Abstract
Arbitrarily-primed DNA markers can be very useful for genetic fingerprinting and for facilitating positional cloning of genes. This class of technologies is particularly important for less studied species, for which genome sequence information is generally not known. The technologies include Randomly Amplified Polymorphic DNA (RAPD), DNA Amplification Fingerprinting (DAF), and Amplified Fragment Length Polymorphism (AFLP). We have modified the DAF protocol to produce a robust PCR-based DNA marker technology called Randomly Amplified DNA Fingerprinting (RAF). While the protocol most closely resembles DAF, it is much more robust and sensitive because amplicons are labelled with either radioactive 33P or fluorescence in a 30-cycle PCR, and then separated and detected on large polyacrylamide sequencing gels. Highly reproducible RAF markers were readily amplified from either purified DNA or alkali-treated intact leaf tissue. RAF markers typically display dominant inheritance. However, a small but significant portion of the RAF markers exhibit codominant inheritance and represent microsatellite loci. RAF compares favorably with AFLP for efficiency and reliability on many plant genomes, including the very large and complex genomes of sugarcane and wheat. While the two technologies detect about the same number of markers per large polyacrylamide gel, advantages of RAF over AFLP include: (i) no requirement for enzymatic template preparation, (ii) one instead of two PCRs, and (iii) overall cost. RAF and AFLP were shown to differ in the selective basis of amplification of markers from genomes and could therefore be used in complementary fashion for some genetic studies.
INTRODUCTION
DNA markers can be used for genetic fingerprinting, estimating genetic diversity, marker-assisted selection in plant and animal breeding, and facilitating the map-based cloning of genes [1]. Southern detection of Restriction Fragment Length Polymorphism (RFLP) was the first DNA marker technology [1], but it has generally been superceded by PCR-based protocols. These PCR-based methods include simple sequence repeats (SSRs) [2] and arbitrarily-primed DNA marker amplification [3, 4, 5, 6, 7, 8, 9]. SSRs (also known as microsatellites) have the advantages of increased levels of polymorphism and codominant inheritance that allow detection of heterozygotes. However, these markers are typically laborious to produce, involving the initial cloning and sequencing of every locus [2]. The advantage of arbitrarily-primed DNA marker technologies, on the other hand, is that no prior knowledge of DNA sequence is required, and therefore markers can be quickly amplified and detected from any organism.
Several protocols based on arbitrary amplification of DNA markers have been reported in the literature. Randomly Amplified Polymorphic DNA (RAPD) [3] and Arbitrarily Primed PCR (AP-PCR) [4] use relatively low concentrations (eg, 0.2 μmol/L) of single short oligonucleotide primers in the PCR. The annealing temperature for these protocols generally ranges from 37-40°C, and up to 20 markers can be simultaneously amplified and detected. While the technologies are simple, not technically demanding and require only agarose gel electrophoresis for detection, a major problem has been lack of reproducibility between laboratories [10, 11]. DNA Amplification Fingerprinting (DAF) also implements a single short oligonucleotide primer but at a higher concentration (5 μmol/L), and higher annealing temperatures (53-57°C) are used in the PCR [5, 6, 7]. In addition, the DAF protocol uses the DNA polymerase Stoffel Fragment, and small silver nitrate-stained polyacrylamide gels for fragment detection [6]. About 40 markers can be detected per PCR [7], and we have found DAF to be much more reliable and transferable between laboratories than RAPD (Waldron et al, unpublished data, December 1999).
Amplified Fragment Length Polymorphism (AFLP) is a PCR-based method for arbitrarily amplifying restriction fragments [9, 12]. In contrast to other protocols, template for AFLP is prepared by digesting purified genomic DNA with two restriction endonucleases, followed by ligation of corresponding adaptors to the digested genomic DNA. Pairs of oligonucleotide primers complementary to the adaptor sequences but with one to four additional 3′ nucleotides, are used in PCR [9, 12]. For AFLP on large genomes, a two-step PCR is implemented with annealing temperatures starting at 65°C and stepping down to 55°C. In the first PCR, the two AFLP primers are composed of the AFLP adaptor sequences plus one additional 3′ nucleotide. The products of the first PCR are then diluted and used as template in the second PCR, wherein the oligonucleotide primers generally have three or four additional 3′ nucleotides. One of the primers is labelled with a radioactive 33P [9, 12] or a fluorescent tag [13] for the detection of the markers after amplification and separation on large polyacrylamide sequencing gels. The major advantage of AFLP over the earlier arbitrarily-primed PCR protocols described above, is that up to 100 markers can be detected per PCR [9, 12]. Major disadvantages are the additional steps involved in AFLP template preparation and the two-step PCR required for large and complex genomes. The additional 3′ nucleotides on AFLP primers are commonly referred to as 3′ “selective” nucleotides, and depending on their sequence are thought to arbitrarily, but selectively, amplify DNA markers from genomes [9].
In this paper, we describe several modifications of the DAF protocol that greatly increase the efficiency of detecting arbitrarily-primed DNA markers. The new protocol is called Randomly Amplified DNA Fingerprinting (RAF) and represents a culmination of the DNA marker technologies based on arbitrarily-primed PCR. We compared the efficiency, robustness, and cost of the RAF and AFLP protocols and demonstrated that the two technologies differ in the selective basis of amplification of DNA markers from plant genomes.
MATERIALS AND METHODS
Plant genotypes
Plant genotypes used in the study included Lycopersicon esculentum (tomato) cultivar Moneymaker [14], Arabidopsis thaliana C24, several Glycine max (soybean) and Gycine soja acessions [15, 16], Triticum aestivum (wheat) cultivar Hartog, sugarcane hybrids Q117 and 79A362, F1 progeny from a cross between sugarcane hybrids 79N1396 and Q161, and macadamia cultivars Keauhou and A16 and their F1 progeny [17].
Preparation of genomic DNA and alkali-treated leaf tissue for PCR
DNA was purified from young leaf tissue of tomato, Arabidopsis, soybean, wheat, and sugarcane by using the method described by Carroll et al [14]. For macadamia, DNA was purified from leaves as described by Peace [17]. Alkali-treated intact leaf tissue [14, 18] from tomato and sugarcane (rather than purified DNA) was also used as a template to generate RAF profiles. A barely visible piece of alkali-treated leaf tissue was used as a crude template in PCR.
All marker profiles were produced at least in duplicate from replicate DNA extractions or replicate alkali-treated intact leaf tissue.
DAF protocol
DNA amplification fingerprinting (DAF) was performed on several plant species/genotypes using the protocol described by [7].
RAF protocol
Randomly Amplified DNA Fingerprinting (RAF) reactions were prepared in 10 μL volumes on ice. Each PCR contained 1x DAF buffer (10 mmol/L Tris pH8, 10 mmol/L KCl, 5 mmol/L MgCl2) [7], 1.5 units of DNA polymerase Stoffel Fragment (Applied Biosystems), 20 μmol/L dNTPs, 1μCi α-labelled 33P-dATP, and 5 μmol/L of a single oligonucleotide primer of ten nucleotides. Oligonucleotides from the Operon Technologies Inc. A and K kits were generally used, however additional ones were designed (sequences listed below) and purchased from Genset Pacific Pty. Ltd. (Lismore, Australia). Undigested genomic DNA (1 to 500 ng) or alkali-treated intact leaf tissue was used as DNA template in the PCR. PCR was performed with a hot start (85°C), followed by a denaturation at 94°C for 5 min, then 30 cycles of 94°C for 30 secs and 60 sec at each of 57°C, 56°C, 55°C, 54°C, and 53°C. The PCR was concluded with a final extension step at 72°C for 5 min. For wheat, which has a considerably larger genome than the other species analyzed [19], the annealing/extension temperatures were 2°C higher (ie, 30 cycles of 94°C for 30 secs and 60 sec at each of 59°C, 58°C, 57°C, 56°C, and 55°C). The radio-labelled PCR products were separated on 4 or 5% polyacrylamide sequencing gels, and the dried gels were then exposed to photographic film for 6 hours or overnight. For some experiments, an FAM (6-carboxy-fluorescein)-tagged oligonucleotide (purchased from Genset Pacific Pty. Ltd., Lismore, Australia) or a fluorescent deoxynucleoside triphosphate (flourescent dCTP R110, Perkin Elmer cat. no. 402175) was used in the PCR instead of radio-labeling. In these cases, the fluorescent RAF profiles were detected on an Applied Biosystem 373A DNA sequencer.
The nature of selectivity of RAF fragment amplification was investigated by assessing the extent of sequence homology between RAF products amplified with primers that varied in one of the last three 3′ nucleotides. For these experiments, the following two series of oligonucleotide primers (K-01 and K-02) were synthesized:
K-01 5′-CATTCGAGCC-3′ K-02 5′-GTCTCCGCAA-3′ K-01a 5′-CATTCGAGCA-3′ K-02a 5′-GTCTCCGCAC-3′ K-01b 5′-CATTCGAGCG-3′ K-02b 5′-GTCTCCGCAG-3′ K-01c 5′-CATTCGAGAC-3′ K-02c 5′-GTCTCCGCCT-3′ K-01d 5′-CATTCGAGTC-3′ K-02d 5′-GTCTCCGCCA-3′ K-01e 5′-CATTCGACCC-3′ K-02e 5′-GTCTCCGCGA-3′ K-01f 5′-CATTCGAACC-3′ K-02f 5′-GTCTCCGAAA-3′ K-01g 5′-CATTCGATCC-3′ K-02g 5′-GTCTCCGTAA-3′ K-02h 5′-GTCTCCGGAA-3′
The oligonucleotides within a series varied in just one of the last three nucelotides (variant nucleotides from the original primer sequence are underlined above). These primers were used to generate the standard radio-labelled RAF profile, but unlabelled RAF products were also amplified in parallel. The unlabelled RAF products were separated on a 2% agarose gel, blotted onto a nylon membrane and then probed with radio-labelled RAF products amplified with the original primer of the series (K-01 or K-02). To determine if the RAF products represented highly repetitive DNA, 1 μg of MseI-digested genomic DNA was usually included on the agarose gel. ØX174 digested with Hinf1 (500 ng) (Promega, Annandale, NSW, Australia, cat. no. E3511) was included on the gel as a size marker. Hybridization of the probe to the membrane was performed as described by [20]. To detect the size marker, dephosphorylated ØX174 digested with Hinf1, was kinased with 33P [20] and included with the RAF probe in the hybridization. Following washes at high stringency (0.1x SSPE and 0.1% SDS at 65°C), the membrane was exposed to photographic film to detect the hybridization of the probe.
AFLP protocols
The original AFLP protocol [9, 12] was modified to investigate the selective nature of the amplification in the first PCR of the protocol. In one modification, the first PCR was the same as the standard protocol with both oligonucleotide primers having one additional 3′ nucleotide (A or T) [9]. However, in the second PCR, while both primers had three additional 3′ nucleotides, the first one did not correspond to the additional nucleotide used in the first PCR. In another modification, three additional 3′ nucleotides (instead of one) were used on the primers in the first PCR, and then the same three or three different additional 3′ nucleotides were used on the primers in the second PCR.
To further investigate the selective nature of the amplification in the first PCR of the AFLP protocol, the extent of homology between first PCR products amplified with different combinations of additional 3′ nucleotides on the primers was assessed by Southern hybridization analysis. For these experiments, the EcoRI oligonucleotide primer E36, with three additional 3′ nucleotides (5′-GACTGCGTACCAATTCACC-3′), was used in a first PCR together with a range of MseI primers, each with a different combination of three additional 3′ nucleotides. The amplified products were separated on a 2% agarose gel and then blotted onto a nylon membrane. The membrane was then hybridized to a probe derived from the first PCR products amplified using the E36 primer with one of the MseI primers. To determine if the first PCR products represented highly repetitive genomic DNA, 500 or 1000 ng of MseI-digested genomic DNA was also included on the gel and blotted onto the membrane. To label the probe, eight standard 20 μL AFLP first PCRs were prepared, except that 2.5 μCi α-labelled 33P-dATP was included in each PCR. The eight PCRs were combined as the probe and hybridized onto the membrane. Hybridization of the probe to the membrane was performed as described by [20]. To detect the size marker, dephosphorylated ØX174 digested with Hinf1, was kinased with 33P [20] and included with the AFLP probe in the hybridization. Following washes at high stringency (0.1x SSPE and 0.1% SDS at 65°C), the membrane was exposed to photographic film to detect hybridization of the probe.
RESULTS
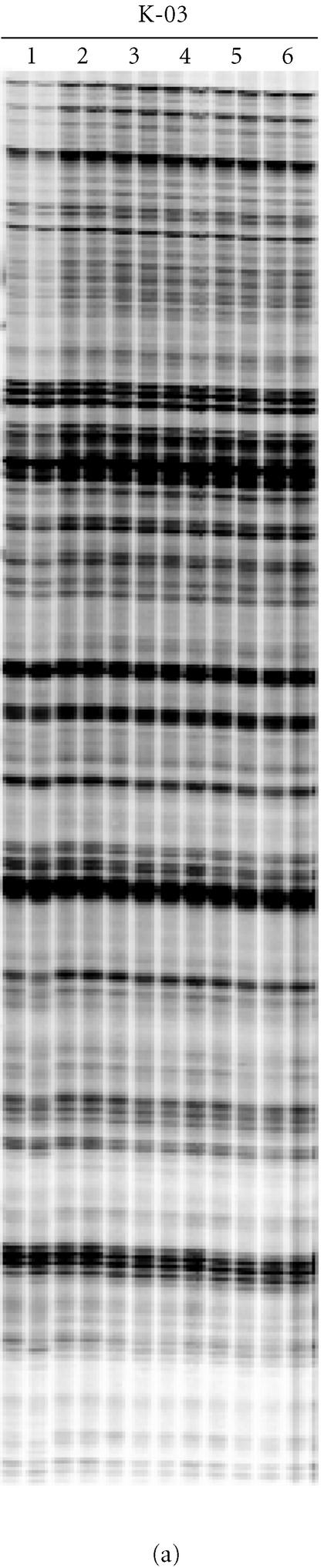
Highly reproducible RAF profiles were generated from replicate DNA extractions for all plant species tested, including tomato, wheat, sugarcane, macadamia, and Arabidopsis. The size of RAF markers detected ranged from 40 to about 1,000 base pair (bp). Examples of the results are shown in Figure 1 for sugarcane, tomato, soybean, and wheat. Reliable and reproducible RAF profiles were also generated for a wide range of DNA template concentrations. For example in sugarcane, identical RAF profiles were generated for genomic DNA template concentrations ranging from 10 to 500 ng (Figure 1a). The standard RAF protocol was used for all plant species, except for wheat. The wheat genome is at least four times larger than the other genomes analyzed [19], and in this case, optimal RAF profiles were obtained by raising the annealing temperatures by 2°C (Figure 1c).
Figure 1.
RAF profiles amplified from sugarcane (a), tomato and soybean (b), and wheat (c). Radio-labelled profiles amplified from replicate ((a) and (b)) or triplicate (c) genomic DNA extractions were run side-by-side on the gel and given the same lane number. The Operon oligonucleotide primers used are also listed above the lanes. (a) RAF profiles amplified from different amounts of genomic DNA extracted from sugarcane hybrid 79A362. Lanes 1, 12.5 ng; lanes 2, 25 ng; lanes 3, 50 ng; lanes 4, 100 ng; lanes 5, 250 ng; and lanes 6, 500 ng. (b) RAF profiles amplified from tomato cv. Moneymaker (lanes 1), Glycine soja (lanes 2), soybean (G. max) cv. Bragg (lanes 3). (c) RAF profiles amplified from wheat cv. Hartog; lanes 1-5 represent individual Hartog plants.
Highly reproducible RAF profiles were also amplified from alkali-treated leaf tissue for both species tested, namely tomato and sugarcane, and the profiles were almost identical to those amplified from genomic template from the same genotype (Figure 2). Thus, DNA purification is not required to generate reproducible RAF profiles.
Figure 2.
RAF profiles amplified from genomic DNA and alkali-treated leaf tissue. Lanes 1 show the profiles from replicate DNA extractions from a sugarcane seedling, and lanes 2 show the profiles from replicate alkali-treated leaf tissue samples from the same seedling. Similarly, lanes 3 show the profiles from genomic DNA extractions of a tomato plant, and lanes 4 show the profiles from alkali-treated leaf tissue samples from the same plant. The Operon oligonucleotide primers used are listed above the lanes.
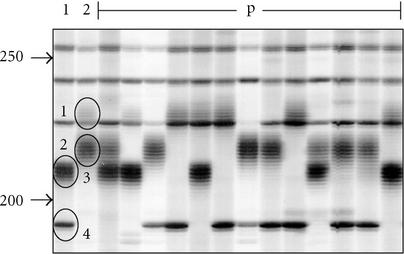
In contrast to the DAF protocol which detected about 40 markers per PCR, each RAF PCR detected between 70 and 100 DNA markers. As reported for all of the other arbitrarily-primed DNA markers [3, 4, 5, 6, 7, 8, 9], dominant Mendelian inheritance of the RAF markers was generally observed in mapping populations of several plant species including soybean, sugarcane, wheat, and macadamia (data not shown). However, about 1% of RAF markers in macadamia were shown to display a stuttering phenotype typical of simple sequence repeat (SSR) markers with codominant inheritance in a mapping population (Figure 3). The cloning and sequencing of several stuttered RAF markers confirmed that they contained simple sequence repeats (J. Neal, written communication, September 2001). RAF markers with stuttered phenotypes were also observed for other species at about the same frequency (at least one marker for every two primers) (see Figure 1).
Figure 3.
Close-up of autoradiograph of an RAF simple sequence repeat locus in macadamia. The first two lanes contain amplification products (Operon primer A-06) of two macadamia cultivars, and their F1 progeny (p) are represented in the rest of the lanes. The four alleles segregating in the cross are circled and numbered. Elsewhere, on the autoradiograph, can be seen some nonstuttering, nonpolymorphic RAF markers. Approximate fragments sizes (in bp) are indicated on the left by arrows.
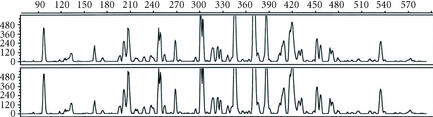
As with radio-labelled RAF profiles, highly reproducible fluorescent RAF profiles were detected from replicate DNA extractions. An example of replicate fluorescent RAF profiles generated with FAM-labelled oligonucleotide primer K-06 (5′-CACCTTTCCC-3′) for sugarcane hybrid Q117 is shown in Figure 4. RAF profiles were also produced by incorporation of R110 fluorescently labelled dCTP (Perkin Elmer, Melbourne, Vic, Australia, cat. no. 402175) into amplicons (data not shown). While these profiles were highly reproducible on replicate DNA extractions, sharper florescent profiles were obtained by incorporating the florescent tag into the oligonucleotide primer.
Figure 4.
Fluorescent detection of RAF profiles. The oligonucleotide primer K-06 (5′-CACCTTTCCC-3′) was synthesized and tagged with FAM (6-carboxy-fluorescein), and then used in the RAF protocol with genomic DNA from sugarcane hybrid Q117. After the PCR, the RAF profiles were separated and detected on an Applied Biosystem 373A DNA sequencer. The two panels represent RAF profiles amplified from replicate DNA extractions. The size of RAF markers (in bp) is shown on the X axis, and the relative intensity of the signals is shown on the Y axis. Larger RAF fragments were also amplified but these are not shown.
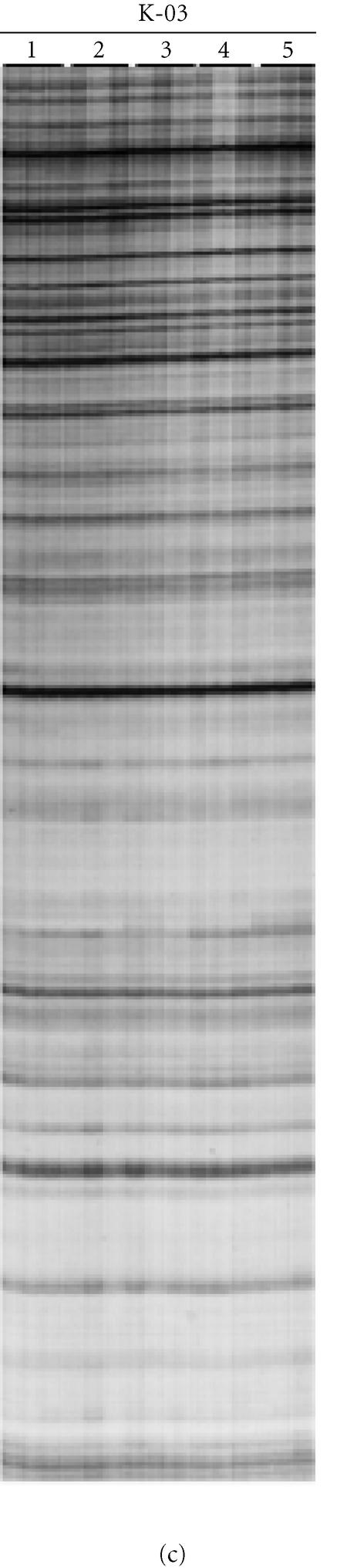
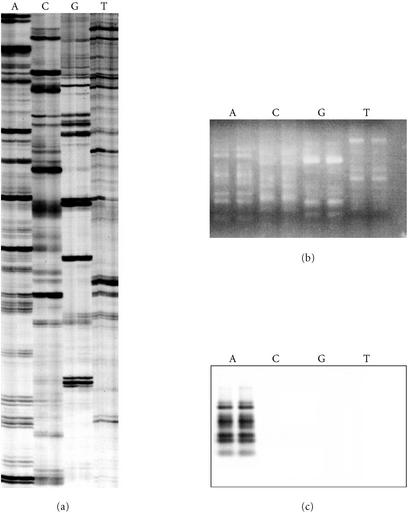
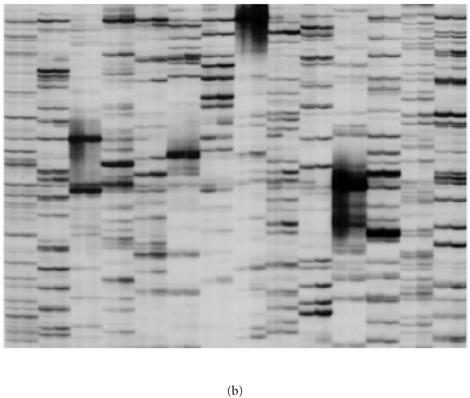
The nature of selectivity of RAF fragment amplification was also investigated with two series of oligonucleotide primers, that varied in one of the last three 3′ nucleotides. Varying the last nucleotide of the primer resulted in new and distinct RAF profiles (Figure 5a) that were non-homologous based on the Southern hybridization assay (Figure 5b). Similarly, varying the second or third last nucleotide resulted in new, distinct, nonhomologous RAF profiles (data not shown). Radio-labelled RAF fragments did not strongly hybridize to MseI-digested DNA, thereby indicating that RAF fragments do not represent highly repetitive DNA in the genome (data not shown).
Figure 5.
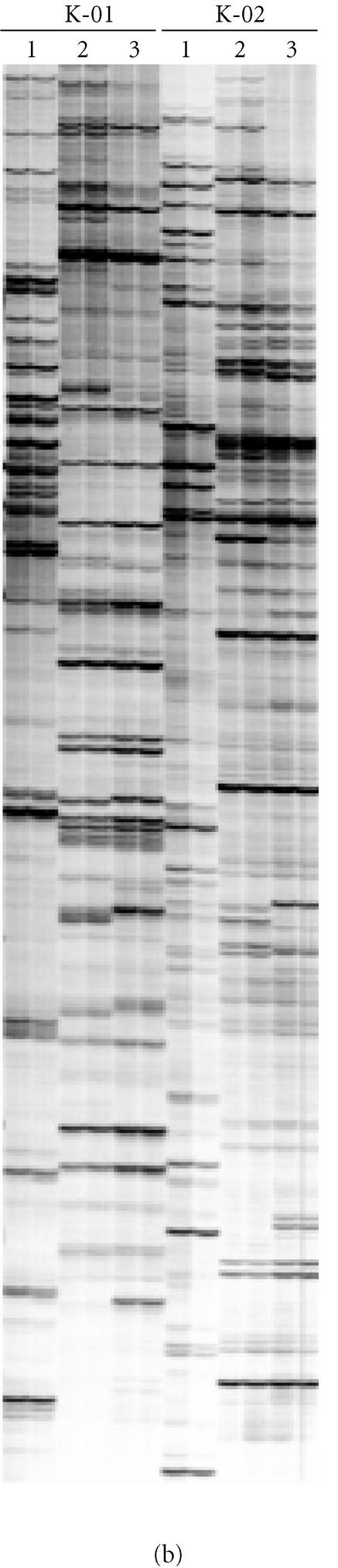
Selective amplification of RAF markers is dependent on the 3′ nucleotide of the oligonucleotide primer. (a) Radio-labelled RAF profiles for sugarcane hybrid 79A362 using oligonucleotide primers K-02 (5′-GTCTCCGCAA-3′) (lanes A), K-02a (5′-GTCTCCGCAC-3′) (lanes C), K-02b (5′-GTCTCCGCAG-3′) (lanes G), and K-02c (5′-GTCTCCGCAT-3′) (lanes T). These primers only varied in the final 3′ nucleotide. The profiles were amplified from replicate DNA extractions of sugarcane 79A362. (b) Identical reactions, also in replicate, were produced without radio-labelling, separated on a 2% agarose gel, ethidium-bromide stained, and photographed. (c) The unlabelled RAF fragments were then blotted from the agarose gel onto a nylon membrane and probed with radio-labelled RAF products amplified with K-02 (lanes A in panel (a)).
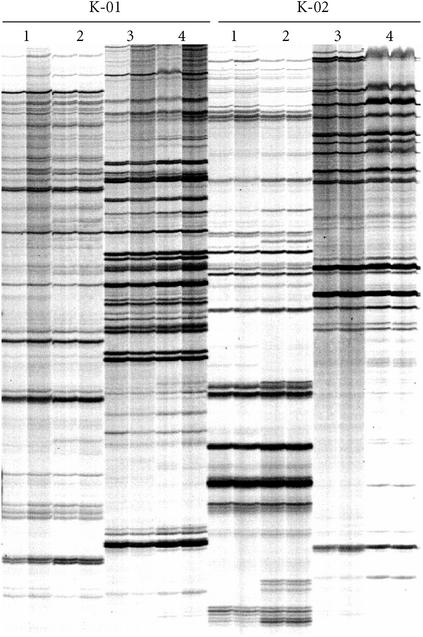
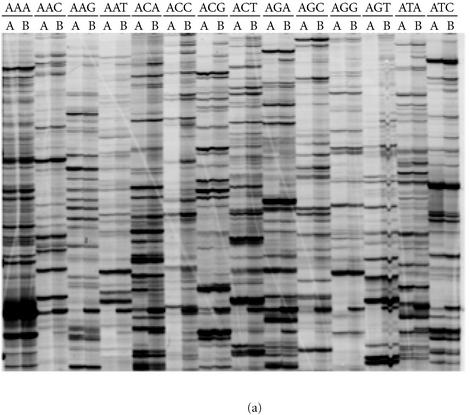
In contrast to the RAF protocol, we demonstrated that in the first PCR of the AFLP protocol, DNA amplification is not dependent on the terminal 3′ nucleotide(s) added to the AFLP primers. This conclusion was supported by results from two different experiments. Firstly, PCR products amplified with EcoRI AFLP primers that varied from each other in the last three 3′ nucleotides were shown to cross-hybridize in a Southern assay (Figure 6). Secondly, the employment of nonmatching combinations of additional 3′ nucleotides on EcoRI primers in the first and subsequent second PCR had little impact on the AFLP profile obtained in the second radioactive PCR (Figure 7). Essentially, the great majority of the AFLP markers that were detected after the second PCR were determined by the terminal 3′ nucleotides used in the second PCR, regardless of the terminal 3′ nucleotides used in the first PCR. The results were the same whether three additional 3′ nucleotides (Figure 7) or one additional 3′ nucleotide (data not shown) were incorporated into the AFLP primers for the first PCR.
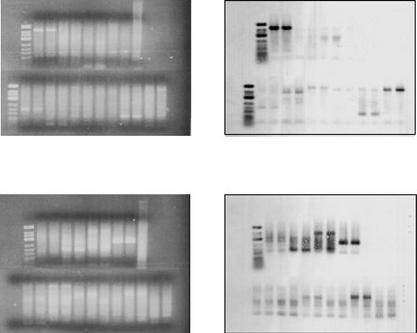
Figure 6.
Evidence for homology between first PCR AFLP products amplified with different oligonucleotide primers. PCR was performed on replicate DNA templates of sugarcane hybrid 79A362 ((a) and (b)) and the nod49 mutant soybean cv. Bragg ((c) and (d)). The EcoRI oligonucleotide primer E36 with three additional 3′ nucleotides (5′-GACTGCGTACCAATTCACC-3′) was used in the first PCR with a range of MseI primers, each with a different combination of three additional 3′ nucleotides. The three additional 3′ nucleotides on the MseI primers were as follows: lanes 1, ACA; lanes 2, ACC; lanes 3, ACG; lanes 4, ACT; lanes 6, AAA, lanes 7, AGA; lanes 8, ATA; lanes 9, CCA; lanes 10, GCA; and lanes 11, TCA. Lane 5 contained 500 ng of genomic DNA digested with MseI. The reactions were separated on 2% agarose gels ((a) and (c)) and blotted onto nylon membranes. A radioactive probe corresponding to the PCR in lanes 1 ((a) and (c)) was prepared (see Experimental procedures) and hybridized to the membrane. After stringent washing, hybridization to the probe was detected by autoradiography ((b) and (d)).
Figure 7.
AFLP amplification is dependent on the terminal 3′ nucleotide sequence of the AFLP primer in the second, but not in the first PCR. The EcoRI oligonucleotide primer E36 with three additional 3′ nucleotides (5′-GACTGCGTACCAATTCACC-3′) was used with a range of MseI primers, each with a different combination of three additional 3′ nucleotides. For all lanes marked A, the same MseI oligonucleotide primer was used in the first and second PCR. For all lanes marked B, one specific MseI primer, M31, was used in the first PCR, and then another MseI primer was used in the second PCR. The additional 3′ nucleotides on M31 were 3′-AAA-3′, and the additional 3′ nucleotides on the primer used in the second PCR to generate the AFLP profile are listed above the lanes (5′ to 3′). The AFLP profiles were generated for (a) sugarcane hybrid 79A362 and (b) soybean cv. Bragg.
The two main experimental applications for DNA marker technologies involve either DNA profiling on individual DNA samples or bulk segregant analysis to identify DNA markers linked to a gene of interest [21]. For both applications, we estimate that RAF is less expensive to perform than AFLP, particularly for fingerprinting individual plants (Table 1).
Table 1.
Comparative costs of template preparation and PCR in RAF and AFLP. The two research applications given are the equivalent of (i) profiling 2,000 DNA markers on 48 individual DNA samples, and (ii) using bulk segregant analysis to map 20,000 DNA markers for linkage to a gene of interest (four DNA samples; two parents and two bulks). Estimated costs are in Australian dollars ($A). Labour costs were estimated at $20 per hour. Electrophoresis and detection costs are the same for both protocols, and these costs were not included.
| Research application | DNA marker technology | Material costs per 20 gels ($A) | Labour costs per 20 gels ($A) | Combined costs ($A) | ||
| Template preparation | PCR | Template preparation | PCR | |||
| (i) DNA profiling on individuals | RAF | 0 | 340 | 0 | 200 | 540 |
| AFLP | 270 | 420 | 110 | 200 | 1000 | |
| (ii) Bulk segregant analysis | RAF | 0 | 340 | 0 | 200 | 540 |
| AFLP | 22 | 420 | 10 | 200 | 652 | |
DISCUSSION
DNA marker technologies based on arbitrarily-primed PCR are particularly important for less studied plant species for which whole genome sequences are not available. The RAF protocol described here represents a culmination of DNA marker protocols based on arbitrarily-primed PCR. Advantages of RAF over previous protocols include robustness and reliability, no requirement for highly-purified DNA template, relatively few steps required, the opportunity for sensitive detection via radio-labeling or fluorescent tagging, more markers being simultaneously detected, and the ability to identify codominant loci. The RAF protocol has been successfully and readily applied to all plant species we have tested thus far.
While the RAF protocol most closely resembles DAF [5, 6, 7, 8], it is much more efficient, robust, and sensitive because amplicons are labelled with either radioactive 33P or fluorescence in a relatively short single PCR, and then separated and detected on large polyacrylamide sequencing gels. The key aspects of the protocol are
(i) inclusion of high concentrations of a single oligonucleotide primer (10 nucleotides long), high annealing temperatures (53-59°C), and DNA polymerase Stoffel Fragment;
(ii) radioactive- or fluorescent-labeling of amplicons in a 30-cycle PCR; and
(iii) separation and detection of amplicons on large polyacrylamide sequencing gels or DNA sequencing machines.
The second two points listed above distinguish RAF from DAF, the latter of which involves more PCR cycles (35-45) and detection of markers on small, silver nitrate-stained polyacrylamide gels [5, 6, 7, 8]. While these may appear to be small modifications to the DAF protocol, they doubled the number of DNA markers detected per PCR. In addition, the modifications enable the detection of simple sequence repeat markers at a low but significant frequency (Figure 3). This capacity of the RAF technology to detect such codominant markers is particularly important for less studied plant species like macadamia.
The RAF protocol compares favorably with AFLP in efficiency and reliability on many plant genomes, including the very large and complex genomes of sugarcane and wheat. While the two technologies detect about the same numbers of markers per large polyacrylamide gel, major advantages of RAF over AFLP include (i) no requirement for enzymatic template preparation, (ii) one instead of two PCRs, and (iii) overall costs.
The selective basis of amplification of DNA markers from genomes is different for RAF and AFLP. Varying any of the last three nucleotides on the RAF primer resulted in the amplification of a nonhomologous set of RAF markers, confirming that amplification of a specific RAF profile is dependent on the 3′ nucleotide sequence of the primer. In contrast, we showed that varying any of the last three nucleotides on AFLP primers in the first PCR had almost no effect on the AFLP profile subsequently detected after the second PCR (Figure 7). Southern analysis also showed that cross hybridization, occurred between first PCR AFLP products, amplified by primers with varying 3′ nucleotides (Figure 6). Our data therefore indicate that the same homologous subset of the genome is amplified in the first PCR of EcoRI/MseI AFLP, regardless of the sequence of the last one to three 3′ nucleotides used in the primer.
While our results using AFLP reactions as probes on MseI-digested genomic DNA indicate that AFLP markers do not represent highly repetitive genomic DNA, it has been demonstrated that these markers represent moderately repeated sequences in Asparagus officinalis [22]. The Asparagus work based on flourescent in-situ hybridization (FISH) analysis with cloned AFLP probes, showed that repetitive signals in the form of clusters were observed on all chromosomes [22]. Not surprisingly, therefore, clustering of AFLP markers on genetic maps has been frequently reported (eg, [23, 24]), much more so with EcoRI-MseI than with PstI-MseI AFLP [25, 26].
At this stage, it is not known whether RAF markers also represent moderately repeated sequences, but we have observed some clustering of RAF markers on a macadamia genetic map [17]. Strong clustering of DNA markers can serendipitously facilitate or impede the identification of markers linked to a gene of interest. For example, we have used bulk segregant analysis [21] in attempts to identify EcoRI-MseI AFLP markers linked to four nodulation loci in soybean, namely nts-1, nod49, nod139-1, and nod139-2 [27]. Eleven out of 2,600 AFLP markers mapped to within about 5 centimorgan of the nts-1 locus, whereas zero out of 1,300 AFLP markers were closely linked to the other three loci [27]. In view of the potential clustering of both AFLP and RAF markers, and the different basis of amplification from the genome, it may be beneficial to use both technologies simultaneously to maximize the chances of identifying DNA markers linked to genes of interest. Such an approach is feasible as both technologies use similar laboratory equipment.
In conclusion, the RAF protocol was demonstrated to be an extremely efficient DNA marker technology, particularly for applications to less-studied plant genomes. More recently, we have been using the RAF technology to (i) identify DNA markers linked to disease resistance genes in tomato and sugarcane, (ii) assess genetic relatedness of genotypes within several plant species (sugarcane, soybean, macadamia, Cassia, and mangosteen), (iii) construct the first genetic map for macadamia, and (iv) demonstrate the loss of DNA in genetically-engineered sugarcane. These investigations using the RAF protocol will be reported in detail elsewhere. While RAF has distinct advantages over AFLP particularly for fingerprinting individual plants, both technologies could be used in a complementary fashion for some genetic studies.
Acknowledgments
ACKNOWLEDGMENTS
The authors are grateful to the Australian Sugar Research and Development Corporation (SRDC), the Queensland Bureau of Sugar Experiment Stations (BSES), the Australian Grains Research and Development Corporation (GRDC), and the Australian Research Council (ARC) for financial support and/or technical assistance.
References
- Tanksley S D, Ganal M W, Martin G B. Chromosome landing: a paradigm for map-based gene cloning in plants with large genomes. Trends Genet. 1995;11(2):63–68. doi: 10.1016/s0168-9525(00)88999-4. [DOI] [PubMed] [Google Scholar]
- Akkaya M S, Shoemaker R C, Specht J E, Bhagwat A A, Cregan P B. Integration of simple sequence repeat DNA markers into a soybean linkage map. Crop Sci. 1995;35:1439–1445. [Google Scholar]
- Williams J G, Kubelik A R, Livak K J, Rafalski J A, Tingey S V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990;18(22):6531–6535. doi: 10.1093/nar/18.22.6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;18(24):7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caetano-Anolles G, Bassam B J, Gresshoff P M. DNA amplification fingerprinting using very short arbitrary oligonucleotide primers. Biotechnology (N Y) 1991;9(6):553–557. doi: 10.1038/nbt0691-553. [DOI] [PubMed] [Google Scholar]
- Bassam B J, Caetano-Anolles G, Gresshoff P M. Fast and sensitive silver staining of DNA in polyacrylamide gels. Anal Biochem. 1991;196(1):80–83. doi: 10.1016/0003-2697(91)90120-i. [DOI] [PubMed] [Google Scholar]
- Bentley S, Bassam B J. A robust DNA amplification fingerprinting system applied to analysis of genetic variation within Fusarium oxysporum fsp cubense. J Phytopath. 1996;144:207–213. [Google Scholar]
- Jiang Q, Gresshoff P M. Classical and molecular genetics of the model legume Lotus japonicus. Mol Plant Microbe Interact. 1997;10(1):59–68. doi: 10.1094/MPMI.1997.10.1.59. [DOI] [PubMed] [Google Scholar]
- Vos P, Hogers R, Bleeker M, et al. AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res. 1995;23(21):4407–4414. doi: 10.1093/nar/23.21.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pooler M R, Scorza R. Aberrant transmission of RAPD markers in haploids, doubled haploids, and F1 hybrids of peach: Observations and speculation on causes. Scientia Horticult. 1995;64:233–241. [Google Scholar]
- Weeden N F, Timmerman G M, Hemmat M, Kneen B E, Lodhi M A. Inheritance and reliability of RAPD markers. In: Hoisington D, McNab A, editors. A proceedings of the symposium on application of RAPD technology to plant breeding. Crop Science Society of America; Madison, WI: 1992. pp. 12–17. [Google Scholar]
- Thomas C M, Vos P, Zabeau M, et al. Identification of amplified restriction fragment polymorphism (AFLP) markers tightly linked to the tomato Cf-9 gene for resistance to Cladosporium fulvum. Plant J. 1995;8(5):785–794. doi: 10.1046/j.1365-313x.1995.08050785.x. [DOI] [PubMed] [Google Scholar]
- Huang J, Sun M. A modified AFLP with fluorescence-labelled primers and automated DNA sequencer detection for efficient fingerprinting analysis in plants. Biotech Techniques. 1999;13:277–278. [Google Scholar]
- Carroll B J, Klimyuk V I, Thomas C M, et al. Germinal transpositions of the maize element Dissociation from T-DNA loci in tomato. Genetics. 1995;139(1):407–420. doi: 10.1093/genetics/139.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll B J, McNeil D L, Gresshoff P M. Mutagenesis of soybean (Glycine max (L.) Merr) and the isolation of nonnodulating mutants. Plant Sci. 1986;47:109–114. [Google Scholar]
- Song L, Carroll B J, Gresshoff P M, Herridge D F. Field assessment of supernodulating genotypes of soybean for yield, N-2 fixation and benefit to subsequent crops. Soil Biol Biochem. 1995;27:563–569. [Google Scholar]
- Peace C P. [PhD thesis] The University of Queensland; Brisbane, Australia: 2002. Genetic characterization of macadamia with DNA markers. [Google Scholar]
- Klimyuk V I, Carroll B J, Thomas C M, Jones J D. Alkali treatment for rapid preparation of plant material for reliable PCR analysis. Plant J. 1993;3(3):493–494. doi: 10.1111/j.1365-313x.1993.tb00169.x. [DOI] [PubMed] [Google Scholar]
- Arumuganathan K, Earle E D. Nuclear DNA content of some important plant species. Molec Gen Genet. 1991;9:208–218. [Google Scholar]
- Sambrook J, Fritsch E F, Maniatis T. 2nd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. Molecular Cloning : A Laboratory Manual. [Google Scholar]
- Michelmore R W, Paran I, Kesseli R V. Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA. 1991;88(21):9828–9832. doi: 10.1073/pnas.88.21.9828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reamon-Buttner S M, Schmidt T, Jung C. AFLPs represent highly repetitive sequences in Asparagus officinalis L. Chromosome Res. 1999;7(4):297–304. doi: 10.1023/a:1009231031667. [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco C, Peeters A J, Koornneef M, et al. Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J. 1998;14(2):259–271. doi: 10.1046/j.1365-313x.1998.00115.x. [DOI] [PubMed] [Google Scholar]
- Bert P F, Charmet G, Sourdille P, Hayward M D, Balfourier F. A high-density molecular map for ryegrass (Lolium perenne) using AFLP markers. Theor Appl Genet. 1999;99:445–452. doi: 10.1007/s001220051256. [DOI] [PubMed] [Google Scholar]
- Castiglioni P, Ajmone-Marsan P, van Wijk R, Motto M. AFLP markers in a molecular linkage map of maize: Codominant scoring and linkage group distribution. Theor Appl Genet. 1999;99:425–431. doi: 10.1007/s001220051253. [DOI] [PubMed] [Google Scholar]
- Young W P, Schupp J M, Keim P. DNA methylation and AFLP marker distribution in the soybean genome. Theor Appl Genet. 1999;99:785–792. [Google Scholar]
- Searle I R. [PhD thesis] The University of Queensland; Brisbane, Australia: 2002. Towards the map-based cloning of the supernodulation (nts-1) locus of soybean. [Google Scholar]