% fetal calf serum, 100 IU of streptomycin, and 100 μg of penicillin at 37°C in 5% CO2.
Human Tissue
Formalin-fixed, paraffin-embedded tissue blocks of 54 primary biopsies of nodular sclerosing HL patients were retrieved from the Department of Pathology, LUMC, The Netherlands. Frozen tissue was retrieved from 24 cases as well. All cases were diagnosed between 1990 and 2001 and classified according to the World Health Organization new classification. The diagnostic immunohistochemical panel always included CD3, CD15, CD20, CD30, CD45, EMA, ALK1, LMP1, and EBER RNA in situ hybridization. This panel was extended if necessary for appropriate diagnosis. Only the first diagnostic biopsy specimen of the patients was investigated. As positive controls for immunohistochemistry, both paraffin-embedded and frozen material from tonsil, thymus, kidney, and testis was used (Table 1).
Table 1.
Antibodies Used in this Study in Combination with Staining Conditions
| Antigen | Antibody | Species | Subtype | Titer | Fluorochrome | Treatment | Control tissue |
|---|---|---|---|---|---|---|---|
| EED | M26 | Mouse | IgG1 | 1:4000 o/n | strepAPC | Citrate ABC/CARD | Tonsil |
| EZH2 | K358 | Rabbit | 1:400 o/n | GaRALEXA 488 | EDTA ABC/CARD | Tonsil | |
| HPC1 | K350 | Rabbit | 1:100 o/n | GaRALEXA 488 | EDTA ABC/CARD | Kidney | |
| HPC2 | K326 | Rabbit | 1:100 o/n | GaRALEXA 488 | EDTA ABC/CARD | Tonsil | |
| HPH1 | K344 | Rabbit | 1:100 o/n | GaRALEXA 488 | EDTA ABC/CARD | Tonsil | |
| RING1 | K320 | Rabbit | 1:100 o/n | GaRALEXA 488 | Citrate ABC/CARD | Tonsil | |
| BMI-1 | K333 | Rabbit | 1:2000 o/n | GaRALEXA 488 | Citrate ABC/CARD | Tonsil | |
| BMI-1 | 6C9 | Mouse | IgG2b | undiluted | Rhodamine/tyramine | Citrate ABC/CARD | Tonsil |
| Ki67 | Mib-1 | Mouse | IgG1 | strepAPC | Citrate ABC/CARD | Tonsil | |
| MEL-18 | Rabbit | 1:400 o/n | GaRALEXA 488 | Citrate ABC/CARD | Tonsil | ||
| YY1 | Mouse | IgG1 | 1:250 o/n | strepAPC | Citrate ABC/CARD | Testis | |
| CtBP | Rabbit | 1:4000 o/n | GaRALEXA 488 | Citrate ABC/CARD | Testis | ||
| P16 | Mouse | IgG2a | 1:500 | Rhodamine/tyramine | Citrate ABC/CARD | Cervix carcinoma |
Abbreviations: o/n, overnight at 4°C; citrate, antigen retrieval using 0.01M citrate; pH 6.0, 15 minute in the autoclave; EDTA, antigen retrieval using 10mM EDTA, pH 8.0, 15 minute in the autoclave; strepAPC, allophycocyanin-conjugated streptavidine (blue signal); GaRALEXA, ALEXA 488-conjugated goat-anti-rabbit (green signal); rhodamine/tyramine (red signal); ABC, horseradish-peroxidase-conjugated streptavidin-biotin-complex; CARD, catalyzed reporter deposition method. For details about isolation and characterization of PcG-specific antisera: see references in Materials and Methods section.
Immunohistochemistry
For immunohistochemistry, 3-μm sections of paraffin-embedded material were used for the detection of various PcG proteins and cell cycle-related proteins Ki67 and p16 (Table 1). After rehydration, endogenous peroxidase was quenched by incubation of the sections in 0.3% H2O2 diluted in methanol for 30 minutes. Thereafter, antigen retrieval was performed using either citrate (pH 6.0) or EDTA (pH 8.0), depending on the antigen (Table 1), for 15 minutes in the autoclave. After cooling, the slides were rinsed with phosphate-buffered saline (PBS) containing 0.5% Triton X-100 (5 minutes), followed by PBS only (3 × 5 minutes). Subsequently, the slides were incubated with 0.1 mol/L glycine (10 minutes) and rinsed in PBS.
After pre-incubation with normal swine serum or normal rabbit serum, for polyclonal and monoclonal antibodies, respectively, the primary antibodies26,27,31,43,50,51 were applied at optimal dilution (Table 1). Secondary antibodies were biotinylated swine anti-rabbit or biotinylated rabbit-anti-mouse (Dako, Glostrup, Denmark). This was followed by application of horseradish peroxidase-conjugated streptavidine-biotin-complex (sABC-HRP) and further signal enhancement was achieved by the CARD-method (catalyzed reporter deposition method, DAKO). The signal was visualized by 3-amino-9-ethylcarbazole (AEC) and counterstained with hematoxylin. Photographs were taken with a Zeiss Axiophot microscope (Zeiss, Oberkochen, Germany) and were digitized using an Agfa duoscan (Agfa, Morstel, Belgium).
Double and Triple Immunofluorescence
To confirm PcG protein staining patterns in neoplastic cells and the surrounding infiltrate, immunofluorescent double and triple staining was performed. This also allowed us to study PcG proteins in relation to other markers, such as p16INK4a, CtBP, and Ki67. Frozen tissue sections were fixed in 1% formaldehyde, followed by quenching of endogenous peroxidase using 1% H2O2 diluted in PBS. After preincubation with 5% bovine serum albumin (BSA), a combination of two or three primary antibodies was applied overnight at 4°C. BMI-1 was detected by incubation with HRP-conjugated goat-anti-mouse IgG2b and subsequent rhodamine/tyramine intensification. Detection of HPC1, HPC2, HPH1, RING1, EZH2, YY1, p16, and CtBP was performed by incubating the slides with ALEXA 488-conjugated goat-anti-rabbit antiserum. The other markers were visualized by incubating the slides with biotinylated goat-anti-mouse IgG1 or IgG2a, depending on Ig-subclass of the primary antibody, followed by allophycocyanin-coupled streptavidin. Cross-reactivity of the antisera was excluded by appropriate controls and for each double- or triple-immunofluorescence experiment; single-staining controls were included. In addition, positive and negative controls were routinely included. Sections were analyzed with a Leica DMR confocal laserscan microscope (Leica, Deerfield, IL). Images were stored digitally and processed using Adobe Photoshop 6.
Evaluation of Immunohistochemical Staining
Percentages of PcG expressing H/RS cells were determined visually and classified according to the percentage of positively staining tumor cells: 0, no staining in H/RS cells; 1, <25% of the H/RS cells positive; 2, 25% and <50% H/RS cells positive; 3, 50% and <75% H/RS cells positive; 4, 75% of the H/RS cells showed positive staining. In all cases, except for HPC1, small reactive lymphocytes served as an internal positive control.
Results
PcG Expression in Neoplastic H/RS Cells in Hodgkin’s Lymphoma
We analyzed the expression of various components of the PcG PRC1/HPC-HPH complex (BMI-1, HPC1, HPC2, HPH1, RING1, MEL-18, and CtBP) and the PRC2/EED-EZH PcG complex (EED, EZH2, and YY1) by immunohistochemistry in 54 cases of HL (all cases classified as nodular sclerosing). For all stainings, appropriate controls were included (Table 1).
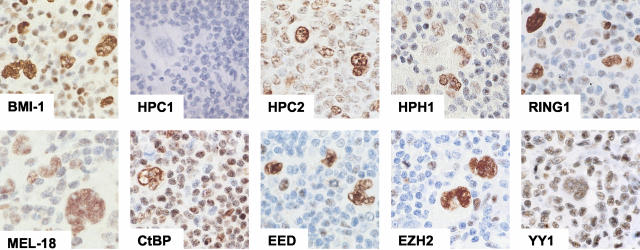
We initially focused on expression of PcG proteins belonging to the PRC1/HPC-HPH complex. Expression of the BMI-1 binding partners, RING1, HPH1, HPC2, and MEL-18, was found as strong nuclear staining in virtually all H/RS cells in HL (Table 2, Figure 1). The HPC1 PcG protein, however, was undetectable by immunohistochemistry in H/RS cells, although it can be part of the HPC-HPH complex and binds to BMI-1. Finally, the non-PcG transcriptional repressor CtBP that interacts with the HPC2 PcG protein, was detectable in the majority of H/RS cells examined in 53 of 54 cases of HL. These results collectively show that virtually all of the PRC1/HPC-HPH PcG complex core proteins are expressed by H/RS cells.
Table 2.
Expression of PcG Genes in Hodgkin’s Lymphoma and HL-Derived Cell Lines
| PcG gene | Hodgkin lymphoma RS cells (n = 54) | Hodgkin lymphoma reactive lymphocytes (n = 54) | HL cell lines* (n = 2) |
|---|---|---|---|
| BMI-1 | 54/54 | 54/54 (30–80% positive cells) | Positive |
| HPH1 | 54/54 | 54/54 (30–80% positive cells) | Positive |
| HPC1 | 7/54** | 0/54 | Positive |
| HPC2 | 54/54 | 54/54 (30–80% positive cells) | Positive |
| RING1 | 54/54 | 54/54 (30–80% positive cells) | Positive |
| MEL-18 | 54/54 | 8/54 (∼5% positive cells) | Positive |
| 46/54 (∼90% positive cells) | |||
| EZH2 | 54/54 | 54/54 (<5% positive cells) | Positive |
| EED | 54/54 | 54/54 (<5% positive cells) | Positive |
| YY1 | 52/54† | 54/54 (>95% positive cells) | Positive |
| CtBP | 53/54‡ | 54/54 (>95% positive cells) | Positive |
Expression of PcG proteins was investigated in HL-derived cell lines L428 and L1236.
HPC1 expression in neoplastic H/RS cells is only observed using immunofluorescence on frozen tissue sections (7 individual cases were studied). In paraffin-embedded HL tissue, HPC1 is not detected, most likely due to low expression in tumor cells.
In 2 of 54 cases YY1 expression was absent from the neoplastic H/RS cells but present in the reactive lymphocytes.
In 1 of 54 cases CtBP expression was absent from the neoplastic H/RS cells but present in the reactive lymphocytes. CtBP-/YY1- cases were not observed.
Figure 1.
Immunohistochemical detection of PcG expression in H/RS cells. Neoplastic cells in HL express various members of the PRC1/HPC-HPH PcG complex (BMI-1, MEL-18, RING1, HPH1, HPC2, and CtBP) and the PRC2/EED-EZH complex (EED, EZH2, and YY1). HPC1, a member of the PRC1/HPH-HPC PcG complex, was undetectable in H/RS cells by immunohistochemistry.
Our previous work on PcG expression in neoplastic cells of HL demonstrated that H/RS cells express BMI-1 in combination with EZH2, belonging to the PRC2/EED-EZH complex and normally present only in cycling B cells.9 We therefore examined whether other components of this complex are also present in H/RS cells. Immunohistochemical staining for EZH2, EED, and YY1 revealed strong nuclear expression of these proteins in the nuclei of virtually all H/RS cells (Table 2, Figure 1). In two instances, YY1 was undetectable in H/RS cells whereas lymphocytes in the surrounding infiltrate stained for this protein. The expression patterns of EZH2, EED, and YY1 confirm the presence of EZH2 in H/RS cells and show widespread expression of its binding partner EED in combination with YY1.
PcG Expression in Reactive Lymphocytes in Hodgkin’s Disease
Variable numbers of reactive lymphocytes, but always more than 50%, expressed PcG proteins belonging to the PRC1/HPC-HPH complex. BMI-1 and its binding partners RING1, HPH1, and HPC2, displayed a similar staining pattern, both in numbers of positive cells as well as intensity of staining (Figure 1). Expression of HPC1, however, was never detected in reactive lymphocytes. Interestingly, MEL-18 expression in H/RS cells was always strongly positive, whereas in 8 of 54 cases (15%), only a small number of reactive lymphocytes stained positive (< 50% of the total number of lymphocytes). In the remaining cases, expression of MEL-18 in reactive lymphocytes was found in virtually every cell. Expression of EZH2 and its binding partner EED on the other hand, was rarely detected in reactive infiltrating lymphocytes. The third PcG protein associated with the PRC2/EED-EZH complex, YY1, was found in virtually all of the reactive lymphocytes in all HL cases tested.
Co-Expression of PcG Complexes PRC2/EED-EZH and PRC1/HPC-HPH in Relation with Proliferation State of H/RS Cells
PcG expression profiles were further explored by double and triple immunofluorescence to gain insight in the (co-)expression of both PcG complexes in relation with cell cycle as determined by Ki67 expression in H/RS cells. Multiple combinations of primary antibodies were applied and co-expression was evaluated by confocal laserscan microscopy. The combinations of primary antibodies chosen included PcG proteins derived from both PRC1/HPC-HPH and the PRC2/EED-EZH complex in combination with Ki67 (Table 3).
Table 3.
Combinations of Antibodies Used for Double/Triple Immunofluorescence on Hodgkin’s Lymphoma
| Green fluorescence* | Red fluorescence† | Blue fluorescence‡ | Coexpression in H/RS cells |
|---|---|---|---|
| HPC1 | BMI-1 | Ki67 | Yes |
| HPC2 | BMI-1 | Ki67 | Yes |
| HPH1 | BMI-1 | Ki67 | Yes |
| RING1 | BMI-1 | Ki67 | Yes |
| EZH2 | BMI-1 | Ki67 | Yes |
| CtBP | BMI-1 | Ki67 | Yes |
| MEL-18 | BMI-1 | Yes | |
| HPC1 | BMI-1 | EED | Yes |
| HPC2 | BMI-1 | EED | Yes |
| HPH1 | BMI-1 | EED | Yes |
| RING1 | BMI-1 | EED | Yes |
| EZH2 | BMI-1 | EED | Yes |
| CtBP | BMI-1 | EED | Yes |
| MEL-18 | Ki67 | Yes |
Polyclonal rabbit-derived antibodies were visualized using an ALEXA 488-conjugated goat-anti-rabbit antiserum.
Antibodies with an IgG2b subtype were visualized with an HRP-conjugated goat-anti-mouse IgG2b specific antibody followed by rhodamine/tyramine intensification.
Antibodies if IgG1 or IgG2a subtype were detected by a biotinylated goat-anti-mouse IgG1 or IgG2a followed by allophycocyanin-coupled streptavidine.
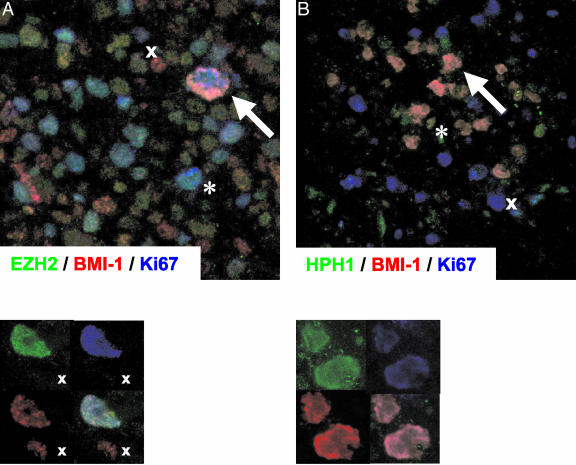
H/RS cells are, in most instances, cycling cells expressing Ki67 and EZH2, and we therefore first performed triple immunofluorescent staining for Ki67 and EZH2 in combination with another PcG protein. We were unable to examine expression of the EZH2 binding partner EED in relation to Ki67, because antisera recognizing these proteins are both of IgG1 subtype. In agreement with our earlier findings, staining for BMI-1 and EZH2 clearly showed co-expression of these proteins in the nuclei of cycling neoplastic Ki67POS H/RS cells (Figure 2A). Normal reactive lymphocytes in the surrounding infiltrate were primarily Ki67NEG resting cells, expressing BMI-1 in the absence of EZH2 (Figure 2A). As previously shown,9,46 we occasionally observed dividing Ki67POS reactive cells, which expressed EZH2 but lacked BMI-1.
Figure 2.
Immunofluorescent detection of BMI-1 in combination with EZH2 and HPH1 in Ki67POS H/RS cells. A: Triple immunofluorescence staining for BMI-1 (red signal), EZH2 (green signal), and Ki67 (blue signal). Large Ki67POS H/RS cells co-express BMI-1 and EZH2 (arrow) whereas BMI-1- and EZH2 expression are separated in healthy infiltrating cells (x denotes a resting (Ki67NEG) BMI-1POS/EZH2NEG cell; * denotes a dividing (Ki67POS) BMI-1NEG/EZH2POS cell). Lower panel: detail of a single H/RS cell, showing single fluorescence signals (upper left and right, and lower left) and the combination of these signals (lower right). B: Triple immunofluorescence staining for BMI-1(red signal), HPH1 (green signal), and Ki67 (blue signal). Large Ki67POS H/RS cells co-express BMI-1 and HPH1 (arrow) whereas dividing healthy infiltrating cells do not express BMI-1 and HPH1 (x). By contrast, BMI-1 and HPH1 are detectable in resting Ki67NEG healthy infiltrating cells (*). Lower panel: as in A.
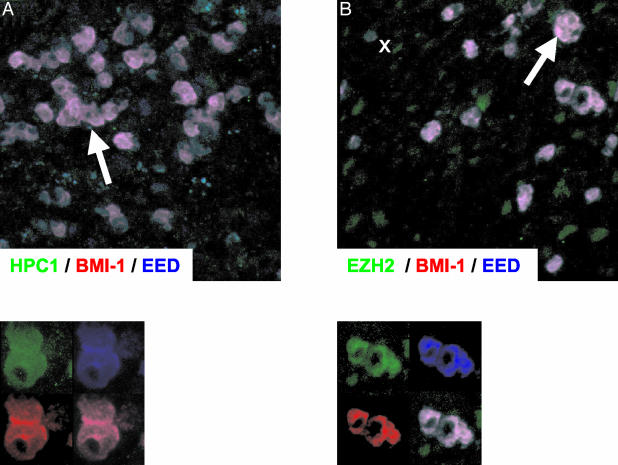
Investigation of BMI-1 and EZH2 binding partners by triple immunofluorence (Table 3) confirmed the staining patterns obtained by immunohistochemistry. Representative examples are shown in Figures 2 and 3. Staining for BMI-1 in combination with HPC2, HPH1, and RING1 revealed that these PcG proteins were co-expressed in the nuclei of large dividing Ki67POS H/RS cells and in resting Ki67NEG cells of the reactive infiltrate. A representative example of triple immunofluorescent staining for BMI-1 in combination with HPH1 and Ki67 is shown in Figure 2B. In contrast to the results obtained by immunohistochemistry, the HPC1 PcG protein was clearly detectable in neoplastic cells of HL by triple immunofluorescence (Table 3). This is illustrated by triple staining for BMI-1 in combination with HPC1 and EED in Figure 3B. Detection of HPC1 by immunofluorescence suggests low level HPC1 expression in the nuclei of malignant cells, because immunofluorescence on frozen sections is generally more sensitive than immunohistochemistry. We did not observe expression of HPC1 in normal reactive lymphocytes. Finally, a combination of EZH2, BMI-1, and EED showed that EZH2 and its binding partner EED are expressed in the nuclei of the malignant cells in combination with BMI-1, which does not bind EZH2 nor EED and is part of another PcG complex.
Figure 3.
Immunofluorescent detection of BMI-1 and EED in combination with HPC1 and EZH2. BMI-1 was detected by a red signal, EED by a blue signal, and HPC1 and EZH2 by a green signal. A: Expression of the BMI-1 binding partner HPC1 in combination with EZH2 and EED. B: Large EZH2POS tumor cells express the EZH2 binding partner EED in combination with BMI-1, which does not bind to EZH2 and is part of another PcG complex.
In summary, proliferating neoplastic H/RS cells co-express PcG core proteins derived from the PRC1/HPC-HPH and the PRC2/EED-EZH complex. The expression pattern of these proteins, BMI-1 in combination with RING1, HPH1, and HPC2, versus EZH2 in combination with EED, is always exclusive in reactive lymphocytes. In addition, H-RS cells express low levels of HPC1, which is undetectable in healthy reactive lymphocytes.
High Levels of BMI-1 Are Not Associated with p16 Down-Regulation
Mouse experimental model systems demonstrated that altered expression of PcG genes has strong effects on regulation of the cell cycle. For instance, overexpression of Bmi-1 in mice leads to down-regulation of p16INK4a and subsequent induction of lymphoproliferation and development of lymphomas.40–42 Therefore, the p16 gene appears to be a downstream target of Bmi-1. Given the expression of BMI-1 in H/RS cells, we questioned whether the presence of BMI-1 in neoplastic cells is associated with absence of p16INK4a. We found that p16INK4a expression was detected as nuclear staining in the H/RS cells of 27 of 52 (52%) cases (Figure 4), which were all BMI-1POS. As an internal positive control, few scattered reactive lymphocytes stained. This demonstrates that expression of BMI-1 in H/RS cells is not necessarily associated with absence of p16INK4a expression.
Figure 4.
Immunohistochemical detection of p16INK4a on two cases of nodular sclerosing HL. Two representative cases for p16INK4a positive and negative cases of HL in A and B, respectively
PcG Expression in HL-Derived Cell Lines L428 and L1236
Cytospin preparations were made from HL-derived cell lines L428 and L1236 and PcG expression was determined using immunohistochemistry. The PcG staining pattern of these cell lines closely resembled the expression pattern of neoplastic H/RS cells in patient material. Both L428 and L1236 expressed all investigated proteins of the PRC1/HPC-HPH complex, including BMI-1, MEL-18, RING1, HPH1, and HPC1/2. In addition, these PRC1/HPC-HPH complex proteins were co-expressed with the EED, EZH2, and YY1 proteins of the PRC2/EED-EZH complex.
Discussion
Classical H/RS cells originate from follicular B cells in the germinal center.2–4 One group of genes that may contribute to malignant transformation and development of HL, belongs to the Polycomb-group (PcG).9 PcG proteins are responsible for preservation of cell identity, and contribute to regulation of lymphoid development at all stages of hematopoiesis.10,12,16,17 We recently showed that healthy follicular B cells in germinal centers display a mutually exclusive expression pattern of PcG genes encoding the PRC1/HPC-HPH- and PRC2/EED-EZH PcG complexes.45,46 Resting Ki67NEG mantle cells and centrocytes preferentially expressed the PRC1/HPC-HPH BMI-1 and RING1 PcG genes, whereas dividing Ki67POS centroblasts were associated with detection of the PRC2/EED-EZH proteins EED and EZH2. Interestingly, from experiments in mutant mice it was concluded that core proteins of the two complexes have opposing functions. For instance, enhanced lymphoid proliferation was observed in transgenic mice overexpressing Bmi-1,40–42 and in knockout mice with diminished expression of Eed.44 These observations collectively suggest that a balance between the different PcG complexes is critical for normal division of hematopoietic cells.
In the current study and in our previous work,9 we demonstrated that this balance is lost in neoplastic H/RS cells and HL-derived cell lines. This is best illustrated by the expression pattern of BMI-1 and its binding partners. BMI-1 is not detected in healthy dividing EZH2POS B cells, whereas H/RS cells and HL-derived cell lines co-express BMI-1 and EZH2 (this study, and9,45,46). This expression pattern suggests abnormal expression of BMI-1 in transformed cells, and is in line with experimental model systems.40 The fact that BMI-1 functions as part of a multiprotein complex, raises the question whether the presence of BMI-1 in neoplastic cells is functionally relevant. One condition that should be met is that the binding partners of BMI-1 are also present in the nucleus. In the current study we showed that H/RS cells and HL-derived cell lines display a highly conserved PcG expression profile of the core proteins that form the PRC1/HPC-HPH- and PRC2/EED-EZH PcG complexes. In all instances, BMI-1 was expressed in combination with its binding partners RING1, HPH1, HPC1/2, and MEL-18. In addition, various proteins constituting the PRC2/EED-EZH complex, including EZH2, EED, and YY1 were also expressed in the same nuclei. These data suggest that both PcG complexes can be assembled in neoplastic H-RS cells at the same time.
Three conclusions can be drawn from the available expression data: firstly, H/RS cells display abnormal expression of PRC1/HPC-HPH proteins. This is best illustrated by the expression of BMI-1 and its binding partners, because normal dividing B lymphocytes do not express BMI-1 in combination with MEL-18, RING1, HPH1, and HPC proteins (this study, and van Galen et al, manuscript submitted). Secondly, expression of this set of PRC1/HPC-HPH PcG genes in combination with the PRC2/EED-EZH complex indicates an additional pattern of abnormal PcG expression, because mature lymphocytes in germinal centers express core proteins of PRC1/HPC-HPH- and PRC2/EED-EZH complexes in a mutually exclusive pattern. Finally, if the PcG proteins that are present in H-RS cells associate to form PcG complexes, we suggest that this will result in formation of a different PRC1/HPC-HPH complex than the one seen in healthy resting B cells. The main difference in complex composition would most likely be related to low-level expression of HPC1 in the nuclei of H/RS cells, because this PcG protein is absent from healthy mature cells in the reactive infiltrate. Whether PRC1/HPC-HPH and PRC2/EED-EZH complexes actually bind to their target genes in tumor cells cannot be determined by this type of study, but we showed that various PcG proteins with DNA-binding activity, such as MEL-1852 and YY1,53,54 are present in the H/RS cells. We propose that abnormal PcG expression in H/RS cells results in altered composition of the PRC1/HPC-HPH PcG complex, and that this contributes to interference with normal gene silencing patterns in H-RS cells and loss of cell identity.
A causative relationship between abnormal expression of PcG genes, altered gene silencing patterns, and loss of cell identity is supported by various experimental model systems. The best-known example is the Bmi-1 transgenic mouse, which identified Bmi-1 as an oncogene capable of down-regulating the cell cycle-associated proteins p16INK4a and p19ARF.41,42 In non-proliferating cells, p16INK4a and p19ARF specifically bind to the cdk4/6 protein complex, thereby preventing cyclin D activation and subsequent transition toward the S-phase. Bmi-1-mediated down-regulation of p16INK4a induces high levels of proliferating hematopoietic cells and formation of lymphomas. The inverse correlation between Bmi-1 and p16INK4a expression in mouse lymphocytes naturally leads to the question whether human p16INK4a is also a downstream target of BMI-1. As reported in other studies,55 we found that p16INK4a is undetectable in a large number of HL cases. Although loss of p16INK4a expression in malignant lymphomas is usually associated with altered methylation patterns or deletion of the gene,55,56 it is formally possible that BMI-1 overexpression is an additional contributing factor. However, our data show that expression of BMI-1 in H/RS tumor cells is not necessarily related to p16INK4a down-regulation, because more than half of the HL cases expressed BMI-1 in combination with p16INK4a. Although this suggests that human p16INK4a may not be regulated by BMI-1, these results should be interpreted with caution because regulation of individual target genes of Bmi-1 may be cell type-specific.57 Indeed, recent studies of lung and penile carcinomas58,59 reported an inverse correlation between the presence of BMI-1 and the absence of p16INK4a. It is theoretically possible that an effect of BMI-1 on p16INK4a in neoplastic cells of HL may be masked by the expression of other PcG genes. Several of these are associated with malignant transformation through mechanisms that do not involve p16INK4a. Enhanced expression of RING-1, for instance, resulted in oncogenic transformation and up-regulation of proto-oncogenes c-jun and c-fos,22 but Mel-18 seems to function as a tumor suppressor gene in the mouse.52 A further complication is that several PcG genes detected in H/RS cells, such as HPC2, are normally expressed by proliferating B cells (this study, and van Galen et al, submitted). However, it is unclear whether the level of HPC2 expression is altered in tumor cells. This is an important issue to resolve because interference with the expression or function of HPC2 is associated with abnormal cell cycle regulation.43 In addition, enhanced expression of EZH2 was recently associated with mantle-cell lymphomas48 and progression of prostate and breast tumors,60–62 suggesting that this PcG gene may also contribute to development of neoplastic H/RS cells.
Concluding Remarks
The importance of PcG proteins in maintenance of cell identity is underscored by the relationship between abnormal expression of PcG genes and malignant transformation in experimental model systems. In the current study we demonstrated that neoplastic cells in HL abnormally express multiple PcG genes. Although this suggests that the individual regulation of several PcG genes is disturbed, it is also possible that a regulatory mechanism shared by multiple PcG genes is affected. A potential application of the observed PcG expression patterns in HL is improved diagnostics. For instance, expression of HPC1 by H-RS cells may possibly be used to distinguish HL from CD30+ ALCL. Preliminary results of PcG expression patterns in T-NHL showed that CD30+ ALCL are HPC1 negative (van Galen et al, in preparation), but this finding needs to be confirmed in a larger patient population. We propose that abnormal PcG expression in HL results in an altered composition of the PRC1/HPC-HPH PcG complex in H/RS cells, which probably affects expression of target genes involved in regulation of apoptosis and/or the cell cycle. Our observations add to the increasing evidence that PcG genes are important contributors to development of hematological malignancies.
Footnotes
Address reprint requests to Frank M. Raaphorst, Ph.D., Department of Pathology, VU Medical Center, De Boelelaan 1117, 1081 HV Amsterdam, The Netherlands. E-mail: fm.raaphorst@vumc.nl.
References
- Diehl V, von Kalle C, Fonatsch C, Tesch H, Juecker M, Schaadt M. The cell of origin in Hodgkin’s disease. Semin Oncol. 1990;17:660–672. [PubMed] [Google Scholar]
- Cossman J, Annunziata CM, Barash S, Staudt L, Dillon P, He WW, Ricciardi-Castagnoli P, Rosen CA, Carter KC. Reed-Sternberg cell genome expression supports a B-cell lineage. Blood. 1999;94:411–416. [PubMed] [Google Scholar]
- Foss HD, Reusch R, Demel G, Lenz G, Anagnostopoulos I, Hummel M, Stein H. Frequent expression of the B-cell-specific activator protein in Reed-Sternberg cells of classical Hodgkin’s disease provides further evidence for its B-cell origin. Blood. 1999;94:3108–3113. [PubMed] [Google Scholar]
- Carbone A, Gloghini A, Gaidano G, Franceschi S, Capello D, Drexler HG, Falini B, Dalla-Favera R. Expression status of BCL-6 and syndecan-1 identifies distinct histogenetic subtypes of Hodgkin’s disease. Blood. 1998;92:2220–2228. [PubMed] [Google Scholar]
- Gruss HJ, Dower SK. Tumor necrosis factor ligand superfamily: involvement in the pathology of malignant lymphomas. Blood. 1995;85:3378–3404. [PubMed] [Google Scholar]
- Oudejans JJ, Jiwa NM, Meijer CJ. Epstein-Barr virus in Hodgkin’s disease: more than just an innocent bystander. J Pathol. 1997;181:353–356. doi: 10.1002/(SICI)1096-9896(199704)181:4<353::AID-PATH782>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Brink AA, Oudejans JJ, van den Brule AJ, Kluin PM, Horstman A, Ossenkoppele GJ, van Heerde P, Jiwa M, Meijer CJ. Low p53 and high bcl-2 expression in Reed-Sternberg cells predicts poor clinical outcome for Hodgkin’s disease: involvement of apoptosis resistance? Mod Pathol. 1998;11:376–383. [PubMed] [Google Scholar]
- Chu WS, Aguilera NS, Wei MQ, Abbondanzo SL. Antiapoptotic marker Bcl-X(L), expression on Reed-Sternberg cells of Hodgkin’s disease using a novel monoclonal marker, YTH-2H12. Hum Pathol. 1999;30:1065–1070. doi: 10.1016/s0046-8177(99)90224-1. [DOI] [PubMed] [Google Scholar]
- Raaphorst FM, van Kemenade FJ, Blokzijl T, Fieret E, Hamer KM, Satijn DP, Otte AP, Meijer CJ. Coexpression of BMI-1 and EZH2 polycomb group genes in Reed-Sternberg cells of Hodgkin’s disease. Am J Pathol. 2000;157:709–715. doi: 10.1016/S0002-9440(10)64583-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaphorst FM, Otte AP, Meijer CJ. Polycomb-group genes as regulators of mammalian lymphopoiesis. Trends Immunol. 2001;22:682–690. doi: 10.1016/s1471-4906(01)02082-8. [DOI] [PubMed] [Google Scholar]
- Simon J. Locking in stable states of gene expression: transcriptional control during Drosophila development. Curr Opin Cell Biol. 1995;7:376–385. doi: 10.1016/0955-0674(95)80093-x. [DOI] [PubMed] [Google Scholar]
- Lessard J, Sauvageau G. Polycomb group genes as epigenetic regulators of normal and leukemic hemopoiesis. Exp Hematol. 2003;31:567–585. doi: 10.1016/s0301-472x(03)00081-x. [DOI] [PubMed] [Google Scholar]
- Caldas C, Aparicio S. Cell memory and cancer: the story of the trithorax and Polycomb group genes. Cancer Metastasis Rev. 1999;18:313–329. doi: 10.1023/a:1006333610078. [DOI] [PubMed] [Google Scholar]
- Jacobs JJ, van Lohuizen M. Cellular memory of transcriptional states by Polycomb-group proteins. Semin Cell Dev Biol. 1999;10:227–235. doi: 10.1006/scdb.1999.0304. [DOI] [PubMed] [Google Scholar]
- Jacobs JJ, van Lohuizen M. Polycomb repression: from cellular memory to cellular proliferation and cancer. Biochim Biophys Acta. 2002;1602:151–161. doi: 10.1016/s0304-419x(02)00052-5. [DOI] [PubMed] [Google Scholar]
- van Lohuizen M. Functional analysis of mouse Polycomb group genes. Cell Mol Life Sci. 1998;54:71–79. doi: 10.1007/s000180050126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takihara Y, Hara J. Polycomb-group genes and hematopoiesis. Int J Hematol. 2000;72:165–172. [PubMed] [Google Scholar]
- Otte AP, Kwaks TH. Gene repression by Polycomb group protein complexes: a distinct complex for every occasion? Curr Opin Genet Dev. 2003;13:448–454. doi: 10.1016/s0959-437x(03)00108-4. [DOI] [PubMed] [Google Scholar]
- Levine SS, Weiss A, Erdjument-Bromage H, Shao Z, Tempst P, Kingston RE. The core of the polycomb repressive complex is compositionally and functionally conserved in flies and humans. Mol Cell Biol. 2002;22:6070–6078. doi: 10.1128/MCB.22.17.6070-6078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardos JI, Saurin AJ, Tissot C, Duprez E, Freemont PS. HPC3 is a new human polycomb orthologue that interacts and associates with RING1 and Bmi1 and has transcriptional repression properties. J Biol Chem. 2000;275:28785–28792. doi: 10.1074/jbc.M001835200. [DOI] [PubMed] [Google Scholar]
- Shao Z, Raible F, Mollaaghababa R, Guyon JR, Wu CT, Bender W, Kingston RE. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell. 1999;98:37–46. doi: 10.1016/S0092-8674(00)80604-2. [DOI] [PubMed] [Google Scholar]
- Satijn DP, Otte AP. RING1 interacts with multiple Polycomb-group proteins and displays tumorigenic activity. Mol Cell Biol. 1999;19:57–68. doi: 10.1128/mcb.19.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto N, Brock HW, Nomura M, Kyba M, Hodgson J, Fujita Y, Takihara Y, Shimada K, Higashinakagawa T. RAE28, BMI1, and M33 are members of heterogeneous multimeric mammalian Polycomb group complexes. Biochem Biophys Res Commun. 1998;245:356–365. doi: 10.1006/bbrc.1998.8438. [DOI] [PubMed] [Google Scholar]
- Kyba M, Brock HW. The Drosophila polycomb group protein Psc contacts ph and Pc through specific conserved domains. Mol Cell Biol. 1998;18:2712–2720. doi: 10.1128/mcb.18.5.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoorlemmer J, Marcos-Gutierrez C, Were F, Martinez R, Garcia E, Satijn DP, Otte AP, Vidal M. Ring1A is a transcriptional repressor that interacts with the Polycomb-M33 protein and is expressed at rhombomere boundaries in the mouse hindbrain. EMBO J. 1997;16:5930–5942. doi: 10.1093/emboj/16.19.5930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunster MJ, Satijn DP, Hamer KM, den Blaauwen JL, de Bruijn D, Alkema MJ, van Lohuizen M, van Driel R, Otte AP. Identification and characterization of interactions between the vertebrate polycomb-group protein BMI1 and human homologs of polyhomeotic. Mol Cell Biol. 1997;17:2326–2335. doi: 10.1128/mcb.17.4.2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkema MJ, Bronk M, Verhoeven E, Otte A, van’t Veer LJ, Berns A, van Lohuizen M. Identification of Bmi1-interacting proteins as constituents of a multimeric mammalian polycomb complex. Genes Dev. 1997;11:226–240. doi: 10.1101/gad.11.2.226. [DOI] [PubMed] [Google Scholar]
- Satijn DP, Hamer KM, den Blaauwen J, Otte AP. The polycomb group protein EED interacts with YY1, and both proteins induce neural tissue in Xenopus embryos. Mol Cell Biol. 2001;21:1360–1369. doi: 10.1128/MCB.21.4.1360-1369.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng J, Hart CM, Morgan K, Simon JA. A Drosophila ESC-E(Z) protein complex is distinct from other polycomb group complexes and contains covalently modified ESC. Mol Cell Biol. 2000;20:3069–3078. doi: 10.1128/mcb.20.9.3069-3078.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie F, Furuyama T, Harte PJ. The Drosophila Polycomb group proteins ESC and E(Z) bind directly to each other and co-localize at multiple chromosomal sites. Development. 1998;125:3483–3496. doi: 10.1242/dev.125.17.3483. [DOI] [PubMed] [Google Scholar]
- Sewalt RG, van d V, Gunster MJ, Hamer KM, den Blaauwen JL, Satijn DP, Hendrix T, van Driel R, Otte AP. Characterization of interactions between the mammalian polycomb-group proteins Enx1/EZH2 and EED suggests the existence of different mammalian polycomb-group protein complexes. Mol Cell Biol. 1998;18:3586–3595. doi: 10.1128/mcb.18.6.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lohuizen M, Tijms M, Voncken JW, Schumacher A, Magnuson T, Wientjens E. Interaction of mouse polycomb-group (Pc-G) proteins Enx1 and Enx2 with Eed: indication for separate Pc-G complexes. Mol Cell Biol. 1998;18:3572–3579. doi: 10.1128/mcb.18.6.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K, Sonoda M, Inokuchi J, Shirasawa S, Sasazuki T. Polycomb group, suppressor of Zeste 12, links heterochromatin protein 1α and enhancer of Zeste 2. J Biol Chem. 2004;279:401–406. doi: 10.1074/jbc.M307344200. [DOI] [PubMed] [Google Scholar]
- Breiling A, Turner BM, Bianchi ME, Orlando V. General transcription factors bind promoters repressed by Polycomb group proteins. Nature. 2001;412:651–655. doi: 10.1038/35088090. [DOI] [PubMed] [Google Scholar]
- Saurin AJ, Shao Z, Erdjument-Bromage H, Tempst P, Kingston RE. A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature. 2001;412:655–660. doi: 10.1038/35088096. [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal polycomb sites. Cell. 2002;111:185–196. doi: 10.1016/s0092-8674(02)00975-3. [DOI] [PubMed] [Google Scholar]
- Czermin B, Imhof A. The sounds of silence: histone deacetylation meets histone methylation. Genetica. 2003;117:159–164. doi: 10.1023/a:1022927725945. [DOI] [PubMed] [Google Scholar]
- Sewalt RG, Lachner M, Vargas M, Hamer KM, den Blaauwen JL, Hendrix T, Melcher M, Schweizer D, Jenuwein T, Otte AP. Selective interactions between vertebrate polycomb homologs and the SUV39H1 histone lysine methyltransferase suggest that histone H3-K9 methylation contributes to chromosomal targeting of polycomb group proteins. Mol Cell Biol. 2002;22:5539–5553. doi: 10.1128/MCB.22.15.5539-5553.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Vlag J, Otte AP. Transcriptional repression mediated by the human polycomb-group protein EED involves histone deacetylation. Nat Genet. 1999;23:474–478. doi: 10.1038/70602. [DOI] [PubMed] [Google Scholar]
- Alkema MJ, Jacobs H, van Lohuizen M, Berns A. Pertubation of B and T cell development and predisposition to lymphomagenesis in Emu Bmi1 transgenic mice require the Bmi1 RING finger. Oncogene. 1997;15:899–910. doi: 10.1038/sj.onc.1201262. [DOI] [PubMed] [Google Scholar]
- Jacobs JJ, Scheijen B, Voncken JW, Kieboom K, Berns A, van Lohuizen M. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 1999;13:2678–2690. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- Satijn DP, Olson DJ, van d V, Hamer KM, Lambrechts C, Masselink H, Gunster MJ, Sewalt RG, van Driel R, Otte AP. Interference with the expression of a novel human polycomb protein, hPc2, results in cellular transformation and apoptosis. Mol Cell Biol. 1997;17:6076–6086. doi: 10.1128/mcb.17.10.6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessard J, Schumacher A, Thorsteinsdottir U, van Lohuizen M, Magnuson T, Sauvageau G. Functional antagonism of the Polycomb-group genes eed and Bmi1 in hemopoietic cell proliferation. Genes Dev. 1999;13:2691–2703. doi: 10.1101/gad.13.20.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaphorst FM, van Kemenade FJ, Fieret E, Hamer KM, Satijn DP, Otte AP, Meijer CJ. Cutting edge: polycomb gene expression patterns reflect distinct B-cell differentiation stages in human germinal centers. J Immunol. 2000;164:1–4. doi: 10.4049/jimmunol.164.1.1. [DOI] [PubMed] [Google Scholar]
- Raaphorst FM, Otte AP, van Kemenade FJ, Blokzijl T, Fieret E, Hamer KM, Satijn DP, Meijer CJ. Distinct bmi-1 and ezh2 expression patterns in thymocytes and mature t cells suggest a role for polycomb genes in human t-cell differentiation. J Immunol. 2001;166:5925–5934. doi: 10.4049/jimmunol.166.10.5925. [DOI] [PubMed] [Google Scholar]
- van Kemenade FJ, Raaphorst FM, Blokzijl T, Fieret E, Hamer KM, Satijn DP, Otte AP, Meijer CJ. Coexpression of BMI-1 and EZH2 polycomb-group proteins is associated with cycling cells and degree of malignancy in B-cell non-Hodgkin lymphoma. Blood. 2001;97:3896–3901. doi: 10.1182/blood.v97.12.3896. [DOI] [PubMed] [Google Scholar]
- Visser HP, Gunster MJ, Kluin-Nelemans HC, Manders EM, Raaphorst FM, Meijer CJ, Willemze R, Otte AP. The Polycomb group protein EZH2 is up-regulated in proliferating, cultured human mantle-cell lymphoma. Br J Haematol. 2001;112:950–958. doi: 10.1046/j.1365-2141.2001.02641.x. [DOI] [PubMed] [Google Scholar]
- Diehl V, Pfreundschuh M, Fonatsch C, Stein H, Falk M, Burrichter H, Schaadt M. Phenotypic and genotypic analysis of Hodgkin’s disease-derived cell lines: histopathological and clinical implications. Cancer Surv. 1985;4:399–419. [PubMed] [Google Scholar]
- Satijn DP, Gunster MJ, van d V, Hamer KM, Schul W, Alkema MJ, Saurin AJ, Freemont PS, van Driel R, Otte AP. RING1 is associated with the polycomb group protein complex and acts as a transcriptional repressor. Mol Cell Biol. 1997;17:4105–4113. doi: 10.1128/mcb.17.7.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer KM, Sewalt RG, den Blaauwen JL, Hendrix T, Satijn DP, Otte AP. A panel of monoclonal antibodies against human Polycomb group proteins. Hybrid Hybridomics. 2002;21:245–252. doi: 10.1089/153685902760213859. [DOI] [PubMed] [Google Scholar]
- Kanno M, Hasegawa M, Ishida A, Isono K, Taniguchi M. mel-18, a Polycomb group-related mammalian gene, encodes a transcriptional negative regulator with tumor suppressive activity. EMBO J. 1995;14:5672–5678. doi: 10.1002/j.1460-2075.1995.tb00254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown JL, Mucci D, Whiteley M, Dirksen ML, Kassis JA. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell. 1998;1:1057–1064. doi: 10.1016/s1097-2765(00)80106-9. [DOI] [PubMed] [Google Scholar]
- Mohd-Sarip A, Venturini F, Chalkley GE, Verrijzer CP. Pleiohomeotic can link polycomb to DNA and mediate transcriptional repression. Mol Cell Biol. 2002;22:7473–7483. doi: 10.1128/MCB.22.21.7473-7483.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia MJ, Martinez-Delgado B, Cebrian A, Martinez A, Benitez J, Rivas C. Different incidence and pattern of p15INK4b and p16INK4a promoter region hypermethylation in Hodgkin’s and CD30-positive non-Hodgkin’s lymphomas. Am J Pathol. 2002;161:1007–1013. doi: 10.1016/S0002-9440(10)64261-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler HG. Review of alterations of the cyclin-dependent kinase inhibitor INK4 family genes p15, p16, p18, and p19 in human leukemia-lymphoma cells. Leukemia. 1998;12:845–859. doi: 10.1038/sj.leu.2401043. [DOI] [PubMed] [Google Scholar]
- Dimri GP, Martinez JL, Jacobs JJ, Keblusek P, Itahana K, van Lohuizen M, Campisi J, Wazer DE, Band V. The Bmi-1 oncogene induces telomerase activity and immortalizes human mammary epithelial cells. Cancer Res. 2002;62:4736–4745. [PubMed] [Google Scholar]
- Vonlanthen S, Heighway J, Altermatt HJ, Gugger M, Kappeler A, Borner MM, Lohuizen M, Betticher DC. The bmi-1 oncoprotein is differentially expressed in non-small cell lung cancer and correlates with INK4A-ARF locus expression. Br J Cancer. 2001;84:1372–1376. doi: 10.1054/bjoc.2001.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreux E, Lont AP, Horenblas S, Gallee MP, Raaphorst FM, Von Knebel DM, Meijer CJ, Snijders PJ. Evidence for at least three alternative mechanisms targeting the p16INK4A/cyclin D/Rb pathway in penile carcinoma, one of which is mediated by high-risk human papillomavirus. J Pathol. 2003;201:109–118. doi: 10.1002/path.1394. [DOI] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, Sabel MS, Livant D, Weiss SJ, Rubin MA, Chinnaiyan AM. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci USA. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaTulippe E, Satagopan J, Smith A, Scher H, Scardino P, Reuter V, Gerald WL. Comprehensive gene expression analysis of prostate cancer reveals distinct transcriptional programs associated with metastatic disease. Cancer Res. 2002;62:4499–4506. [PubMed] [Google Scholar]