Abstract
Treatment of Saccharomyces cerevisiae cells with the immunosuppressive drug rapamycin results in a variety of cellular changes in response to perceived nutrient deprivation. Among other effects, rapamycin treatment results in the nuclear localization of the global nitrogen activators Gln3p and Nil1p/Gat1p, which leads to expression of nitrogen assimilation genes. The proline utilization (Put) pathway genes were shown to be among the genes induced by rapamycin. Having previously shown that the Put pathway activator Put3p is differentially phosphorylated in response to the quality of the nitrogen source, we examined the phosphorylation status of Put3p after rapamycin treatment. Treatment with rapamycin resulted in the hyperphosphorylation of Put3p, which was independent of Gln3p, Nil1p, and Ure2p. The relative contributions of global nitrogen (Gln3p and Nil1p) and pathway-specific (Put3p) activators to rapamycin-induced expression of the target gene PUT1 were also examined. We found that Nil1p and Put3p, but not Gln3p, play major roles in rapamycin-induced PUT1 expression. Our findings show that perceived nitrogen deprivation triggered by rapamycin treatment and steady-state growth in nitrogen-derepressing conditions are associated with hyperphosphorylation of Put3p and increased PUT1 expression. Rapamycin treatment and nitrogen derepression may share some, but not all, regulatory elements, since Gln3p and Nil1p do not participate identically in both processes and are not required for hyperphosphorylation. A complex relationship exists among the global and pathway-specific regulators, depending on the nature and quality of the nitrogen source.
Utilization of nonpreferred nitrogen sources in the yeast Saccharomyces cerevisiae is the result of the interplay between two types of regulatory systems, global and specific. The global system represses the utilization of alternative nitrogen sources when preferred nitrogen sources are present and relieves this repression when the preferred nitrogen sources are depleted (reviewed in reference 45). It includes four GATA factor family members that bind GATAA sequences (UASNTR), consisting of two positively acting regulators, Gln3p (47) and Nil1p/Gat1p (17, 58), two negatively acting regulators, Dal80p/Uga43p (19, 23) and Nil2p/Deh1p/Gzf3p (18, 50, 56), and a global nitrogen repressor, Ure2p (31, 34, 35). The GATA factors are involved in a complex interplay of cross- and autoregulation and competition for GATAA sites (18, 22, 24, 56, 57). Gln3p is thought to be kept in an inactive complex with Ure2p in the cytoplasm under nitrogen-rich conditions (4, 7).
In addition to the global regulators, pathway-specific regulators activate permeases and catabolic enzymes in response to induction by their substrates or intermediates. In several of the best-characterized systems, e.g., allantoin utilization (49) and γ-aminobutyrate (GABA) utilization (2, 59), analysis of cis-acting sequences demonstrates that both global and specific factors bind to the promoters of specific transporter and enzyme genes. Few studies to date address the question of the nature and kinds of interactions that exist between global and specific nitrogen-regulatory proteins.
Recent studies of the effects of rapamycin on yeast cells demonstrated that transcription of many genes in nutrient-sensing pathways, including those of the proline utilization pathway, was altered (14, 36). Treatment with rapamycin resulted in the dephosphorylation and subsequent nuclear localization of Gln3p, mimicking what occurs during growth under nitrogen-limiting conditions (4, 5).
In this study, we present an analysis of interactions between two GATA factors, Gln3p and Nil1p, and Put3p, the pathway-specific activator, in controlling the regulation of the proline utilization pathway in response to rapamycin treatment or steady-state growth under nitrogen-poor conditions. Proline is the most abundant source of nitrogen for yeast cells growing in the wild (6) and a poor nitrogen source for laboratory strains. Transcription of the proline transporter genes GAP1 and PUT4 and the genes that encode the enzymes of the pathway, PUT1 and PUT2, is sensitive to nitrogen regulation (15, 16, 26, 38, 50, 56-58, 61), and the latter two are induced by proline (12, 13). The Put3p pathway-specific activator is absolutely required for induction by proline (10, 27, 54), binds at UASPUT sites to the promoters of its target genes under all conditions (3), and undergoes differential phosphorylation as a function of the quality of the nitrogen source (37) and conformational changes in response to proline (28).
The induction of PUT gene expression by rapamycin led us to examine which factors were required for the increase in expression and what effect the drug had on phosphorylation of Put3p. We report that the induction of PUT1 gene expression by rapamycin is mediated through Nil1p and Put3p, with little involvement by Gln3p. Rapamycin treatment results in the hyperphosphorylation of Put3p. We observed only partial congruence between factors important for elevated gene expression due to rapamycin treatment and those required for nitrogen derepression caused by growth of yeast cells in nitrogen-poor media.
MATERIALS AND METHODS
Strains and plasmids.
Protease-deficient strains BJ2168 (MATa pep4-3 prb1-1122 prc1-451 ura3-52 leu2 trp1) and DB1000, a put3Δ::LEU2 derivative of BJ2168, have been described previously (37, 39). Strain BJ2168ure2Δ was constructed by transformation of strain BJ2168 with a 3.5-kb EagI-ClaI fragment carrying ure2Δ11::LEU2 from plasmid 1C-CSΔU (20). Leu+ transformants were screened by DNA hybridization analysis to verify that the genomic URE2 gene had been replaced by the disrupted copy. Strain BJ2168nil1Δ was constructed by transformation of strain BJ2168 with a 7.4-kb XhoI fragment carrying nil1::hisG::URA3::hisG from plasmid pMS61 (58). Ura+ transformants were screened by DNA hybridization to determine that the genomic NIL1 gene had been disrupted. These transformants were then streaked on 5-fluoroorotic acid plates, and resistant clones were isolated. Loss of URA3 and formation of the nil1::hisG disruption were again confirmed by DNA hybridization. Strain BJ2168gln3Δ was constructed by transformation of strain BJ2168 with a 3.4-kb AatII fragment carrying gln3::LEU2 from plasmid pPM35 (47). Leu+ transformants were screened by DNA hybridization to determine that the genomic GLN3 gene had been disrupted. Strain BJ2168nil1Δgln3Δ was constructed by transformation of strain BJ2168nil1Δ with the same 3.4-kb AatII fragment carrying gln3::LEU2 as described for BJ2168gln3Δ. Leu+ transformants were screened by DNA hybridization to determine that both the genomic GLN3 and NIL1 genes had been disrupted. All protease-deficient strains were derived from the S288C background; their regulation is similar, although not identical, to that of the Σ1278b strains described below. Protease-deficient strains were used to enhance the detection of phosphoforms of Put3p.
Strains DB26-3A (MATa ade2 ura3-52 TRP1-PUT1-lacZ), DB26-2B (MATa put3Δ::ADE2 ade2 ura3-52 TRP1-PUT1-lacZ), and DB27-5B (MATa ade2 ura3-52 TRP1-PUT2-lacZ) were transformed with the leu2::hisG-URA3-hisG cassette from plasmid pNKY85 (1) to introduce, after selection for 5-fluoroorotic acid resistance, the leu2::hisG mutation, yielding strains DB200, DB201, and DB202, respectively. Strain DB200 (MATa ade2 ura3-52 leu2::hisG TRP1-PUT1-lacZ) is the parent of strains DB1200 (gln3Δ::LEU2), DB2000 (nil1Δ::URA3), and DB2200 (gln3Δ::LEU2 nil1Δ::URA3). Strain DB201 (MATa put3Δ::ADE2 ade2 ura3-52 leu2::hisG TRP1-PUT1-lacZ) is the parent of strains DB1201 (gln3Δ::LEU2), DB2201 (nil1Δ::URA3), and DB2300 (gln3Δ::LEU2 nil1Δ::URA3). Strain DB202 (MATa ade2 ura3-52 leu2::hisG TRP1-PUT2-lacZ) is the parent of strains DB1202 (gln3Δ::LEU2), DB2001 (nil1Δ::URA3), and DB2202 (gln3Δ::LEU2 nil1Δ::URA3). Strain DB27-3A (MATa put3Δ::ADE2 ade2 ura3-52 leu2::hisG TRP1-PUT2-lacZ) is the parent of strains DB1273A (gln3Δ::LEU2), DB2273 (nil1Δ::URA3), and DB2301 (gln3Δ::LEU2 nil1Δ::URA3). All DB strains are isogenic and were derived from the Σ1278b background.
Plasmid YEp13 (high-copy vector carrying LEU2) (8), pWB36 (low-copy vector carrying PUT1-lacZ (60), and pABC4 (low-copy vector carrying PUT2-lacZ) (53) have been previously described. Plasmid pMDB4 carries a 3.6-kb EagI-XhoI fragment containing PUT3 from plasmid pBS-PUT3A (28) ligated to plasmid YEp24 cut with EagI and SalI. Plasmid pMDB9 carries a 3.6-kb SnaBI fragment containing PUT3 from plasmid pBS-PUT3A inserted at the SmaI site of plasmid YEp24 (8).
Media, growth conditions, extract preparation, and β-galactosidase assays.
The minimal medium used in this study has been described previously (11). Glucose (2%) was the carbon source. Nitrogen sources were glutamine (0.1%), ammonium sulfate (0.2%), glutamate (0.1%), GABA (0.1%), or proline (0.1%). Supplements of tryptophan, uracil, leucine, adenine, and glutamine were added when required; strains with related genotypes were grown under identical conditions. gln3 strains were always grown in the presence of glutamine (30 mg/liter).
The methods for growth of cells and extract preparation have been described previously (46). The units of specific activity are nanomoles of o-nitrophenol formed per minute per milligram of protein. The numbers represent the average of two to four determinations; variation is listed in the figures. Protein concentrations of crude extracts were determined by the method of Bradford (9), using crystalline bovine serum albumin as the standard.
For experiments in which rapamycin treatment was used, cells were grown on a glucose-glutamine minimal medium until they reached a cell density of 50 Klett units (blue filter). A control sample of 10 ml was removed, and rapamycin (dissolved in 90% ethanol-10% Tween 20) or the drug vehicle alone was added to a final concentration of 200 ng/ml. Cells (10 ml) were removed at various time points, and extracts were made as described above for β-galactosidase activity.
Preparation of extracts for phosphorylation profiles, dephosphorylation, and immunodetection.
Protease-deficient strains BJ2168, DB1000, BJ2168ure2Δ, BJ2168nil1Δ, and BJ2168nil1Δgln3Δ were used for the analyses. Cell extracts were prepared as described previously (37) in modified IP buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 5 mM EDTA, 5 mM EGTA, 10 mM dithiothreitol) with protease inhibitors. For dephosphorylation of extracts with calf intestinal phosphatase, 25 μg of total protein was treated with 15 U of calf intestinal phosphatase (New England Biolabs) with or without phosphatase inhibitors for 1 h at 37°C (37). The reaction was stopped by addition of 5× Laemmli sample buffer (42) to a final concentration of 1× and boiling for 5 min. For maximum separation of Put3p isoforms, longer gels (18 by 24 cm) were used and electrophoresis was carried out at 77 V at room temperature for 18 to 20 h under modified sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) conditions (37). Proteins were transferred to polyvinylidene difluoride membrane and probed with anti-Put3p serum diluted 1:4,000.
RESULTS
Rapamycin treatment results in hyperphosphorylation of Put3p.
Put3p is a phosphoprotein whose ability to activate transcription of its target genes is correlated with the extent of its phosphorylation and with decreased quality of the nitrogen source in the medium (37). The poorer the nitrogen source in the growth medium, the more slowly Put3p migrates in denaturing gels. Since Put3p is large, with a molecular mass of 117 kDa, the difference in migration of hyper- and hypophosphorylated forms of Put3p is small but reproducible; separation of the isoforms requires modification of the standard SDS running buffer (37).
To determine if rapamycin treatment resulted in hyperphosphorylation of Put3p, extracts of glutamine-grown (protease-deficient) cells were separated by SDS-PAGE and subjected to immunoblotting with anti-Put3p antiserum. The Put3p band migrated more slowly in extracts of cells treated with the drug than in those that were untreated (Fig. 1A, lanes 1 and 2), and treatment with phosphatase converted the slower-migrating species to a faster-migrating form (lanes 3 and 4). Similar results were obtained after rapamycin treatment of cells grown on ammonia-containing medium (data not shown). These results demonstrate that Put3p is hyperphosphorylated upon rapamycin treatment, similar to our previous observations for cells grown in nitrogen-derepressing conditions (37).
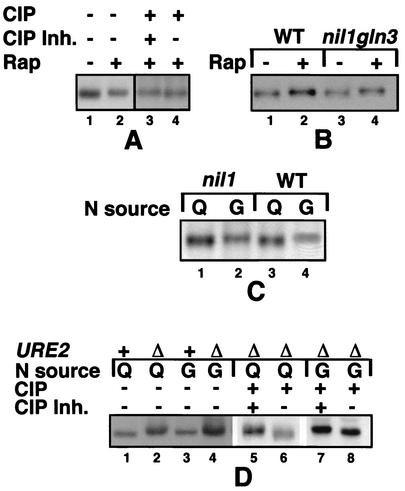
FIG. 1.
Phosphorylation profiles of Put3p in wild-type and GATA factor-deficient yeast strains. (A) Hyperphosphorylation of Put3p after rapamycin treatment in a wild-type strain. Protease-deficient strain DB1000 carrying high-copy PUT3-containing plasmid pMDB4 was grown in minimal glucose-glutamine medium supplemented with tryptophan. Rapamycin (lanes +; 200 ng/ml) or the drug vehicle alone (lanes −; 90% ethanol-10% Tween 20) was added, and the cells were harvested 60 mins later. Lanes:1 and 2, extracts were prepared and separated by SDS-PAGE followed by immunoblotting with anti-Put3p antibody; 3 and 4, rapamycin-treated extracts were incubated with calf intestinal phosphatase (CIP) in the presence or absence of phosphatase inhibitors (CIP Inh.), as described in Materials and Methods. (B) Hyperphosphorylation of Put3p after rapamycin treatment in GATA factor-deficient yeast strains. Strains BJ2168 (wild type [WT]) and BJ2168 nil1Δgln3Δ (nil1::hisG gln3::LEU2) carrying high-copy plasmid pMDB9 (PUT3) were grown in a minimal glucose-glutamine medium supplemented with tryptophan and leucine and treated with rapamycin (lanes 2 and 4) or drug vehicle (lanes 1 and 3) as in panel A. Extracts were prepared and separated by SDS-PAGE followed by immunoblotting with anti-Put3p antibody as in panel A. (C) Hyperphosphorylation of Put3p in wild-type and nil1 strains grown in nitrogen-derepressing media. Strains BJ2168 and BJ2168 nil1Δ (nil1::hisG) carrying plasmid pMDB9 were grown in a minimal glucose medium supplemented with tryptophan and leucine and containing either glutamine (Q; lanes 1 and 3) or GABA (G; lanes 2 and 4) as the sole nitrogen source. Extracts were prepared and separated by SDS-PAGE followed by immunoblotting with anti-Put3p antibody as in panel A. (D) Hyperphosphorylation of Put3p in a ure2Δ strain. Extracts of strains DB1000 or BJ2168 ure2Δ (ure2::LEU2) carrying plasmid pMDB4 (PUT3) grown in minimal media with either glutamine (Q; lanes 1, 2, 5, and 6) or GABA (G; lanes 3, 4, 7, and 8) as the nitrogen source were subjected to modified SDS-PAGE and immunoblotting with anti-Put3p antiserum. In lanes 5 to 8, the same extracts were incubated with calf intestinal phosphatase in the presence or absence of phosphatase inhibitors followed by SDS-PAGE and immunoblotting.
To determine if rapamycin-induced hyperphosphorylation of Put3p is dependent (directly or indirectly) on the positively acting GATA factors, we assayed the migration of Put3 phosphoforms in strains in which Gln3p and Nil1p were deleted. In the nil1Δgln3Δ strain, Put3p is hyperphosphorylated comparable to the level in the wild-type strain (Fig. 1B, compare lanes 1 and 2 with lanes 3 and 4). Thus, rapamycin-induced hyperphosphorylation of Put3p does not require the presence of these GATA factors.
Hyperphosphorylation of Put3p is independent of Nil1p in steady-state growth.
To compare features of rapamycin-induced nitrogen limitation with those of steady-state growth on nitrogen-poor media, we examined the involvement of Nil1p in hyperphosphorylation of Put3p under steady-state nitrogen-derepressing conditions. Cells of the wild-type and nil1 strains were grown in nitrogen-rich (glutamine) and nitrogen-poor (GABA) conditions, and proteins were extracted and separated to identify Put3p phosphoforms. As with the rapamycin-treated cells, removal of Nil1p did not appear to affect the nitrogen-sensitive hyperphosphorylation of Put3p (Fig. 1C). Unfortunately, the protease-deficient gln3Δ and nil1Δ gln3Δ strains grew so poorly on GABA-containing media that it was impossible to assay the phosphoforms under those conditions.
Mutations in URE2 cause hyperphosphorylation of Put3p.
One of the roles of Ure2p is to prevent the expression of alternative nitrogen source utilization pathways when preferred nitrogen sources are present; under these conditions, Ure2p binds to Gln3p in the cytoplasm and prevents its nuclear localization (4, 7). In ure2 strains, genes of these alternative pathways are inappropriately expressed in nitrogen-rich conditions (15, 21, 30-33, 35, 61). Since the PUT genes are controlled by Ure2p, we examined the phosphorylation pattern of wild-type Put3p in a ure2Δ strain grown under either nitrogen-rich or nitrogen-poor conditions. Like the previously described PUT3c constitutive (nitrogen-insensitive) alleles (37), wild-type Put3p in a ure2 background is aberrantly hyperphosphorylated in cells grown in nitrogen-rich conditions (Fig. 1D). This hyperphosphorylation is again correlated with aberrantly high levels of PUT1 and PUT2 expression (15, 61).
Roles of Put3p, Gln3p, and Nil1p in rapamycin-induced expression of PUT1.
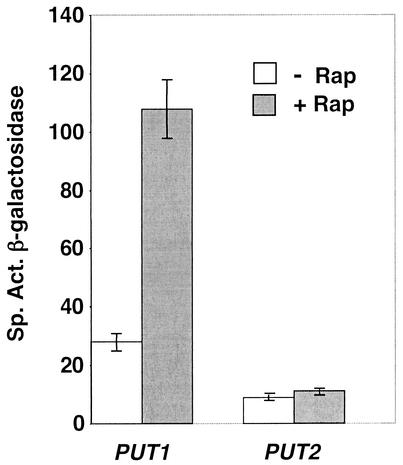
Recent studies from three laboratories on the effects of rapamycin on yeast cells demonstrated that transcription of the PUT genes was induced (4, 14, 36). To confirm these results for strains and growth conditions in use in our laboratory, we first determined that laboratory yeast strains of two different backgrounds (Σ1278b and S288C) grown in a minimal glutamine medium responded to rapamycin in a similar manner to the strains grown in YPD medium used by the Schreiber (36) and Heitman (14) laboratories. We measured the β-galactosidase expression of protease-proficient Σ1278b-derived and protease-deficient S288C-derived strains (used for the phosphoform analysis) carrying the reporter genes PUT1-lacZ and PUT2-lacZ, exposed to rapamycin for various times. We found that PUT1 expression increased approximately fourfold after a 1-h rapamycin treatment (Fig. 2) and was observed within 15 min of treatment (data not shown). These results are consistent with the 4.6-fold increase in steady-state PUT1 RNA levels after a 1-h treatment reported by Hardwick et al. (36). We did not see an increase in PUT2-lacZ expression comparable to the twofold effect on PUT2 RNA levels observed by this group, which may reflect the down-regulation of overall protein synthesis by rapamycin. Given these results, we used expression of PUT1-lacZ in cells growing in a minimal medium with glutamine as the sole nitrogen source to monitor rapamycin-induced expression of the proline utilization pathway.
FIG. 2.
PUT gene expression after treatment with rapamycin. Protease-deficient strain BJ2168 carrying plasmids pYEp13 (LEU2) and either pWB36 (PUT1-lacZ) or pABC4 (PUT2-lacZ) was grown in minimal glucose-glutamine medium supplemented with tryptophan. Rapamycin (+; 200 ng/ml) or the drug vehicle alone (−; 90% ethanol-10% Tween 20) was added, and the cells were harvested 60 min later and assayed for β-galactosidase activity. Results are the average of three independent experiments with standard deviation as indicated.
To determine which activators were playing a role in the rapamycin-induced expression of PUT1, we assayed the effect of removal of Put3p, Gln3p, and Nil1p, singly and in combination, on β-galactosidase expression in cells growing in the presence or absence of rapamycin treatment in a set of eight strains carrying PUT1-lacZ integrated into the genome. Compared to the wild-type strain, removal of Nil1p resulted in the loss of rapamycin induction. Removal of either Gln3p or Put3p affected the overall level of expression but did not affect the magnitude of rapamycin induction; rapamycin treatment of gln3Δ or put3Δ strains induced PUT1 expression 3.6- and 3.5-fold, respectively, compared with the 4-fold induction observed in the wild-type strain (Fig. 3).
FIG. 3.
Roles of Put3p, Gln3p, and Nil1p in rapamycin induction of PUT1 expression. Strains DB26-3A (wild type [WT]), DB1200 (gln3Δ), DB2000 (nil1Δ), DB2200 (gln3Δnil1Δ), DB26-2B (put3Δ), DB1201 (put3Δgln3Δ), DB2201 (put3Δnil1Δ), and DB2300 (put3Δgln3Δnil1Δ) carrying a genomic copy of PUT1-lacZ were grown in minimal glucose-glutamine medium supplemented with adenine, uracil, and leucine and treated as described in the legend to Fig. 2. Rapamycin induced PUT1 expression in wild-type, gln3Δ, and put3Δ strains by 4-, 3.6-, and 3.5-fold, respectively.
In the absence of Put3p, the level of PUT1 expression was reduced to about one-third of that observed in the wild type, in agreement with previous studies (10, 27, 37). In the gln3Δ nil1Δ, put3Δ gln3Δ, put3Δ nil1Δ, and put3Δ gln3Δ nil1Δ strains, expression was at or near background levels (those of the triple knockout) and rapamycin-induced PUT1 expression was twofold or less. These results suggest that rapamycin induction is mediated mainly through Nil1p and Put3p, with a small contribution by Gln3p.
Nitrogen regulation of the PUT genes by the GATA factors and Put3p.
Rapamycin treatment of yeast cells is thought to mimic nitrogen deprivation (4, 5). In a shift from a rich to a poor nitrogen source, the GATA factors Gln3p and Nil1p/Gat1p enter the nucleus and activate genes in nitrogen-assimilatory pathways (4). Having demonstrated that Nil1p is the major nitrogen activator in rapamycin-induced expression of PUT1, we compared the participation of both Nil1p and Gln3p in nitrogen derepression of PUT1 during steady-state growth. The eight PUT1-lacZ strains with single or multiple combinations of mutations in PUT3, GLN3, and NIL1 were assayed during steady-state balanced growth in nitrogen-repressing (glutamine or ammonia as the sole nitrogen source), nitrogen derepressing (glutamate or GABA as the sole nitrogen source), and nitrogen-derepressing, proline-inducing (proline as the sole nitrogen source) conditions. Glutamine and glutamate were specifically chosen so that we could compare the results with previous studies of GAP1 encoding the general amino acid permease (58). A systematic study of PUT1 expression in wild-type and various mutant strains grown with these five nitrogen sources has not been reported previously.
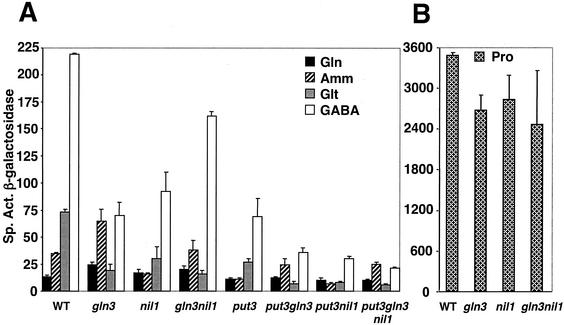
The behavior of PUT1 expression in the wild-type strain grown on nitrogen sources of different quality gives the classic pattern for a nitrogen-derepressible gene: as the quality of the nitrogen source decreased, PUT1 expression increased dramatically (Fig. 4A). Compared to expression in a medium containing the richest nitrogen source, glutamine, expression increased approximately 3-fold in an ammonia-containing medium, 6-fold in a glutamate-containing medium, and 18-fold in a GABA-containing medium. When the medium contained proline as the sole source of nitrogen, the combination of full nitrogen derepression and proline induction led to the highest expression, an increase of almost 300-fold (Fig. 4B; note difference in scales between Fig. 4A and B).
FIG. 4.
Effects of removal of Put3p, Gln3p, and Nil1p on nitrogen regulation of PUT1. (A) Nitrogen regulation of PUT1 under noninducing conditions. (B) Expression of PUT1 with proline as the sole source of nitrogen. The strains listed in Fig. 3 were grown in minimal medium containing glutamine (Gln, 0.1%), ammonia (Amm, 0.2%), glutamate (Glt, 0.1%), GABA (0.1%), or proline (Pro, 0.1%) as the nitrogen source. Extracts were made and assayed for β-galactosidase activity. Results are the average of two to five independent measurements with variations as indicated. WT, wild type.
Removal of either Gln3p or Nil1p decreased the expression of PUT1 in derepressing conditions (glutamate or GABA) by about two-thirds compared with wild-type levels. A comparison of PUT1 levels in glutamine- and GABA-containing media demonstrates that the gene is still subject to nitrogen derepression in the single-mutant strains although it is reduced from 18-fold in the wild type to 3- to 7-fold in the mutants. In steady-state growth, Gln3p and Nil1p contribute equally to the activation of PUT1 in both glutamate- and GABA-containing media. These findings are in agreement with previous results obtained for a different strain background by Coffman et al. (17). Unexpectedly, removal of both GATA factors resulted in surprisingly high PUT1 levels in GABA-containing media: 75% of the levels observed in the wild-type strain. The possibility that Gln3p and/or Nil1p can (directly or indirectly) play negative roles, perhaps by interfering with the activity of Put3p, is presented in the Discussion.
Put3p appears to work synergistically with the GATA factors in PUT1 expression. Removal of the pathway-specific activator Put3p reduced the expression of PUT1 by two-thirds in glutamate- and GABA-containing media (Fig. 4A), prevented growth on proline as the sole nitrogen source, but did not affect nitrogen derepression: results we have reported previously (25, 61). In the absence of Put3p, further removal of either Gln3p, Nil1p or both resulted in very low expression levels.
Expression of PUT1 in nitrogen-rich conditions (glutamine or ammonia) was low, as expected, since the GATA factors are reported to be cytoplasmic under these conditions. One anomaly was observed: on an ammonia-containing medium, PUT1 expression in the gln3Δ strain was elevated above that observed in the wild-type strain. Similar findings for the GAP1 gene have been reported previously (58); it is possible that this increase is due to reduction in the amount of the negatively acting GATA factor Dal80p which requires Gln3p for its production (18, 26, 56). This explanation requires that some Gln3p be present in the nucleus under these conditions; the relative amounts of cytoplasmic and nuclear Gln3p are not currently known under any conditions.
Steady-state levels of PUT2 expression in cells grown in glutamate versus glutamine as the nitrogen source did not show significant nitrogen derepression (data not shown). Gln3p does not play a role in its regulation under either nitrogen-repressing or -derepressing conditions, confirming previous data reported by us (61) and others (26). Removal of Nil1p resulted in 25 to 50% reduction in expression in both types of media (data not shown), indicating that Nil1p activates PUT2.
Maximum expression of PUT1 (Fig. 4B) and PUT2 (data not shown) was observed when proline was the sole nitrogen source, since proline is the inducer for the pathway as well as one of the most derepressing nitrogen sources for S. cerevisiae. Under these conditions, the gln3Δ and gln3Δ nil1Δ strains grew more slowly than the wild type (doubling times of 4 and 8 h, respectively, compared to 3 h for the wild-type and nil1Δ strains) and reduced PUT1 (Fig. 4) and PUT2 (data not shown) expression levels by approximately 25% compared to the levels observed in the wild-type strain. The small effect of removal of the GATA factors on the expression of both genes with proline as the nitrogen source reflects the predominance of Put3p as the major activator; the slow growth of the gln3Δnil1Δ strain is most probably due to a reduction in proline transport (58).
We conclude that in steady-state nitrogen-derepressing, noninducing conditions (glutamate or GABA as the nitrogen source), Gln3p, Nil1p, and Put3p each contribute to the expression of PUT1. In rapamycin-induced expression, Nil1p and Put3p are the major regulators. Under fully inducing and nitrogen-derepressing conditions (proline as the nitrogen source), Put3p is the major transcriptional activator. Although Nil1p and Gln3p are responsible for nitrogen derepression of PUT1, Put3p works synergistically with them in activation. Activation of PUT1 expression is correlated with hyperphosphorylation of Put3p, a process that does not involve the positive GATA factors.
DISCUSSION
In this report, we demonstrate that rapamycin treatment results in hyperphosphorylation of the activator of the proline utilization pathway, Put3p. To date, this is only the second report of rapamycin-induced hyperphosphorylation of a regulatory protein in yeast; Rtg3p, a regulator of the retrograde signaling pathway that activates nuclear genes in response to mitochondrial defect (43, 44), is hyperphosphorylated and translocated into the nucleus after rapamycin treatment (40). In most cases so far reported, regulators or kinases undergo dephosphorylation on rapamycin treatment. Examples include Gln3p (4), Ure2p (14, 36), Npr1p (51), and Mks1p (29). The rapamycin-induced hyperphosphorylation of Put3p resembles that previously described in yeast cells grown under nitrogen-derepressing conditions (37). Furthermore, the hyperphosphorylation of Put3p, induced either by rapamycin or by nitrogen derepression, does not require the presence of the GATA factors Gln3p and Nil1p/Gat1p or the global nitrogen repressor Ure2p. Hyperphosphorylation of Put3p is correlated with increased expression of its target genes, PUT1 and PUT2 (37); phosphorylation does not affect its subcellular localization, which is constitutively nuclear (3). Whether a member of the TOR signaling pathway or some other pathway is responsible for the hyperphosphorylation is unknown.
The PUT1 gene responds to (at least) two types of signals: the presence of the pathway inducer proline and the absence of preferred nitrogen sources. Put3p is the transducer of the proline signal; in the absence of the regulator or its binding site, there is no proline induction (10, 12, 37, 53, 54). The involvement of the global nitrogen repressor Ure2p on the transcription of both PUT1 and PUT2 confirmed that nitrogen regulation also operated at the level of the structural genes of the Put pathway, rather than solely on the proline transporters, as originally believed (12, 15, 61). Using RNA hybridization analysis, Cooper and coworkers established that both Gln3p and Nil1p/Gat1p contribute to the nitrogen responsiveness of PUT1 in cells grown on derepressing sources of nitrogen (16, 17, 26). Since Gln3p is an activator of Nil1p synthesis (17, 18, 50, 56), it is difficult to determine whether Gln3p acts directly, or indirectly through Nil1p. The largely Gln3p-independent behavior of PUT1 in transcription profiles reported by Schreiber and colleagues (41, 52) supports the more predominant role of Nil1p in PUT1 expression.
Our observations of steady-state PUT1 expression under nitrogen-derepressing conditions extend those of the Cooper laboratory. In contrast, rapamycin-induced PUT1 expression is mediated predominantly by Nil1p, with Gln3p playing a minor role. Cooperation between Nil1p and Put3p still plays an important part, since the absence of Put3p caused PUT1 gene expression to drop by two-thirds, although rapamycin induction remained. PUT2, a gene previously shown to be less sensitive to nitrogen derepression, is regulated by Ure2p but not by Gln3p (26, 48, 61). PUT2 expression is only slightly affected by the removal of Nil1p (our unpublished results) and has a much smaller response to rapamycin-induced nutrient deprivation.
In the absence of both positive GATA factors, Put3p can activate transcription of the PUT genes by itself in GABA- and proline-containing media to levels about 70% of those observed in the wild-type strain. The significant increase in PUT1 expression in the gln3Δ nil1Δ strain grown in a GABA-containing medium compared to that of either of the single-mutant strains suggests that Gln3p and Nil1p may hinder Put3p from activating PUT1. In the wild-type strain, the GATA factors may prevent Put3p from activating in the absence of proline, keeping the levels of PUT1 expression 15-fold lower than their maximum when proline is present. Such an arrangement in the wild-type strain would save cellular energy by preventing the synthesis of the proline-degrading enzymes when they are not needed. It is also important to note that the hyperphosphorylation of Put3p in the gln3Δ nil1Δ strain growing on a GABA-containing medium correlates with the elevated target gene activity observed.
What is clear from our comparative nitrogen source study is the synergy that exists among the GATA factors and Put3p in the expression and responsiveness of the Put pathway. Rai et al. (48) suggested a form of cooperation between Put3p and Gln3p, before the existence of Nil1p was demonstrated, based on an analysis of mutations at the Put3p- and GATA-binding sites. They identified two functional GATA sites separated by about 25 bp that are located about 25 and 50 bp, respectively, downstream (3′) of the proximal of two Put3p-binding sites in the PUT1 promoter (48). This layout potentially places a GATA factor in close proximity to Put3p under conditions in which these activators are binding their respective upstream activation sites.
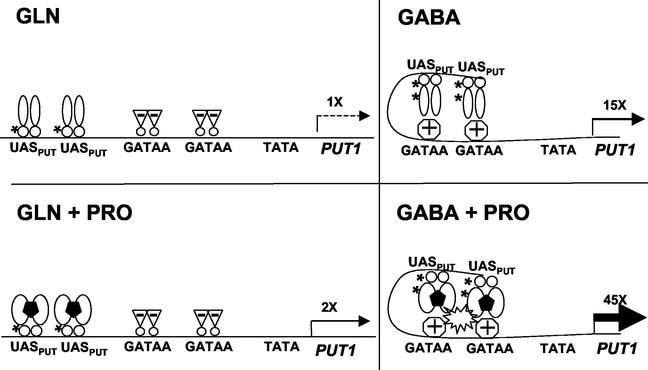
Our current working model based on their promoter analysis and our studies of Put3p is shown in Fig. 5. Put3p increases transcription of the PUT genes when proline is added to the medium, even in the presence of a good nitrogen source (glutamine in this example). The addition of proline converts Put3p into a more active conformation (28). We speculate here that Put3p can bind proline, but this has not yet been demonstrated. The GATA sites are not occupied by the positively acting GATA factors, since Gln3p and Nil1p are cytoplasmic under these conditions (4). It is possible that the negatively acting GATA factors Dal80p/Uga43p and Nil2p/Deh1p/Gzf3p may bind these sites under these conditions, but this has not been established. In a nitrogen-poor environment (GABA in this example), Nil1p and/or Gln3p binds to the GATA sites and may interact with a hyperphosphorylated form of Put3p to increase transcription of the PUT genes, even in the absence of proline. On addition of proline, Put3p undergoes a conformational change, which, together with the hyperphosphorylation, results in dramatic enhancement in transcriptional activation by Put3p. Our model suggests some interaction between Gln3p or Nil1p and Put3p; whether Put3p interacts with a GATA factor directly, by mutual binding of a common coactivator or adaptor molecule (e.g., Ada1p and Gln3p [55]), by changes in DNA bending, or by some other means remains to be determined.
FIG. 5.
Working model for the regulation of PUT1 transcription by nitrogen derepression and proline induction. See the text for details. Put3p binding to proline, binding of negatively acting GATA factors, DNA bending, and Put3p-GATA factor interactions are speculative at present. Abbreviations: UASPUT, binding sites for Put3p (bp −307 to −293 and −280 to −265 from the ATG of PUT1); GATAA, binding sites for GATA factors (bp −235 and −204); TATA, TATA box (bp −118). Symbols: →, extent and levels of PUT1 transcription relative to level on Gln; *, phosphorylation sites;  , Put3p dimers;
, Put3p dimers;  , proline;
, proline;  , positive GATA factors;
, positive GATA factors;  , negative GATA factors.
, negative GATA factors.
Acknowledgments
We are grateful for the excellent technical assistance of D. Barber in strain construction and M. D'Alessio in plasmid construction. We thank H. Huang for instruction in phosphoforms gel analysis, K. Arndt and M. Hall for technical advice and initial supplies of rapamycin, and B. Magasanik and S. Garrett for comments on the manuscript.
This research was funded by grants from the National Institute of General Medical Sciences (GM40751) and the Foundation of the University of Medicine and Dentistry of New Jersey (15-02).
REFERENCES
- 1.Alani, E., L. Cao, and N. Kleckner. 1987. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics 116:541-545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andre, B., D. Talibi, S. Soussi Boudekou, C. Hein, S. Vissers, and D. Coornaert. 1995. Two mutually exclusive regulatory systems inhibit UASGATA, a cluster of 5′-GAT(AT)A-3′ upstream from the UGA4 gene of Saccharomyces cerevisiae. Nucleic Acids Res. 23:558-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Axelrod, J. D., J. Majors, and M. C. Brandriss. 1991. Proline-independent binding of PUT3 transcriptional activator protein detected by footprinting in vivo. Mol. Cell. Biol. 11:564-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck, T., and M. N. Hall. 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402:689-692. [DOI] [PubMed] [Google Scholar]
- 5.Bertram, P. G., J. H. Choi, J. Carvalho, W. Ai, C. Zeng, T.-F. Chan, and X. F. S. Zheng. 2000. Tripartite regulation of Gln3p by TOR, Ure2p and phosphatases. J. Biol. Chem. 275:35727-35733. [DOI] [PubMed] [Google Scholar]
- 6.Bisson, L. F. 1991. Influence of nitrogen on yeast and fermentation of grapes, p. 78-89. In J. Rantz (ed.), Proceedings of the International Symposium on Nitrogen in Grapes and Wine. American Society of Enology and Viticulture. Davis, Calif.
- 7.Blinder, D., P. E. Coschigano, and B. Magasanik. 1996. Interaction of the GATA factor Gln3p with the nitrogen regulator Ure2p in Saccharomyces cerevisiae. J. Bacteriol. 178:4734-4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Botstein, D., S. C. Falco, S. E. Stewart, M. Brennan, S. Scherer, D. T. Stinchcomb, K. Struhl, and R. W. Davis. 1979. Sterile host yeast (SHY): a eukaryotic system of biological containment for recombinant DNA experiments. Gene 8:17-24. [DOI] [PubMed] [Google Scholar]
- 9.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 10.Brandriss, M. C. 1987. Evidence for positive regulation of proline utilization in Saccharomyces cerevisiae. Genetics 117:429-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brandriss, M. C. 1979. Isolation and preliminary characterization of Saccharomyces cerevisiae proline auxotrophs. J. Bacteriol. 138:816-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brandriss, M. C., and B. Magasanik. 1979. Genetics and physiology of proline utilization in Saccharomyces cerevisiae: enzyme induction by proline. J. Bacteriol. 140:498-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brandriss, M. C., and B. Magasanik. 1979. Genetics and physiology of proline utilization in Saccharomyces cerevisiae: mutation causing constitutive enzyme expression. J. Bacteriol. 140:504-507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cardenas, M. E., N. S. Cutler, M. C. Lorenz, C. J. Di Como, and J. Heitman. 1999. The TOR signaling cascade regulated gene expression in response to nutrients. Genes Dev. 13:3271-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coffman, J. A., H. M. El Berry, and T. G. Cooper. 1994. The URE2 protein regulates nitrogen catabolic gene expression through the GATAA-containing UASNTR element in Saccharomyces cerevisiae. J. Bacteriol. 176:7476-7483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coffman, J. A., R. Rai, and T. G. Cooper. 1995. Genetic evidence for Gln3p-independent, nitrogen catabolite repression-sensitive gene expression in Saccharomyces cerevisiae. J. Bacteriol. 177:6910-6918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Coffman, J. A., R. Rai, T. Cunningham, V. Svetlov, and T. G. Cooper. 1996. Gat1p, a GATA family protein whose production is sensitive to nitrogen catabolite repression, participates in transcriptional activation of nitrogen-catabolic genes in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:847-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coffman, J. A., R. Rai, D. M. Loprete, T. Cunningham, V. Svetlov, and T. G. Cooper. 1997. Cross regulation of four GATA factors that control nitrogen catabolic gene expression in Saccharomyces cerevisiae. J. Bacteriol. 179:3416-3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coornaert, D., S. Vissers, B. Andre, and M. Grenson. 1992. The UGA43 negative regulatory gene of Saccharomyces cerevisiae contains both a GATA-1 type zinc finger and a putative leucine zipper. Curr. Genet. 21:301-307. [DOI] [PubMed] [Google Scholar]
- 20.Coschigano, P. W., and B. Magasanik. 1991. The URE2 gene product of Saccharomyces cerevisiae plays an important role in the cellular response to the nitrogen source and has homology to glutathione S-transferases. Mol. Cell. Biol. 11:822-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Courchesne, W. E., and B. Magasanik. 1988. Regulation of nitrogen assimilation in Saccharomyces cerevisiae: roles of the URE2 and GLN3 genes. J. Bacteriol. 170:708-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cunningham, T. S., R. Andhare, and T. G. Cooper. 2000. Nitrogen catabolite repression of DAL80 expression depends on the relative levels of Gat1p and Ure2p production in Saccharomyces cerevisiae. J. Biol. Chem. 275:14408-14414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cunningham, T. S., and T. G. Cooper. 1991. Expression of DAL80, whose product is homologous to the GATA factors and is a negative regulator of multiple nitrogen catabolic genes in Saccharomyces cerevisiae, is sensitive to nitrogen catabolite repression. Mol. Cell. Biol. 11:6205-6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cunningham, T. S., R. S. Corrington, and T. G. Cooper. 1994. The UGA4 UASNTR site required for GLN3-dependent transcriptional activation also mediates DAL80-responsive regulation and DAL80 protein binding in Saccharomyces cerevisiae. J. Bacteriol. 176:4718-4725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D'Alessio, M., and M. C. Brandriss. 2000. Cross-pathway regulation in Saccharomyces cerevisiae: activation of the proline utilization pathway by Gal4p in vivo. J. Bacteriol. 182:3748-3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daugherty, J. R., R. Rai, H. El Berry, and T. G. Cooper. 1993. Regulatory circuit for responses of nitrogen catabolic gene expression to the GLN3 and DAL80 proteins and nitrogen catabolite repression in Saccharomyces cerevisiae. J. Bacteriol. 175:64-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.des Etages, S. G., D. A. Falvey, R. J. Reece, and M. C. Brandriss. 1996. Functional analysis of the PUT3 transcriptional activator of the proline utilization pathway in Saccharomyces cerevisiae. Genetics 142:1069-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.des Etages, S. G., D. Saxena, H. L. Huang, D. Barber, and M. C. Brandriss. 2001. Conformational changes play a role in regulating the activity of the proline utilization pathway-specific regulator in Saccharomyces cerevisiae. Mol. Microbiol. 40:890-899. [DOI] [PubMed] [Google Scholar]
- 29.Dilova, I., C.-Y. Chen, and T. Powers. 2002. Mks1 in concert with TOR signaling negatively regulates RTG target gene expression in S. cerevisiae. Curr. Biol. 12:389-395. [DOI] [PubMed] [Google Scholar]
- 30.Drillien, R., M. Aigle, and F. Lacroute. 1973. Yeast mutants pleiotropically impaired in the regulation of the glutamate dehydrogenases. Biochem. Biophys. Res. Commun. 53:367-372. [DOI] [PubMed] [Google Scholar]
- 31.Drillien, R., and F. Lacroute. 1972. Ureidosuccinic acid uptake in yeast and some aspects of its regulation. J. Bacteriol. 109:203-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dubois, E., and M. Grenson. 1974. Absence of involvement of glutamine synthetase and of NAD-linked glutamate dehydrogenase in the nitrogen catabolite repression of arginase and other enzymes in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 60:150-157. [DOI] [PubMed] [Google Scholar]
- 33.Dubois, E., S. Vissers, M. Grenson, and J. M. Wiame. 1977. Glutamine and ammonia in nitrogen catabolite repression of Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 75:233-239. [DOI] [PubMed] [Google Scholar]
- 34.Grenson, M. 1969. The utilization of exogenous pyrimidines and the recycling of uridine-5′-phosphate derivatives in Saccharomyces cerevisiae, as studied by means of mutants affected in pyrimidine uptake and metabolism. Eur. J. Biochem. 11:249-260. [DOI] [PubMed] [Google Scholar]
- 35.Grenson, M., E. Dubois, M. Piotrowska, R. Drillien, and M. Aigle. 1974. Ammonia assimilation in Saccharomyces cerevisiae as mediated by the two glutamate dehydrogenases. Mol Gen. Genet. 128:73-85. [DOI] [PubMed] [Google Scholar]
- 36.Hardwick, J. S., F. G. Kuruvilla, J. K. Tong, A. F. Shamji, and S. L. Schreiber. 1999. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. USA 96:14866-14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang, H. L., and M. C. Brandriss. 2000. The regulator of the yeast proline utilization pathway is differentially phosphorylated in response to the quality of the nitrogen source. Mol. Cell. Biol. 20:892-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jauniaux, J.-C., M. Vandenbol, S. Vissers, K. Broman, and M. Grenson. 1987. Nitrogen catabolite regulation of proline permease in Saccharomyces cerevisiae. Cloning of the PUT4 gene and study of PUT4 RNA levels, in wild-type and mutant strains. Eur. J. Biochem. 164:601-606. [DOI] [PubMed] [Google Scholar]
- 39.Jones, E. W. 1991. Tackling the protease problem in Saccharomyces cerevisiae. Methods Enzymol. 194:428-453. [DOI] [PubMed] [Google Scholar]
- 40.Komeili, A., K. P. Wedaman, E. K. O'Shea, and T. Powers. 2000. Mechanism of metabolic control: target of rapamycin signaling links nitrogen quality to the activity of the Rtg1 and Rtg3 transcription factors. J. Cell Biol. 151:863-878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuruvilla, F. G., A. F. Shamji, and S. L. Schreiber. 2001. Carbon- and nitrogen-quality signaling to translation are mediated by distinct GATA-type transcription factors. Proc. Natl. Acad. Sci. USA 98:7283-7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 227:680-685. [DOI] [PubMed] [Google Scholar]
- 43.Liao, X., and R. A. Butow. 1993. RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell 72:61-71. [DOI] [PubMed] [Google Scholar]
- 44.Liu, Z., and R. A. Butow. 1999. A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in the response to a reduction or loss of respiratory function. Mol. Cell. Biol. 19:6720-6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Magasanik, B., and C. A. Kaiser. 2002. Nitrogen regulation in Saccharomyces cerevisiae. Gene 290:1-18. [DOI] [PubMed] [Google Scholar]
- 46.Marczak, J. E., and M. C. Brandriss. 1991. Analysis of constitutive and noninducible mutations of the PUT3 transcriptional activator. Mol. Cell. Biol. 11:2609-2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Minehart, P. L., and B. Magasanik. 1991. Sequence and expression of GLN3, a positive nitrogen regulatory gene of Saccharomyces cerevisiae encoding a protein with a putative zinc finger DNA-binding domain. Mol. Cell. Biol. 11:6216-6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rai, R., J. R. Daugherty, and T. G. Cooper. 1995. UASNTR functioning in combination with other UAS elements underlies exceptional patterns of nitrogen regulation in Saccharomyces cerevisiae. Yeast 11:247-260. [DOI] [PubMed] [Google Scholar]
- 49.Rai, R., J. R. Daugherty, T. S. Cunningham, and T. G. Cooper. 1999. Overlapping positive and negative GATA factor binding sites mediate inducible DAL7 gene expression in Saccharomyces cerevisiae. J. Biol. Chem. 274:28026-28034. [DOI] [PubMed] [Google Scholar]
- 50.Rowen, D. W., N. Esiobu, and B. Magasanik. 1997. Role of GATA factor Nil2p in nitrogen regulation of gene expression in Saccharomyces cerevisiae. J. Bacteriol. 179:3761-3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidt, A., T. Beck, A. Koller, J. Kunz, and M. N. Hall. 1998. The TOR nutrient signalling pathway phosphorylates NPR1 and inhibits turnover of the tryptophan permease. EMBO J. 17:6924-6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shamji, A. F., F. G. Kuruvilla, and S. L. Schreiber. 2000. Partitioning the transcriptional program induced by rapamycin among the efforts of the Tor proteins. Curr. Biol. 10:1574-1581. [DOI] [PubMed] [Google Scholar]
- 53.Siddiqui, A. H., and M. C. Brandriss. 1988. A regulatory region responsible for proline-specific induction of the yeast PUT2 gene is adjacent to its TATA box. Mol. Cell. Biol. 8:4634-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Siddiqui, A. H., and M. C. Brandriss. 1989. The Saccharomyces cerevisiae PUT3 activator protein associates with proline-specific upstream activation sequences. Mol. Cell. Biol. 9:4706-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Soussi-Boudekou, S., and B. Andre. 1999. A co-activator of nitrogen-regulated transcription in Saccharomyces cerevisiae. Mol. Microbiol. 31:753-762. [DOI] [PubMed] [Google Scholar]
- 56.Soussi-Boudekou, S., S. Vissers, A. Urrestarazu, J.-C. Jauniaux, and B. Andre. 1997. Gzf3p, a fourth GATA factor involved in nitrogen-regulated transcription in Saccharomyces cerevisiae. Mol. Microbiol. 23:1157-1168. [DOI] [PubMed] [Google Scholar]
- 57.Stanbrough, M., and B. Magasanik. 1996. Two transcription factors, Gln3p and Nil1p, use the same GATAAG sites to activate the expression of GAP1 of Saccharomyces cerevisiae. J. Bacteriol. 178:2465-2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stanbrough, M., D. W. Rowen, and B. Magasanik. 1995. Role of the GATA factors Gln3p and Nil1p of Saccharomyces cerevisiae in the expression of nitrogen-regulated genes. Proc. Natl. Acad. Sci. USA 92:9450-9454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Talibi, D., M. Grenson, and B. Andre. 1995. cis- and trans-acting elements determining induction of the genes of the γ-aminobuytrate (GABA) utilization pathway in Saccharomyces cerevisiae. Nucleic Acids Res. 23:550-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, S.-S., and M. C. Brandriss. 1987. Proline utilization in Saccharomyces cerevisiae: sequence regulation and mitochondrial localization of the PUT1 gene product. Mol. Cell. Biol. 7:4431-4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xu, S., D. A. Falvey, and M. C. Brandriss. 1995. Roles of URE2 and GLN3 in the proline utilization pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 15:2321-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]