Abstract
β-Oxidation of long-chain fatty acids and branched-chain fatty acids is carried out in mammalian peroxisomes by a multifunctional enzyme (MFE) or d-bifunctional protein, with separate domains for hydroxyacyl coenzyme A (CoA) dehydrogenase, enoyl-CoA hydratase, and steroid carrier protein SCP2. We have found that Dictyostelium has a gene, mfeA, encoding MFE1 with homology to the hydroxyacyl-CoA dehydrogenase and SCP2 domains. A separate gene, mfeB, encodes MFE2 with homology to the enoyl-CoA hydratase domain. When grown on a diet of bacteria, Dictyostelium cells in which mfeA is disrupted accumulate excess cyclopropane fatty acids and are unable to develop beyond early aggregation. Axenically grown mutant cells, however, developed into normal fruiting bodies composed of spores and stalk cells. Comparative analysis of whole-cell lipid compositions revealed that bacterially grown mutant cells accumulated cyclopropane fatty acids that remained throughout the developmental stages. Such a persistent accumulation was not detected in wild-type cells or axenically grown mutant cells. Bacterial phosphatidylethanolamine that contains abundant cyclopropane fatty acids inhibited the development of even axenically grown mutant cells, while dipalmitoyl phosphatidylethanolamine did not. These results suggest that MFE1 protects the cells from the increase of the harmful xenobiotic fatty acids incorporated from their diets and optimizes cellular lipid composition for proper development. Hence, we propose that this enzyme plays an irreplaceable role in the survival strategy of Dictyostelium cells to form spores for their efficient dispersal in nature.
Membrane-mediated cellular functions are crucial for the life of the cell. For such functions, cellular lipid composition must be strictly regulated. The cellular lipid composition is under the control of the peroxisomal β-oxidation, which degrades very-long-chain fatty acids and branched-chain fatty acids (4). In fact, impaired β-oxidation in peroxisomes causes serious diseases with the accumulation of nonmetabolized fatty acids (25, 27, 28). One of the β-oxidation steps of these fatty acids is catalyzed by d-bifunctional protein (DBP) (23, 25, 26), whose counterpart in rat is called multifunctional enzyme 2 (MFE2) (18). Little is known, however, about how the accumulation of nonmetabolizable fatty acids affects cellular physiology.
Dictyostelium discoideum, one of the simplest multicellular eukaryotes, might serve as a suitable model system for understanding the role of peroxisomal β-oxidation in various cellular functions. The life cycle of Dictyostelium is composed of unicellular proliferating and multicellular developing stages. In the natural environment, Dictyostelium cells multiply their genome by feeding on microorganisms such as bacteria and yeasts that grow on dung or rotten leaves on the forest floor. In laboratories, Dictyostelium cells can grow by feeding upon bacteria such as Escherichia coli or Klebsiella aerogenes on an agar plate. Previous studies on the fatty acid composition of these prey showed that CFAs (cyclopropane fatty acids) are the constitutive components of their lipids (9, 17). Although vegetative Dictyostelium cells take up CFAs from bacteria, these xenobiotic fatty acids do not remain long in Dictyostelium cells (12). Upon exhaustion of nutrients, Dictyostelium amoebae initiate multicellular development to form fruiting bodies. At this stage, CFAs are barely detectable (12), indicating that fatty acid composition is regulated during development.
In order to understand how lipid abnormalities affect cellular functions, we aimed to perturb fatty acid composition in Dictyostelium and examine the consequences. We first searched the Dictyostelium cDNA database (15) to find Dictyostelium counterparts of peroxisomal MFE2 or DBP (MFE2/DBP). We found two genes, mfeA (GenBank accession number AB042104) and mfeB (accession number AB100096), encoding MFE1 and MFE2, respectively. In this study, we analyzed the in vivo functions of MFE1 in multicellular development of Dictyostelium by disrupting the mfeA gene and showed that mfeA plays a crucial role in optimization of cellular lipid composition necessary for multicellular development of bacterially grown Dictyostelium cells. We propose that MFE1 is essential for survival of Dictyostelium cells in nature. This enzyme might also be essential for the survival of other soil amoebae and animals such as Caenorhabditis elegans.
MATERIALS AND METHODS
Strains, cultures, and morphogenesis.
D. discoideum Ax2 (8A) (subcloned from the Ax2 strain at Y. Maeda's lab in Tohoku University) and various mutant strains were used. Cells were cultivated at 21°C either axenically or with E. coli B/r. For axenic culture, HL5 (29) supplemented with 5 ng of vitamin B12 and 100 ng of folic acid per ml was used. In the case of the mfeA-null cells, 10 μg of blasticidin S/ml was added to HL5. For analyses of bacterially grown cells, cultures were carried out for various periods with E. coli B/r on 5LP plates (1% agar containing 0.5% lactose and 0.5% peptone). For multicellular development, harvested cells were washed twice with cold 12 mM NaK2-phosphate buffer (pH 6.1) (PB), resuspended at a density of 1× 107 to 8 × 107 cells/ml in PB, and then allowed to develop on 1.5% nonnutrient agar or a Millipore filter. The mfeA-null strain expressing intact, truncated, or green fluorescent protein (GFP)-conjugated MFE1 was selected and maintained in HL5 containing 20 μg of G418/ml.
Disruption and overexpression of the mfeA gene.
Clone SLA480 (accession no. AB042104 for DDBJ/GenBank) was obtained from the Dictyostelium cDNA project (15). SLA480 contains the open reading frame (ORF) of mfeA encoding MFE1 at the SalI and NotI sites of pSPORT. In order to create a construct for the disruption of mfeA, mfeA was recloned into the SalI and NotI sites of pBluescript II KS(−) after amplification by PCR by using the primers GGTCGACATGGCATTAAATTTTAAAGATAA and AGCGGCCGCTTATAATTTTGAACCTTGCAT. The BSR cassette that provides blasticidin S resistance (1, 22) was inserted into the unique NdeI site of mfeA. After digestion with SalI and NotI, the plasmid was introduced into exponentially growing Ax2 (8A) cells by electroporation, according to the method of Howard et al. (8). For ectopic expression of MFE1 under the control of the actin 15 promoter, mfeA was recloned into the extrachromosomal vector HK12 designed by H. Kuwayama as follows: the ORF of mfeA was amplified by PCR with the primers CGGATCCAAAAATGGCATTAAATTTTAAAG and GTCTAGATTATAATTTTGAACCTTGCATTA. The amplified fragment was digested with BamHI and XbaI and was then cloned into the BglII and SpeI sites of HK12. For ectopic expression of three truncated forms of MFE1, each fragment obtained by PCR as described below was similarly cloned into HK12. The HCD domain was amplified by using the primers CGGATCCAAAAATGGCATTAAATTTTAAAG and GTCTAGATTAACCTGATGGAGTGGCAGCAAG. To add the SKL peroxisome-targeting signal at the C terminus of the HCD domain, CGGATCCAAAAATGGCATTAAATTTTAAAG and GTCTAGATTATAATTTTGAACCTGATGGAGTGGCAGCAAG were used for amplification. Similarly, the SCP2 domain was amplified by using the primers CGGATCCAAAAATGTCTGTCGTTGTCGATGGTTAC and GTCTAGATTATAATTTTGAACCTTGCATTA. For the expression of GFP-MFP1, the ORF of mfeA was also amplified by PCR by using CGCGGATCCAATGGCATTAAATTTTAAAG and the T7 promoter primer. After digestion with BamHI and SacI, the amplified mfeA gene was cloned into the BamHI and SacI sites of pBIGΔBam in order to fuse MFE1 at the C terminus of GFP. pBIGΔBam was a generous gift from T. Uyeda (National Institute of Advanced Industrial Science and Technology of Japan).
Southern blotting.
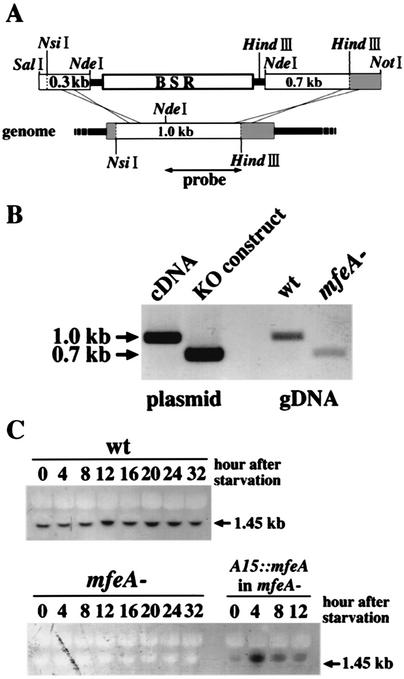
Genomic DNA was extracted according to the method of Nellen et al. (16). For Southern blotting, about 10 μg of DNA was digested with HindIII and NsiI. The NdeI and HindIII fragment obtained from mfeA cDNA (SLA480) (Fig. 2A) was used as a probe after preparation according to the manufacturer's directions (digoxigenin labeling kit; Roche Diagnostics).
FIG. 2.
Generation of the mfeA-null mutant. (A) A schematic view of the disruption construct and the parental gene. The disruption construct was made by the insertion of a BSR cassette into mfeA at the NdeI site. The NdeI and HindIII fragment of mfeA cDNA was used as the probe for both Southern and Northern blotting. The shaded boxes indicate the regions undetected by the probe. (B) Southern blotting was performed. Genomic DNA of wild-type and mfeA-null cells and mfeA cDNA and the disruption construct were analyzed after digestion with NsiI and HindIII. A 0.3-kb shift in band size occurred in mfeA-null cells. (C) Northern blotting was performed. Total RNA samples were extracted from wild-type (wt), mfeA-null (mfeA−), and mfeA-null cells expressing mfeA under the actin 15 promoter (A15::mfeA in mfeA−) at the time points indicated during the starvation. In the wild type, a single 1.45-kb transcript was constitutively expressed during development. In mfeA-null cells, this transcript was missing. In mfeA-null cells expressing mfeA under the actin 15 promoter, a slightly larger transcript (1.85 kb) was expressed depending on the actin 15 promoter activity. Ten micrograms of genomic DNA and total RNA was analyzed for each sample.
Northern blotting.
In order to extract total RNA from cells at various developmental stages, both wild-type and mfeA-null strains that had been cultivated for 2 days on an E. coli lawn were allowed to develop at 2.5 × 106 cells/cm2 on a filter (Whatman no. 50) supported by nonnutrient agar. Under these conditions, bacterially grown mfeA-null cells developed to the mound stage. The extraction of total RNA and Northern blotting were performed as described previously (11). An NdeI and HindIII fragment obtained from SLA480 (Fig. 2A) was used as a probe after preparation according to the manufacturer's directions (digoxigenin labeling kit; Roche Diagnostics).
Preparation of lipids and fatty acids.
Total lipids were extracted from wet E. coli, K. aerogenes, and Dictyostelium cells by the method of Bligh and Dyer (2). After being concentrated in a centrifuge evaporator, the extractable lipid was stored at −20°C. For analysis of the simple lipid composition, the total lipids were separated by thin-layer chromatography (TLC) on a silica plate (silica gel 60; Merck, Darmstadt, Germany) with a solvent system of hexane-diethyl ether-acetic acid (70:30:1 or 80:20:1 [vol/vol/vol]). Authentic cholesterol, cholesterol oleate, triolein, oleic acid, and methyl oleate were purchased from Nu-Check-Prep, Inc. (Elysian, Minn.). Lipids were visualized by heating the plate at 110°C for 10 min after spraying with 15% H2SO4. For fatty acid analysis, the plate was sprayed with acetone-water (4:1 [vol/vol]) containing 0.01% primuline. Each spot detected under UV light was scraped from the plate and was then transmethylated as described previously (19). The fatty acid moiety of each lipid was quantified by gas chromatography (GC) as described below, and methyl heneicosanoic acid was used as an internal standard.
GC and GC/MS.
The fatty acid methyl ester (FAME) residues were then redissolved in an appropriate volume of hexane. Analysis of the FAMEs was performed with a gas chromatograph (GC1700; Shimadzu, Kyoto, Japan) equipped with a capillary column (BPX70, 50 m by 0.22 mm [internal diameter]; SGE Inc., Austin, Tex.). The injector and the detector were maintained at 250°C, and the temperature of the oven was programmed to rise from 185 to 225°C at a rate of 2°C/min and was then maintained at 225°C for 10 min. Peak area was quantified by using a Shimadzu C-R8A (Shimadzu, Kyoto, Japan). GC-mass spectrometry (GC/MS) was performed with a Saturn 2000 ion trap mass spectrometer (Varian Inc., Walnut Creek, Calif.) connected to a Varian 3800 gas chromatograph equipped with a BPX70 capillary column (25 m by 0.22 mm [internal diameter]). The oven temperature was programmed to increase from 80 to 240°C at a rate of 4°C per minute. Helium was used as the carrier gas. GC/MS was operated at 70 eV and at 230°C with a mass range of 40 to 400 atomic mass units.
Nile red staining.
Intracellular lipid droplets were detected by staining cells with Nile red according to the method described previously (7). The cells were observed with an Olympus microscope model AX70 and were photographed with a PM-C35DX camera equipped with U-PHOTO, a fully automatic exposure photo tube (Olympus, Tokyo, Japan).
Incubation with bacterial PE.
Bacterial total lipid was extracted from E. coli B/r that had been cultivated for 2 days at 22°C on 5LP plates according to the method of Bligh and Dyer (2) and was subjected to TLC with a solvent system of chloroform-methanol-water (65:25:4 [vol/vol/vol]). After visualization with primuline, the spot corresponding to phosphatidylethanolamine (PE) was scraped from the plate and reextracted by the method of Bligh and Dyer (2). The effect of bacterial PE was examined by incubating axenically grown mutant cells for 20 h in HL5 supplemented with bacterial PE at 0.16 mg/ml. For an experimental control, dipalmitoyl PE (Sigma, St. Louis, Mo.) was used at the same concentration. After harvesting, cells were washed, resuspended in PB, and allowed to develop on the nitrocellulose filter (Millipore Co., Bedford, Mass.) at a density of 2.5 × 106 cells/cm2. The developmental process was photographed with an HC-300Z/OL digital camera equipped for an Olympus SZX12 (Olympus).
In situ hybridization.
Whole-mount in situ hybridization analysis was performed according to the method described previously (5, 14, 24). Digoxigenin-labeled RNA probe was prepared by in vitro transcription by using SP6 RNA polymerase. SLA480 linearized with SalI digestion was used as a template.
Fluorescent microscopic observation.
To determine the intracellular localization of MFE1, cells expressing GFP-MFE1 were observed under an agar overlay (6) after fixation with 100% ethyl alcohol containing 1% (vol/vol) formaldehyde at −10°C. Bonner's salt solution (10 mM NaCl, 10 mM KCl, and 3 mM CaCl2) (3) was used in this experiment instead of PB, and phosphate-buffered saline containing 0.2% Triton X-100 was used to remove the fixative. Images were photographed with an Olympus AX70.
RESULTS
Structure of peroxisomal MFE1 of Dictyostelium.
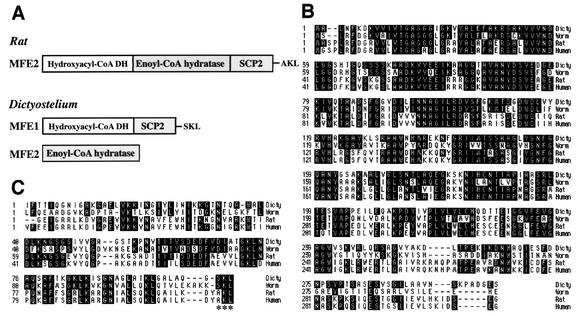
By searching the Dictyostelium cDNA database, we found that Dictyostelium has two genes, mfeA and mfeB, with homology to the mammalian enzyme termed MFE or DBP, which consists of separate domains for hydroxyacyl coenzyme A (CoA) dehydrogenase (HCD), enoyl-CoA hydratase (ECH), and steroid carrier protein SCP2 (Fig. 1A). mfeA encodes MFE1, a protein with homology to the HCD and SCP2 domains, while mfeB encodes MFE2, a protein with homology to the ECH (Fig. 1A). The N-terminal and C-terminal domains of MFE1 show about 57 and 41% identity to the HCD (Fig. 1B) and SCP2 (Fig. 1C) domains, respectively. MFE2 is about 40% identical to the ECH domain (data not shown). A tripeptide sequence SKL, consensus peroxisomal targeting signal 1 (PTS1), exists at the C terminus of MFE1 (Fig. 1C, asterisks). We also found that C. elegans has two enzymes with homology to Dictyostelium MFE1 and MFE2. The worm counterpart of Dictyostelium MFE1 is 54.6% identical to the Dictyostelium enzyme (Fig. 1B and C).
FIG. 1.
(A) Structures of Rat MFE2 and Dictyostelium MFE1. Rat MFE2 consists of three domains, as do other mammalian MFE2/DBPs, while Dictyostelium MFE1 consists of two domains, also found in the C. elegans counterpart. The ECH domain exists as a distinct protein MFE2. (B and C) Comparison in amino acid sequence of the HCD domain (B) and SCP2 domain (C) among Dictyostelium MFE1 (accession number: BAA94961) and worm, rat, and human counterparts (accession numbers T16638, NP077368, and P51659, respectively) by using the MegAlign program of DNASTAR.
Role of MFE1 in multicellular development of Dictyostelium.
In order to understand the role of MFE1 in Dictyostelium development, mfeA, the gene which encodes MFE1, was disrupted by homologous recombination (Fig. 2A). Eight independent transformants were obtained; six were arrested at the preaggregation stage on bacterial lawns (Fig. 3A), and the other two transformants developed normally (data not shown). The plaque sizes of the wild-type cells and the mutant cells on E. coli lawns were indistinguishable, but the mutant cells did not grow in suspension in PB in the presence of E. coli, where wild-type cells grow well (data not shown). Both Southern and Northern blotting confirmed that mfeA was disrupted in the six developmentally arrested clones (Fig. 2B and C) but not in the other two strains (data not shown). Northern blotting also revealed that a single 1.45-kb transcript of mfeA was constitutively expressed throughout development in the wild type. In situ hybridization indicates that mfeA was expressed in all cell types (data not shown).
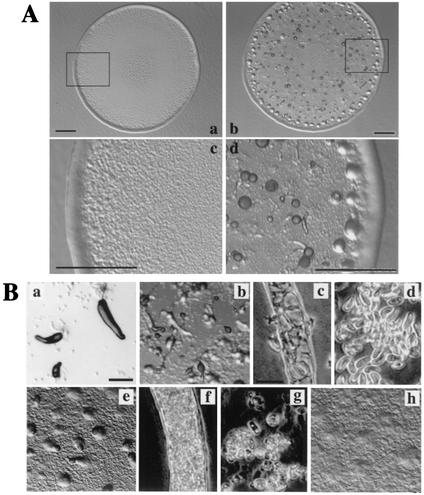
FIG. 3.
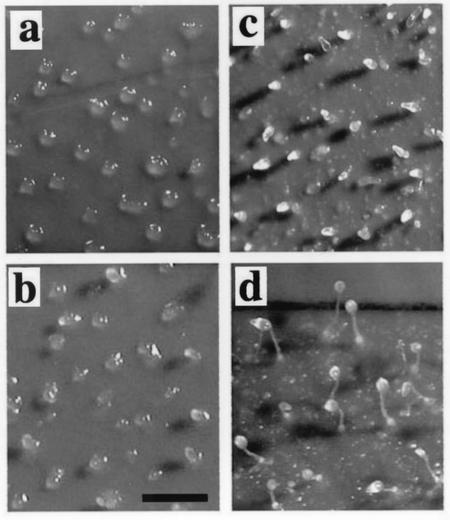
The development of mfeA-null cells cultivated in HL5 or on E. coli B/r. (A) Plaques formed from mfeA-null (a and c) and wild-type cells (b and d) on an E. coli lawn. Panels c and d give higher magnification of the images in the rectangles in panels a and b, respectively. Scale bars, 1 mm. All plaques formed from mfeA-null cells did not show any sign of multicellular development as seen in panels a and c. (B) Effects of culture conditions on development of the mutant cells on nonnutrient agar. The axenically grown mutant cells formed slugs (a) and fruiting bodies (b) with normal stalk cells (c) and spores (d). mfeA-null cells that had been cultivated on bacteria for 3 h often formed aberrant multicellular structures (e) that eventually developed into fruiting body-like structures with a stalk but filled with abnormal cells (f) and a globular mass atop the stalk, which consisted of a mixture of ellipsoidal cells or unhealthy amoebae (g). mfeA-null cells that had been cultivated for 24 h on bacterial lawn did not develop into multicellular structures on nonnutrient agar plates (h). Scale bars, 0.2 mm and 20 μm.
Surprisingly, mfeA-null cells developed into fruiting bodies with normal stalk cells and spores when axenically cultured and were then allowed to develop on nonnutrient agar (Fig. 3B). This suggests that certain bacterial substances exert deleterious effects on development. In order to test this possibility, development of mfeA− cells was monitored on nonnutrient agar after cultivation for various periods with bacteria. Development was severely affected depending on the period of bacterial cultivation. mfeA-null cells that had been cultured for 8 or more hours with E. coli failed to aggregate properly on nonnutrient agar (Fig. 3B). When cultivated with E. coli for 3 h and then transferred to nonnutrient agar, they formed a few fruiting body-like structures. Such structures contained neither intact stalk cells nor intact spores (Fig. 3B). These results suggest that certain bacterial substances inhibit both multicellular development and cell differentiation of the mutant.
HCD domain and PTS1 have essential roles in Dictyostelium development.
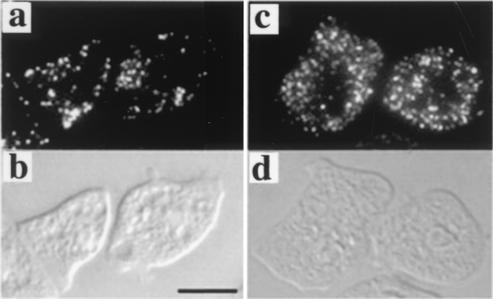
The intracellular localization of MFE1 was examined by expressing GFP-conjugated MFE1 in mfeA-null cells. When GFP was fused to the C terminus of MFE1, MFE1-GFP was distributed throughout the cytoplasm and transformed cells remained developmentally arrested (data not shown), perhaps as a consequence of GFP masking PTS1 at the C terminus. In contrast, GFP-MFE1 in which GFP was fused to the N terminus of MFE1 localized to particulate structures (Fig. 4), as seen in cultured human fibroblasts (10). This fusion protein rescued the mfeA-null phenotype. These results suggest that MFE1 is a peroxisomal enzyme.
FIG. 4.
Subcellular localization of MFE1 was visualized in mfeA-null cells expressing GFP-MFE1 after axenic (a) or bacterial (c) cultivation. (b and d) Differential interference contrast images of panels a and c, respectively. Scale bar, 10 μm.
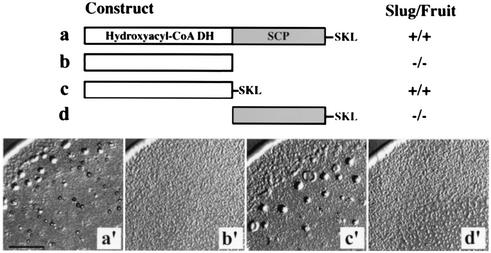
As mentioned previously, MFE1 consists of the N-terminal HCD and C-terminal SCP2 domains and has a tripeptide SKL as PTS1 at the C terminus (Fig. 1A). We next examined which domain is responsible for the MFE1 function. In order to do this, each domain was expressed under the control of the actin 15 promoter in mfeA-null cells and their complementation ability of the mutant phenotype was examined (Fig. 5). Developmental defects of the bacterially grown mutant were rescued by expressing either full-length MFE1 (Fig. 2C and 5) or the SKL-tagged HCD domain (Fig. 5). Therefore, we conclude that MFE1 plays an indispensable role within peroxisomes for development of bacterially grown Dictyostelium cells, which is mediated by the HCD domain.
FIG. 5.
Deletion analysis of each domain of MFE1. The construct to express the gene coding for MFE1 or truncated proteins (lanes a to d) was introduced into mfeA-null cells. Phenotypes of the resulting transformants are shown (a′ to d′). Scale bar, 1 mm.
Altered lipid composition in mfeA-null cells.
The amino acid sequence of MFE1 indicates that it is likely to be involved in β-oxidation of fatty acids. Therefore, intracellular lipid content could be altered in mfeA-null cells. To test this possibility, cells were stained with Nile red, an excellent vital stain for lipid droplets (7). When bacterially grown cells were stained, brilliant green granular structures were detectable in mfeA-null but not in wild-type cells (Fig. 6A), suggesting that nonmetabolized fatty acids might be sequestered in lipid droplets in the mutant. Lipid droplets were not observed in either axenically grown mfeA-null or wild-type cells (data not shown).
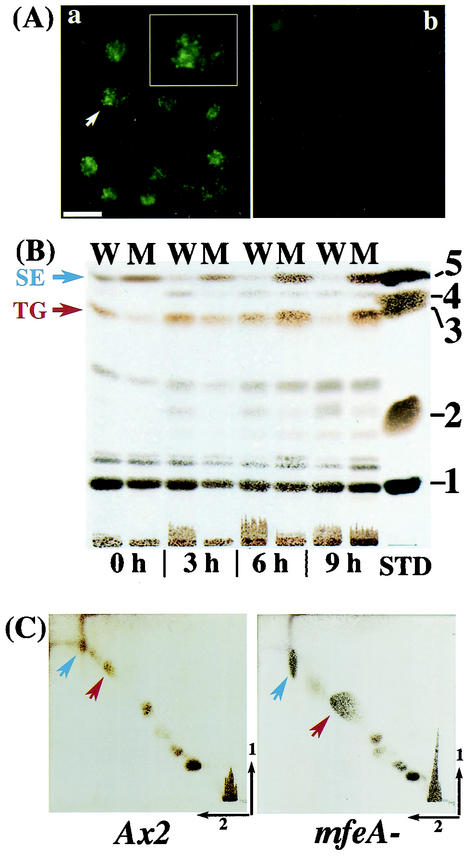
FIG. 6.
Lipid analyses. (A) Lipid droplets in the bacterially grown mfeA-null (a) and wild-type (b) cells were visualized with Nile red. Inset in panel a shows higher magnification of a single point with a white arrow. Scale bar: 20 μm. (B) TLC of total lipids in wild-type (W) and mfeA-null (M) cells after various periods of bacterial cultivation. Lipid moieties were visualized as described in Materials and Methods. The standard lipids (STD), cholesterol oleate (1), methyl oleate (2), triolein (3), oleic acid (4), and cholesterol (5) were tested. (C) Two-dimensional TLC of total lipids extracted from Dictyostelium cells cultivated for 24 h on an E. coli lawn. Left panel, wild-type cells; right panel, mfeA-null cells. Blue arrows, SEs; red arrows, TGs.
Total lipids were extracted from bacterially grown Ax2 and mfeA-null cells and were subjected to TLC. Although the content of total lipids varied depending on culture periods and experiments, it was roughly twofold higher in the mutant than in wild-type cells after 9 h of cultivation with bacteria. The most remarkable change was an increase in triacylglycerides (TGs) and sterolesters (SEs) in the mutant, and this change was dependent on the length of time that the cells were feeding on bacteria (Fig. 6B and C). These results suggest that certain fatty acids might have been sequestered in these neutral lipids due to the loss of MFE1.
Altered fatty acid composition in mfeA-null cells.
In order to understand the function of MFE1 in diet lipid metabolism, fatty acid composition was analyzed in E. coli and Dictyostelium cells (Table 1). The major fatty acid constituents of E. coli were C16:0, C18:1 (11), cy17:0 (cis-9,10-methylenehexadecanoic acid), and cy19:0 (cis-9,10-methyleneoctadecanoic acid). Among these fatty acids, C16:0 was rapidly metabolized in not only bacterially grown wild-type cells but also in bacterially grown mfeA-null cells, while C18:1 remained at a constant level. On the other hand, both cy17:0 and cy19:0 were barely detectable in axenically grown Dictyostelium cells. However, a substantial amount of these fatty acids became detectable in bacterially grown cells. The most remarkable difference between the mutant and wild-type cells was observed in cy17:0 and cy19:0, CFAs, which were more enriched in the mutant than in wild-type cells. These CFAs were the major fatty acids in TGs, SEs, and phospholipids (PLs) in the mutant cells (Table 2). It is noteworthy that these CFAs decreased in wild-type cells during starvation but not in the mutant cells (Table 3), indicating that MFE1 is involved in CFA metabolism. On the other hand, the proportion of C18:2 to total lipid was about 2.5-fold lower in bacterially grown mutant cells than in wild-type cells (Table 1). C18:2 was the major fatty acid constituent of TGs, SEs, and PLs in wild-type cells but not in mfeA mutant cells (Table 2). C18:2 continued to rise in the wild type during starvation but not in the mutant (Table 3). These results suggest that the mutant phenotype might have resulted either from an increase in CFAs or a decrease in C18:2 or both.
TABLE 1.
Fatty acid composition in total lipids of Ax2, mfeA-null cells, and E. colia
| Fatty acid | Composition in:
|
||||
|---|---|---|---|---|---|
| E. coli | Wild type
|
mfeA−
|
|||
| E. coli (%) | HL5 (%) | E. coli (%) | HL5 (%) | ||
| C16:0 | 32.7 | 5.1 | 6.3 | 7.4 | 8.0 |
| C16:1(9) | 4.2 | 2.3 | 4.0 | 6.3 | 7.5 |
| C16:2(5,9) | 5.4 | 1.7 | 4.9 | 3.7 | |
| C18:1(11) | 23.8 | 21.8 | 31.8 | 18.5 | 29.2 |
| C18:2(5,9) | 2.9 | 15.2 | 1.1 | 11.1 | |
| C18:2(5,11) | 41.4 | 32.7 | 17.2 | 14.0 | |
| cy17:0 | 19.5 | 7.6 | 20.7 | ||
| cy19:0 | 13.0 | 8.9 | 15.0 | ||
Fatty acid composition was analyzed for the total lipids extracted from E. coli and Dictyostelium cells that had been grown for 24 h on an E. coli lawn as described in Materials and Methods. Also, it was notable that the ratio of polyunsaturated fatty acids increased to 6.1% in axenically grown mfeA-null cells. In addition to these major fatty acids, C14:0, C16:1(5), and C18:0 were detected as minor components that constituted less than 5% of total fatty acids of bacterially grown wild-type and mfeA-null cells.
TABLE 2.
Major fatty acid composition in TGs, SEs, and PLs of bacterially grown Dictyostelium cellsa
| Fatty acid | Composition in:
|
|||||
|---|---|---|---|---|---|---|
| TGs (%)
|
SEs (%)
|
PLs (%)
|
||||
| Wild type | mfeA-null | Wild type | mfeA-null | Wild type | mfeA-null | |
| C16:1(9) | 2.5 | 3.3 | 1.5 | 2.9 | 3.2 | 5.3 |
| C16:2(5,9) | 4.0 | 2.1 | 3.3 | 2.9 | 7.8 | 5.8 |
| C18:1(11) | 20.7 | 13.5 | 8.0 | 8.0 | 25.0 | 19.2 |
| C18:2(5,11) | 22.6 | 10.0 | 33.7 | 17.8 | 36.7 | 19.9 |
| cy17:0 | 18.4 | 31.8 | 5.8 | 23.3 | 7.0 | 17.3 |
| cy19:0 | 9.5 | 21.7 | 3.2 | 18.0 | 5.5 | 15.0 |
After separation of total lipids by TLC, the plate was visualized by using primuline. Each spot was scraped off from the plate and was then transmethylated. FAMEs were analyzed by GC and GC/MS as described in Materials and Methods.
TABLE 3.
Changes in major fatty acid composition during starvation in bacterially grown wild-type and mfeA-null cellsa
| Fatty acid | Change in:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Wild type (%)
|
mfeA-null (%)
|
|||||||
| 0 h | 3 h | 6 h | 9 h | 0 h | 3 h | 6 h | 9 h | |
| C16:1 | 3.4 | 2.3 | 1.6 | 1.6 | 5.6 | 5.6 | 6.0 | 6.2 |
| C16:2 | 7.0 | 7.2 | 7.3 | 6.7 | 5.4 | 5.2 | 4.9 | 4.8 |
| C18:1 | 20.1 | 20.7 | 20.8 | 20.9 | 22.3 | 22.0 | 22.3 | 22.7 |
| C18:2 | 39.1 | 43.7 | 47.8 | 50.5 | 22.6 | 22.6 | 22.6 | 23.2 |
| cy17:0 | 12.3 | 9.0 | 6.9 | 5.2 | 20.4 | 20.0 | 19.4 | 19.6 |
| cy19:0 | 8.3 | 7.8 | 7.0 | 5.9 | 10.2 | 10.3 | 10.5 | 11.0 |
Dictyostelium cells were grown for 24 h on an E. coli lawn. Cells were allowed to develop on 2% agar plates at a density of 1.5 × 106/cm2. At 3-h intervals total lipids were extracted and their fatty acid compositions were analyzed as described in Materials and Methods. C16:2 and C18:1 were kept at constant ratios during starvation. A big change was observed in C16:1, C18:2, cy17:0, and cy19:0 in wild-type cells. C18:2 gradually increased, but C16:1, cy17:0, and cy19:0 decreased during starvation. On the other hand, such change was not detected in the fatty acid compositions of mfeA-null cells.
Causal relation between accumulated CFAs and developmental defect in mfeA-null cells.
To test whether impaired CFA metabolism caused the developmental defect, the effects of adding bacterial PE, which contains abundant CFAs, were examined in axenically grown mutant cells. Dipalmitoyl PE, which does not contain CFAs, was used as a control. We found that bacterial PE but not dipalmitoyl PE significantly suppressed the multicellular development of the axenically grown mutant cells (Fig. 7). After being cultivated for 20 h in HL5 supplemented with dipalmitoyl PE and then being starved, the mutant cells developed almost normally into fruiting bodies (Fig. 7). However, when cultured in the presence of bacterial PE, mfeA-null cells did not develop beyond the tipped aggregate stage after starvation (Fig. 7). It appears that the developmental arrest of bacterially grown mfeA-null cells results from abnormal accumulation of CFAs.
FIG. 7.
Effect of bacterial PE on the morphogenesis of mfeA-null cells. Morphogenesis of mfeA-null cells was monitored on a filter after cultivation in axenic medium HL5 supplemented with bacterial PE (a and b) or dipalmitoyl PE (c and d). It is clearly shown that bacterial PE that contains abundant CFAs suppressed the mutant development. The pictures were taken at 16 (a and c) and 22 (b and d) h of starvation. Scale bar, 0.5 mm.
DISCUSSION
In the present study, we showed that peroxisomal MFE1 plays an essential role in the survival strategy of Dictyostelium grown on bacteria. Bacterially grown mfeA-null cells cannot develop beyond the aggregation stage, and spores are not made. The amino acid sequence of MFE1 suggests that it is involved in β-oxidation of fatty acids, such as CFAs, that are the major components of bacterial lipids (9, 17). In fact, there was a remarkable increase in TGs and SEs with CFAs in bacterially grown Dictyostelium cells lacking MFE1. Concomitantly, there was a decrease in the proportion of dienoic acids such as C18:2 in bacterially grown mutant cells, suggesting that inhibition of Δ5-desaturases occurred in these cells (13, 20, 21). Thus, either an increase in CFAs or a decrease in these dienoic acids might have led to the mutant phenotype. However, a decrease in the proportion of C18:2 to total lipid also occurred in axenically grown mutant cells, which develop normally. Moreover, we found that development of mfeA-null cells was inhibited when cultured in the presence of bacterial PE, which abundantly contained CFAs. Therefore, it appears that the problems result from abnormal accumulation of CFAs.
How might accumulation of CFAs inhibit multicellular development of Dictyostelium? Since PLs and probably also SEs are the major components of the cell membrane, the accumulation of CFAs in these lipids might cause a reduction of membrane fluidity and/or an alteration of membrane microdomain structures, which might perturb membrane physiology and membrane protein functions. This notion is supported by our preliminary results demonstrating that both cell-cell contact mediated by gp80 and cyclic AMP signaling are impaired in bacterially grown mfeA-null cells (S. Matsuoka et al., unpublished data). We have also noticed that the mutant cells could not grow under shaking culture conditions in PB supplemented with washed bacteria (H. Kuwayama, unpublished data). These observations suggest that the unusual accumulation of CFAs seen in bacterially grown mutant cells interferes with normal membrane functions.
The present study demonstrates that the optimization of the cellular fatty acid composition is essential for the multicellular development of Dictyostelium. MFE1 plays an irreplaceable role in this process. Although the mechanism by which excess CFAs inhibit multicellular development is still unsolved, our study suggests that critical regulation of lipid and fatty acid composition is absolutely necessary for the survival of Dictyostelium in nature.
Acknowledgments
We greatly thank W. F. Loomis of University of California at San Diego for his critical reading of the manuscript and K. Nishio and M. Yokoyama for their technical assistance.
S. Matsuoka and T. Saito contributed equally to this study.
This study was supported by a research grant from Research for the Future (Japan Society for the Promotion of Science) to M.M. as a member of S. Kuhara's project, by Kyushu University (JSPS-RFTF96L00105), and by Grants-in-Aid for Scientific Research on Priority Areas to M.M. (nos. 13024248 and 12640633).
REFERENCES
- 1.Adachi, H., T. Hasebe, K. Yoshinaga, T. Ohta, and K. Sutoh. 1994. Isolation of Dictyostelium discoideum cytokinesis mutants by restriction enzyme-mediated integration of the blasticidin S resistance marker. Biochem. Biophys. Res. Commun. 205:1808-1814. [DOI] [PubMed] [Google Scholar]
- 2.Bligh, E. G., and W. J. Dyer. 1959. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37:911-917. [DOI] [PubMed] [Google Scholar]
- 3.Bonner, J. T., and M. K. Slifkin. 1949. A study of the control of differentiation: the proportion of stalk and spore cells in the slime mold Dictyostelium discoideum. Am. J. Bot. 36:727-734. [Google Scholar]
- 4.Breitling, R., Z. Marijanovic, D. Perovic, and J. Adamski. 2001. Evolution of 17β-HSD type 4, a multifunctional protein of β-oxidation. Mol. Cell. Endocrinol. 171:205-210. [DOI] [PubMed] [Google Scholar]
- 5.Escalante, R., and W. F. Loomis. 1995. Whole-mount in situ hybridization of cell-type-specific mRNAs in Dictyostelium. Dev. Biol. 171:262-266. [DOI] [PubMed] [Google Scholar]
- 6.Fukui, Y., S. Yumura, and T. K. Yumura. 1987. Agar-overlay immunofluorescence: high-resolution studies of cytoskeletal components and their changes during chemotaxis. Methods Cell Biol. 28:347-356. [DOI] [PubMed] [Google Scholar]
- 7.Greenspan, P., E. P. Mayer, and S. D. Fowler. 1985. Nile red: a selective fluorescent stain for intracellular lipid droplets. J. Cell Biol. 100:965-973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Howard, P. K., K. G. Ahern, and R. A. Firtel. 1988. Establishment of a transient expression system for Dictyostelium discoideum. Nucleic Acids Res. 16:2613-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jackson, M. B., and J. E. Cronan, Jr. 1978. An estimate of the minimum amount of fluid lipid required for the growth of Escherichia coli. Biochim. Biophys. Acta 512:472-479. [DOI] [PubMed] [Google Scholar]
- 10.Jiang, L. L., S. Miyazawa, and T. Hashimoto. 1996. Purification and properties of rat d-3-hydroxyacyl-CoA dehydratase: d-3-hydroxyacyl-CoA dehydratase/d-3-hydroxyacyl-CoA dehydrogenase bifunctional protein. J. Biochem. 120:633-641. [DOI] [PubMed] [Google Scholar]
- 11.Kuwayama, H., M. Oyama, Y. Kubohara, and M. Maeda. 2000. A novel role of differentiation-inducing factor-1 in Dictyostelium development, assessed by the restoration of a developmental defect in a mutant lacking mitogen-activated protein kinase ERK2. Dev. Growth Differ. 42:531-538. [DOI] [PubMed] [Google Scholar]
- 12.Long, B. H., and E. L. Coe. 1977. Fatty acid compositions of lipid fractions from vegetative cells and mature sorocarps of the cellular slime mold Dictyostelium discoideum. Lipids 12:414-417. [DOI] [PubMed] [Google Scholar]
- 13.Los, D. A., and N. Murata. 1998. Structure and expression of fatty acid desaturases. Biochim. Biophys. Acta 1394:3-15. [DOI] [PubMed] [Google Scholar]
- 14.Maeda, M., H. Kuwayama, M. Yokoyama, K. Nishio, T. Morio, H. Urushihara, M. Katoh, Y. Tanaka, T. Saito, H. Ochiai, K. Takemoto, H. Yasukawa, and I. Takeuchi. 2000. Developmental changes in the spatial expression of genes involved in myosin function in Dictyostelium. Dev. Biol. 223:114-119. [DOI] [PubMed] [Google Scholar]
- 15.Morio, T., H. Urushihara, T. Saito, Y. Ugawa, H. Mizuno, M. Yoshida, R. Yoshino, B. N. Mitra, M. Pi, T. Sato, K. Takemoto, H. Yasukawa, J. Williams, M. Maeda, I. Takeuchi, H. Ochiai, and Y. Tanaka. 1998. The Dictyostelium developmental cDNA project: generation and analysis of expressed sequence tags from the first-finger stage of development. DNA Res. 5:335-340. [DOI] [PubMed] [Google Scholar]
- 16.Nellen, W., S. Datta, C. Reymond, A. Sivertsen, S. Mann, T. Crowley, and R. A. Firtel. 1987. Molecular biology in Dictyostelium: tools and applications. Methods Cell Biol. 28:67-100. [DOI] [PubMed] [Google Scholar]
- 17.O'Leary, W. M. 1962. S-Adenosylmethionine in the biosynthesis of bacterial fatty acids. J. Bacteriol. 84:967-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin, Y.-M., M. H. Poutanen, H. M. Helander, A.-P. Kvist, K. M. Siivari, W. Schmitz, E. Conzelmann, U. Hellman, and J. K. Hiltunen. 1997. Peroxisomal multifunctional enzyme of β-oxidation metabolizing d-3-hydroxyacyl-CoA esters in rat liver: molecular cloning, expression and characterization. Biochem. J. 321:21-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saito, T., and H. Ochiai. 1996. Identification of a novel all-cis-5,9,12-heptadecatrienoic acid in the cellular slime mold Polysphondylium pallidum. Lipids 31:445-447. [DOI] [PubMed] [Google Scholar]
- 20.Saito, T., and H. Ochiai. 1999. Identification of Δ5-fatty acid desaturase from the cellular slime mold Dictyostelium discoideum. Eur. J. Biochem. 265:809-814. [DOI] [PubMed] [Google Scholar]
- 21.Saito, T., T. Morio, and H. Ochiai. 2000. A second functional delta5 fatty acid desaturase in the cellular slime mold Dictyostelium discoideum. Eur. J. Biochem. 267:1813-1818. [DOI] [PubMed] [Google Scholar]
- 22.Sutoh, K. 1993. A transformation vector for Dictyostelium discoideum with a new selectable marker bsr. Plasmid 30:150-154. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki, Y., L. L. Jiang, M. Souri, S. Miyazawa, S. Fukuda, Z. Zhang, M. Une, N. Shimozawa, N. Kondo, T. Orii, and T. Hashimoto. 1997. d-3-Hydroxyacyl-CoA dehydratase/d-3-hydroxyacyl-CoA dehydrogenase bifunctional protein deficiency: a newly identified peroxisomal disorder. Am. J. Hum. Genet. 61:1153-1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsujioka, M., M. Yokoyama, K. Nishio, H. Kuwayama, T. Morio, M. Katoh, H. Urushihara, T. Saito, H. Ochiai, Y. Tanaka, I. Takeuchi, and M. Maeda. 2001. Spatial expression patterns of genes involved in cyclic AMP responses in Dictyostelium discoideum development. Dev. Growth Differ. 43:275-283. [DOI] [PubMed] [Google Scholar]
- 25.van Grunsven, E. G., E. van Berkel, L. Ijlst, P. Vreken, J. B. C. de Klerk, J. Adamski, H. Lemonde, P. T. Clayton, D. A. Cuebas, and R. J. A. Wanders. 1998. Peroxisomal d-hydroxyacyl-CoA dehydrogenase deficiency: resolution of the enzyme defect and its molecular basis in bifunctional protein deficiency. Proc. Natl. Acad. Sci. USA 95:2128-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van Grunsven, E. G., E. van Berkel, P. A. W. Mooijer, P. A. Watkins, H. W. Moser, Y. Suzuki, L. L. Jiang, T. Hashimoto, G. Hoefler, J. Adamski, and R. J. A. Wanders. 1999. Peroxisomal bifunctional protein deficiency revisited: resolution of its true enzymatic and molecular basis. Am. J. Hum. Genet. 64:99-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wanders, R. J. A., P. Vreken, S. Ferdinandusse, G. A. Jansen, H. R. Waterham, C. W. T. van Roermund, and E. G. van Grunsven. 2001. Peroxisomal fatty acid α- and β-oxidation in humans: enzymology, peroxisomal metabolite transporters and peroxisomal diseases. Biochem. Soc. Trans. 29:250-267. [DOI] [PubMed] [Google Scholar]
- 28.Watkins, P. A., W. W. Chen, C. J. Harris, G. Hoefler, S. Hoefler, D. C. Blake, Jr., A. Balfe, R. I. Kelly, A. B. Moser, M. E. Beard, and H. W. Moser. 1989. Peroxisomal bifunctional enzyme deficiency. J. Clin. Investig. 83:771-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Watts, D. J., and J. M. Ashworth. 1970. Growth of myxamoebae of the cellular slime mould Dictyostelium discoideum in axenic culture. Biochem. J. 119:171-174. [DOI] [PMC free article] [PubMed] [Google Scholar]