Candida albicans represents the most pervasive fungal pathogen colonizing humans (5, 36). Its success stems in part from its capacity to live as a benign commensal in a majority of healthy individuals in one or more of a variety of body locations, most notably the oral cavity, genitalia, and gastrointestinal tract. As an opportunistic pathogen, it lies in wait for a change in some aspect of the host physiology or microflora that normally suppresses growth and invasiveness. In response to this change, it increases in number and invades tissue. Its versatility as a pathogen is reflected in the variety of tissues it can colonize, the variety of disease states it is responsible for, and the ever increasing number of individuals, usually immunocompromised, who die from bloodstream and disseminated infections.
Until the mid-1980s, it was believed that even though its success reflected phenotypic plasticity, the only developmental program C. albicans possessed for generating multiple phenotypes was the bud-hypha transition. Although a variety of different observations supported the conclusion that the hyphal phenotype facilitated tissue invasion (13, 28, 43), there was still the nagging feeling that the bud-hypha transition was not complex enough to account for the remarkable versatility of this pathogen. Therefore, it should have come as no great surprise when Slutsky et al. (41) and Pomes et al. (37) reported in 1985 that different strains of C. albicans switched reversibly and at high frequency between a limited number of different colony morphologies. In the case of the common laboratory strain 3153A, switching occurred among at least seven colony phenotypes, each dictated by differences in the temporal dynamics and spatial distributions of budding cells, pseudohyphae, and hyphae in the colony dome (41, 44). As more strains of C. albicans were analyzed for switching, it became apparent that the majority switched reversibly and at high frequency between a variety of phenotypes but that the repertoires of switch phenotypes were different in different strains. Several reports also demonstrated reversible, high-frequency switching in Candida tropicalis (46), Candida parapsilosis (9), Candida glabrata (24, 26), and even Cryptococcus neoformans (10, 11).
One switching system in C. albicans consisted of a reversible transition between two phases: a white hemispherical colony morphology, referred to as the “white phase,” and a grey flat colony morphology, referred to as the “opaque phase” (42). This switching system, referred to as “the white-opaque transition,” was initially identified in strain WO-1, isolated in 1986 from the bloodstream of a bone marrow transplant patient at The University of Iowa Hospitals and Clinics (42). The patient died from this infection. Strains exhibiting a similar phase transition were identified in 1987 in a survey of switching in roughly 125 independent C. albicans isolates (D. R. Soll, unpublished data). It was estimated at that time that approximately 7% of C. albicans strains underwent the white-opaque transition. Because the white-opaque transition could be discriminated on a variety of agars, involved only two alternative phenotypes, and had dramatic pleiotropic effects on cellular phenotype (2, 42), it was selected as an experimental model for delving into the molecular mechanisms regulating switching in C. albicans and related species. However, in considering the basic biology of the white-opaque transition, there was always the reservation that it occurred in only a minority of natural strains and that it differed mechanistically from the more common switching systems characterized in strain 3153A. There was also the more extreme reservation by some that it simply represented a unique, unrepresentative switching system resulting from a chromosomal rearrangement that had been identified in strain WO-1 (6, 7). As we shall see, the reservation that white-opaque switching was restricted to one or a few strains proved to be unfounded.
Cellular and molecular biology of white-opaque switching.
Conventional microscopy immediately revealed that opaque-phase cells were morphologically unique (1, 2, 40, 42). While white-phase cells of strain WO-1 were round to ovoid and budded like most other strains of C. albicans, opaque-phase cells were twice as large as white-phase cells, asymmetric, elongated, and bean shaped. Transmission electron microscopy revealed that opaque-phase cells contained a large vacuole that in turn contained vesicles, and unique cell wall pimples with channels traversing them (1, 2). An opaque-phase-specific 14.5-kDa antigen was demonstrated to be associated with these pimples (1).
Since it was obvious from early experiments on cellular phenotype that the white-opaque transition had to involve differential gene expression, strategies to identify phase-specific genes were developed in the early 1990s. Differential hybridization screens of opaque-phase cDNA libraries identified the first phase-specific gene, PEP1 (SAP1), which was verified by Northern blot analysis to be expressed exclusively in the opaque phase (34). This was followed by the identification of a number of additional opaque-phase-specific genes (OP4, SAP3, CDR3, EFG1-2.2 kb, and HOS3-2.3 kb) (3, 14, 35, 50, 52, 53) and white-phase-specific genes (WH11, EFG1-3.2 kb, and HOS3-2.5 kb) (48, 50, 51, 52). The list of phase-specific genes continued to grow through the 1990s and has recently been greatly expanded by use of expression array technology (27).
Functional analyses of the promoters of phase-regulated genes supported the idea that switching involved complex regulatory mechanisms. For instance, the promoters of the white-phase-specific genes WH11 and EFG1-3.2 kb were demonstrated to contain unrelated white-phase activation sequences, indicating that the regulation of these coordinately expressed genes involved trans-acting factors containing different DNA-binding proteins (25, 49, 50). In the case of opaque-phase-specific gene promoters, while some contained a MADS box binding site sequence at their activation sites (e.g., OP4 and CDR3), others did not (e.g., SAP1) (31; S. Lockhart and D. R. Soll, unpublished data). Hence, just as in the case of white-phase-specific gene expression, opaque-phase-specific gene expression involves multiple trans-acting factors containing different DNA-binding proteins. It was, therefore, enigmatic that so complex a switching system involving genes present in all strains of C. albicans was expressed in only a small minority of strains.
Discovery of MTL loci in C. albicans.
The idea that switching in C. albicans may have something to do with mating was frequently entertained, in part because switching represented two heritable but metastable states (44). Indeed, the mating type cassette system in Saccharomyces cerevisiae was frequently considered a candidate mechanism for switching. However, the idea was just as frequently dismissed because of the absence of mating type-like (MTL) genes. Things changed, however, when Hull and Johnson (15) reported the discovery of MTLa and MTLα alleles on homologous chromosomes at the same locus in the common laboratory strain of C. albicans, SC5314.
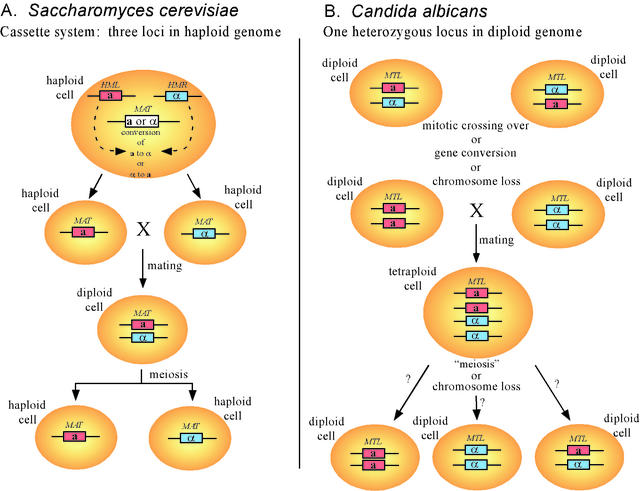
In S. cerevisiae, there are three loci containing mating type genes (Fig. 1A) (12). Two of these loci are silent (HML and HMR). One contains the MATa genes (MATa1 and MATa2), and the other contains the MATα genes (MATα1 and MATα2). The third locus (MAT) contains either the MATa or the MATα genes and is expressed. The mating type of a haploid cell is defined by the genotype (a or α) of the MAT locus. In S. cerevisiae, the MAT locus switches reversibly from a to α or from α to a by site-specific recombination with a copy of the silent HML or HMR gene (Fig. 1A). Mating occurs only between an a and an α cell and results in the diploid heterozygote a/α. In MAT-heterozygous diploids, expression of both MATa1 and MATα2 in the same cell suppresses mating and facilitates meiosis, under proper conditions, through the binding of a Mata1p-Matα2p repressor complex (19).
FIG. 1.
The configuration of mating type loci and the mechanisms for generating cells of the opposite mating type are different for S. cerevisiae and C. albicans. Note that while S. cerevisiae contains a cassette system that includes two silent loci and one expressed locus, C. albicans is normally heterozygous for mating type at one locus. While S. cerevisiae changes mating type at the expression locus with no loss of the alternate mating type information, C. albicans loses the information of one of the two mating types when it expresses a mating type. HML, homothallic mating locus left; HMR, homothetic mating locus right; MAT, mating type locus; MTL, mating type-like locus.
Hull and Johnson (15) demonstrated that in contrast to S. cerevisiae, C. albicans strain SC5314, which is diploid, possesses only one MTL locus on chromosome 5. The locus is heterozygous, containing on one homolog the MTLa gene MTLa1, which is homologous to S. cerevisiae MATa1, and on the other homolog MTLα1 and MTLα2, which are homologous to S. cerevisiae MATα1 and MATα2 (Fig. 1B). Both the MTLa and MTLα loci contain three additional genes not found in the MATa and MATα loci of S. cerevisiae. The MTLa locus of C. albicans is missing a homolog of the S. cerevisiae gene MATa2. Hence, C. albicans does not possess an S. cerevisiae-like cassette system for mating type switching. Rather, it carries opposing MTLa and MTLα alleles at the same locus on the chromosome 5 homologs. This discovery implied that to become a functional homozygote, C. albicans would have to become genetically MTL homozygous (Fig. 1B).
First indications of mating type-dependent fusion.
Hypothesizing that mating may take place between homozygous MTLa and homozygous MTLα cells in nature, as in the case of S. cerevisiae, Hull et al. (17) generated hemizygous MTLa (a/−) and hemizygous MTLα (α/−) strains from the laboratory strain SC5314 and infected mice with mixtures of the two. Alternative adenine and uridine auxotrophy was introduced into the two strains, so that the MTLa and MTLα strains were ade2 URA3 or ADE2 ura3, respectively. Fusants could hence be selected through complementation by plating mixed populations on a medium lacking adenine and uridine. The same in vivo experiment was performed with MTL heterozygotes alone, which were not expected to fuse, if their hypotheses were correct. Yeast cells from the kidneys of infected mice were then plated on selection medium and rich medium. While 44 of 103 total CFU retrieved from the kidneys of mice infected with mixtures of a and α cells proved to be ADE2 URA3 (4.4 × 10−2), none of 103 total CFU retrieved from mice infected with mixtures of ade2 URA3 a/α cells and ADE2 ura3 a/α cells proved to be ADE2 URA3 (<10−3). Hence, mixtures of homozygous a and homozygous α cells fused, but mixtures of heterozygous a/α cells did not, or did so at undetectable rates. Selected ADE2 URA3 cells contained more DNA than parental diploids and also contained both MTLa and MTLα genes, demonstrating fusion. Images of 4′,6′-diamidino-2-phenylindole (DAPI)-stained cells presented in a website supplement to the report of Hull et al. (17) in Science (www.sciencemag.org/future/data/1049869.html) further suggested that the fusants were mononuclear. These data were consistent with a mating system in which homozygous a/a and homozygous α/α strains fused to form tetraploids (Fig. 1B). However, meiosis and recombination were not demonstrated.
In a report published in parallel with that of Hull et al. (17), Magee and Magee (32) also presented evidence that fusion between hemizygous MTLa and MTLα strains occurred in vitro. Taking advantage of the observation by Janbon et al. (18) that growth on sorbose leads to loss of one of the two chromosome 5 homologs that harbor the MTL loci, they generated hemizygous MTLa and MTLα strains from auxotrophic strains and performed crosses on selective agar. Fusants were identified by complementation. Prototrophs arose in a × α crosses but not in a/α × a/α crosses. Again, it was reported that the fusants contained approximately twice the DNA content of either parent strain, but no evidence was presented in that study that cells were mononucleate, and again, neither meiosis nor recombination was demonstrated. Both the in vivo and in vitro demonstrations of mating type-based fusion indicated for the first time that, as in S. cerevisiae, heterozygosity at the mating type locus in C. albicans suppressed mating and homozygosity facilitated it. In both the in vivo and in vitro studies, fusion occurred between auxotrophic hemizygotes, was selected for, and appeared to be an infrequent event. In neither study was it demonstrated how resulting tetraploid cells returned to the diploid state. The obvious alternatives were meiosis and random chromosome loss.
Switching and mating: discovery of a dependent relationship.
In examining the colony morphologies of the hemizygous a and α strains, Miller and Johnson (33) made the remarkable discovery of opaque-phase sectors. This discovery was indeed unexpected, since the parental MTL-heterozygous strain SC5314, which they used to generate the alternative hemizygotes, had previously been demonstrated to undergo 3153A-like switching, not the white-opaque transition (51). Miller and Johnson (33) demonstrated that cells of both the hemizygous a and α derivatives of SC5314 switched between white and opaque colony-forming phenotypes and that cells from the white and opaque phases shared the morphological features of white- and opaque-phase cells, respectively, of strain WO-1. In addition, they demonstrated that the four WO-1 phase-specific genes EFG1, WH11, OP4, and SAP1 were similarly phase regulated during white-opaque switching in hemizygous a and α strains. These observations led Miller and Johnson (33) to conclude that in heterozygotes, switching, like mating, was suppressed. The ramifications of the observations of Miller and Johnson (33) were obvious. Since it appeared that a majority of naturally occurring C. albicans strains were a/α, it was quite possible that the white-opaque transition was suppressed in the majority of C. albicans strains and that any C. albicans strain could undergo the white-opaque transition if it became homozygous at the MTL locus.
Even more surprising and exciting was Miller and Johnson's (33) discovery that mating between opaque-phase hemizygous a and α cells was many orders of magnitude more efficient than mating between white-phase hemizygous a and α cells. The frequency of mating between opaque-phase a and opaque-phase α cells was 105 to 106 times higher than that between white-phase a and white-phase α cells and 103 to 104 times higher than that between white-phase and opaque-phase cells of opposite mating types. These results suggested that the opaque-phase phenotype represented a mating-competent form of C. albicans.
To test the generality of the relationship between MTL homozygosity and white-opaque switching among natural homozygous strains, Lockhart et al. (29) identified six new white-opaque switchers among a large collection of clinical isolates and tested them as well as strain WO-1 (a total of seven white-opaque switchers) for allelism at the MTL locus. They found that all seven white-opaque-switching strains were homozygous at the MTL locus; three were a/a and four were α/α. They next assessed the MTL zygosity of 220 clinical C. albicans isolates. These isolates were distributed among four of the five major C. albicans clades, which were recently identified in cluster analyses with the complex DNA fingerprinting probe Ca3 (4, 38, 39). Of this broad collection, 213 (97%) were MTL heterozygous and 7 (3%) were MTL homozygous. Of the seven homozygous strains, five switched unambiguously between the white and opaque phases, while the remaining two switched with associated color changes but without the distinct cellular phenotype of the opaque phase. These results indicated that the great majority of homozygous strains in nature, if not all, undergo the white-opaque transition. Of 20 MTL-heterozygous strains randomly selected from the general collection, 18 did not undergo the white-opaque transition, while 2 did. However, further examination revealed that the latter two strains spontaneously generated MTL-homozygous progeny at a high frequency, and these homozygous offspring in turn underwent the white-opaque transition. The results of Lockhart et al. (29), therefore, generalized the finding of Miller and Johnson (33) that strains heterozygous at the MTL locus do not undergo white-opaque switching while the great majority of strains homozygous at the MTL locus do.
To test the generality of the relationship between switching and mating in natural homozygous strains, Lockhart et al. (30) performed crosses between natural MTLα- and MTLa-homozygous strains expressing the white or opaque phenotype. In this analysis, they directly assessed fusion at the cellular level without selection (see the description of the cell biology of mating in the next section). They demonstrated that in 12 crosses between different opaque MTLa homozygous strains and opaque MTLα-homozygous strains, fusion occurred. No fusions were observed in three crosses between different opaque MTLα-homozygous strains, in five crosses between different opaque MTLa-homozygous strains, or in four crosses between different MTLa-homozygous and different MTLα-homozygous strains when one strain was white and the other was opaque. By vitally staining opaque homozygous MTLa and opaque homozygous MTLα cells with fluorescein isothiocyanate-concanavalin A (green) and rhodamine-concanavalin A (red), respectively, Lockhart et al. (30) also demonstrated that fusions included exclusively one red and one green cell, never a green-green or red-red combination. These results confirmed the generality of the observations by Miller and Johnson (33) on engineered hemizygous a and α strains from a single strain. The combined observations demonstrate that MTL homozygosity alone does not confer the ability to mate on C. albicans, as it does on S. cerevisiae (Fig. 2A). Rather, in C. albicans both homozygous MTLa and homozygous MTLα cells must first switch to opaque in order to mate (Fig. 2B). These results indicate that the low levels of fusion observed by Hull et al. (17) in crosses between white cells of opposite mating types were not the result of white-white fusion but rather resulted from fusions between opaque-phase cells generated at low levels in each white-phase population through switching.
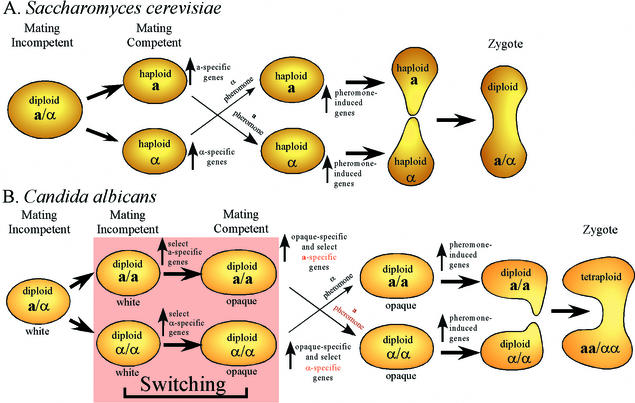
FIG. 2.
C. albicans has inserted an extra developmental step, the switch from white to opaque, into the mating process. In S. cerevisiae, a and α cells are immediately mating competent, and all a-specific and α-specific genes are upregulated. In C. albicans, a homozygous a or α cell is not mating competent unless it switches to the opaque phenotype. In C. albicans, upregulation of a-specific and α-specific gene expression is divided between the transition to a homozygous state and the transition from white to opaque.
Visualizing the mating process.
Mating in S. cerevisiae involves a carefully orchestrated developmental program that includes a specific sequence of morphological steps (8). In response to a pheromone released from the opposite mating type, a haploid S. cerevisiae cell bulges to form a “shmoo” shape and then extends a wide, unconstricted projection (“conjugation tube”). When tubes from opposite mating types meet, they fuse at their apices. Parent cell nuclei, which are arrested in G1, migrate to the ends of the tubes, and after tube fusion, they associate and fuse, forming a diploid nucleus. A daughter cell then forms from the conjugation bridge of the zygote. The nucleus, positioned at the junction of bridge and daughter cell, divides, and one of the two diploid nuclei enters the daughter cell. The daughter cell then divides as a diploid, returning to a haploid state when transferred to starvation medium through the process of sporulation, which includes meiosis.
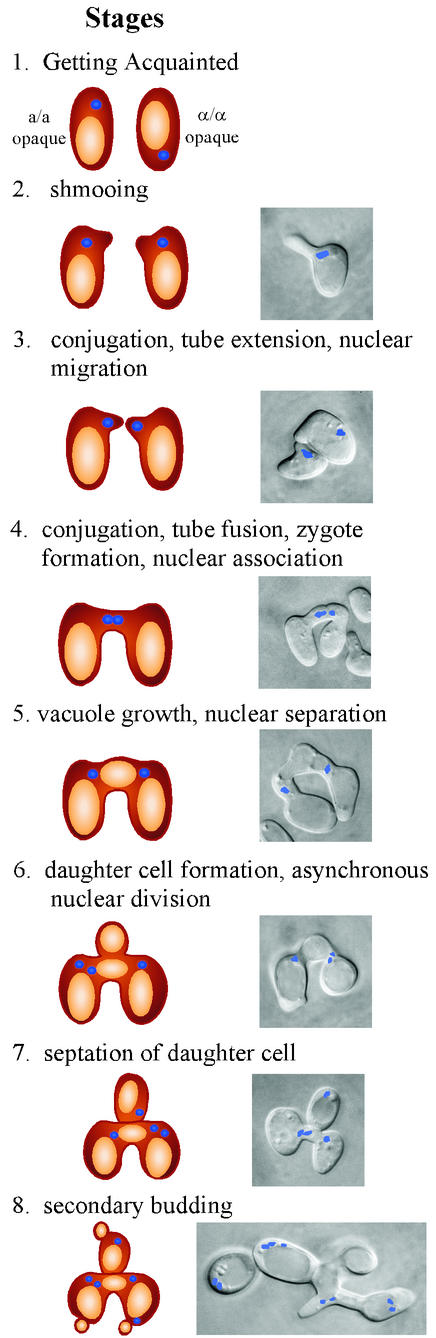
Employing continuous videomicroscopy, computer-assisted three-dimensional reconstruction of living cells, and fluorescence microscopy, Lockhart et al. (30) have demonstrated that the morphological stages in the C. albicans mating process parallel those in the S. cerevisiae mating process. In mixed a/a and α/α cultures, cells form unconstricted projections (conjugation tubes) that continue to elongate (Fig. 3). If tubes of the opposite mating types do not undergo fusion, they continue to grow, reaching lengths up to the equivalent of 5 cell diameters. These conjugation tubes do not contain septa, and in the absence of fusion, they form buds at their apices, in essence reverting to the budding growth form. In the process of reversion, the nucleus then migrates from the mother cell to the junction of conjugation tube and daughter bud, where it divides, and the daughter cell and mother cell each receive a nucleus (30). The tube ends up nucleus free, and a septum forms only at the tube-daughter cell junction.
FIG. 3.
Cell biology of mating in C. albicans. Models are drawn of the stages in the left-hand vertical column, and combined-phase images of the cell bodies and zygotes and fluorescent images of the nuclei are presented in the right-hand vertical column.
If the conjugation tubes of an a/a cell and an α/α cell attract each other through chemotropism, the tubes extend until their apices meet and then fuse (Fig. 3). The nuclei of the a and α cells migrate to the tube apices. Upon tube fusion (formation of a “zygote”), the nuclei end up physically associated but do not fuse (Fig. 3) under the conditions employed by Lockhart et al. (30). In each zygote, the conjugation bridge swells as a vacuole forms in it. As the vacuole enlarges, it separates the two nuclei, which return to the tube-mother cell junctions and undergo asynchronous nuclear division (Fig. 3). A daughter cell then forms from the conjugation bridge and receives one or two nuclei. The daughter cell nuclei can originate from either or both mother cells. A septum then forms exclusively at the conjugation bridge-daughter cell junction and provides a landmark for identifying the daughter cell in fixed preparations (30). The daughter cell, tube, and mother cell then form secondary buds (Fig. 3). The sequence of morphological stages and fusion events observed by Lockhart et al. (30) was not rare. Indeed, a majority of cells in mixed cultures of homozygous MTLa and homozygous opaque MTLα cultures shmooed and formed conjugation tubes, and a significant minority (10%) underwent fusion. Except for the lack of nuclear fusion, the stages of cellular fusion and zygote development clearly resembled mating in S. cerevisiae.
So far, no demonstrated meiosis.
Both Hull et al. (17) and Magee and Magee (32) demonstrated mixing of markers upon mating type-based fusion of a and α cells. They also demonstrated associated increases in cellular DNA content. As noted, Hull et al. (17) also provided evidence in a website supplement to their publication that fusion progeny were mononucleate. These observations indicated that karyogamy had occurred. Lockhart et al. (30), however, did not observe nuclear fusion (karyogamy) in the conjugation bridges of zygotes. Their nuclear staining experiments suggested that after cellular fusion, the nuclei from the original a/a and α/α cells did not undergo karyogamy. Rather, the nuclei translocated back to the tube-cell junctions through expansion of the conjugation bridge vacuole. Lockhart et al. (30) also failed to find mixing or segregation of mating type and other genetic markers, which would have been proof of karyogamy. The apparent difference between the results of Lockhart et al. (30) and those of Hull et al. (17), Magee and Magee (32), and Miller and Johnson (33) may be due to the absence of selection, the high frequency of observed fusions, and possibly the natural origin of true homozygous rather than hemizygous strains in the experiments performed by Lockhart et al. (30). While the studies by Johnson and colleagues (17, 33) and by Magee and Magee (32) involved crosses between MTL-hemizygous auxotrophs and the selection of rare prototrophs, the studies by Lockhart et al. (30) involved natural MTL-homozygous strains and the microscopic identification of fusions. The proportion of cells observed by Lockhart et al. (30) that underwent fusion was far higher than the proportions observed by Hull et al. (17), Magee and Magee (32), and Miller and Johnson (33), leaving open the likely possibility that karyogamy may have occurred in a minority of fusants in the Lockhart study but may have been missed both cytologically and in random genetic analysis of only a limited number of daughter cells. Lockhart et al. (30) did find that a minority of daughter cells formed by zygotes were binucleate and gave rise to binucleate a/α as well as mononucleate a/a and α/α cells. However, there was no mixing or segregation of markers on different chromosomes in their limited analysis, suggesting that binucleate daughter cells did not readily undergo karyogamy. In more-recent experiments using different incubation conditions, Lockhart and colleagues (S. Lockhart, R. Zhao, K. Daniels, and D. R. Soll, unpublished data) found evidence for rare but relatively stable tetraploid fusants.
Gene regulation during mating.
While C. albicans generates homozygous a/a and homozygous α/α cells by a mechanism different from the cassette replacement mechanisms of S. cerevisiae (Fig. 1), the MTL genes are homologous to MAT genes of the S. cerevisiae mating system (15). Because of this homology, one can hypothesize the regulatory roles of the MTL genes from those of their MAT homologs. In a/α diploid cells of S. cerevisiae, the gene products of MATa and MATα are both expressed (19). Mata1p and Matα2p form a heterodimer which represses the transcription of haploid-specific genes, including HO, which encodes the nuclease for mating type switching; RME1, a repressor of early meiosis; and several genes encoding components of the mating-specific mitogen-activated protein kinase pathway. In a/α diploid cells, a-specific genes are repressed by the Matα2p-Mcm1p repressor complex (20, 21). MATα1, which encodes an activator of α-specific genes, is also repressed by the Mata1p-Matα2p repressor complex. Hence, no a-specific or α-specific genes are expressed in a diploid a/α cell. In haploid MATa cells, a-specific genes are expressed due to the absence of the Matα2p-Mcm1p repressor complex, and α-specific genes are not expressed due to the absence of the Matα1p-Mcm1p activator complex. In haploid MATα cells, α-specific genes are expressed through activation by the Matα1p-Mcm1p complex, and a-specific genes are repressed by the Matα2p-Mcm1p repressor complex. When a or α haploid S. cerevisiae cells are exposed to the pheromone of the opposite mating type, the mating-specific mitogen-activated protein kinase pathway is activated through the respective pheromone receptors, resulting in the activation of a number of additional mating-type-specific genes, including those involved in shmooing and fusion (47).
Therefore, in S. cerevisiae, a cells express a-specific genes and α cells express α-specific genes (Fig. 2A). In S. cerevisiae, a and α cells are immediately mating competent. When treated with pheromones, both a and α cells express additional genes involved in fusion and zygote development (Fig. 2A). In C. albicans, MTL a/α heterozygotes are unable to switch, suggesting that an Mtla1p-Mtlα2p complex, homologous to the Mata1p-Mat1α2p complex, may be involved in the suppression of switching. Because C. albicans has added an additional step to the acquisition of mating, the possibility arises that some genes turned on in a and α cells of S. cerevisiae (Fig. 2A) may not be turned on in white a/a or in white α/α cells of C. albicans. Instead, these genes may be turned on after the transition from white to opaque (Fig. 2B) and hence may be regulated by an opaque-specific rather than a homozygous-specific mechanism. Indeed, Zhao and colleagues (R. Zhao, W. Wu, S. Lockhart, and D. R. Soll, unpublished data) recently demonstrated that although MTLα1 is expressed in both homozygous α/α white-phase and homozygous α/α opaque-phase cells, the α pheromone gene MFα is expressed in homozygous α/α opaque-phase but not in homozygous α/α white-phase cells. Neither is expressed in heterozygous a/a cells. Hence, MTLα1 must be activated when a white a/α cell becomes a white α/α cell, but MFα is not activated. MFα is activated upon a subsequent switch to the opaque phase. Therefore, it is likely that genes coordinately activated when S. cerevisiae becomes haploid will separate into two groups in C. albicans: those similarly activated when white cells become homozygous, and those activated by the transition to the opaque phase (Fig. 2B).
The functional reason for the insertion of an additional complex developmental program (Fig. 2B) into the mating process of C. albicans is not immediately obvious, given that it is not an apparent component of mating in other fungi (16). Presumably, there is an advantage specific for C. albicans that may relate to its commensalism and pathogenesis. The unique phenotypic characteristics of opaque-phase cells may shed light on this enigma and are therefore worth reviewing. First, opaque-phase cells form a cell wall that is morphologically distinct from that of white-phase cells. The opaque-phase cell wall possesses unique pimples with channels from the plasma membrane to the wall surface (1, 2). Until now, there were no good suggestions for the role pimples played in the basic biology of C. albicans. Given the role of secretion and adhesion in yeast mating, we can now suggest that pimples may play a role in chemotropism and/or fusion.
Second, opaque-phase cells differentially express and secrete two aspartyl proteinases (14, 34, 53). Although these proteinases have been assumed to be involved in tissue penetration and have been shown to facilitate opaque-phase-specific cavitation on newborn-mouse skin (23), we must now consider their possible involvement in the mating process.
Third, and perhaps most intriguing, is the relationship between the white-opaque transition and temperature. Opaque-phase cells are stable at 25°C but not at 37°C, which is body temperature (42). Transferring opaque-phase cells from 25 to 37°C induces conversion en masse to the white phase (35, 40, 42, 45, 48). The phenotypic transition, assessed as the point of phenotypic commitment to the white phase, occurs semisynchronously at the second cell doubling at 37°C (45, 48). Since mating requires expression of the opaque phenotype, it may be limited to locations outside the human body, such as the surface of skin or environmental reservoirs, where the temperature is below body temperature. It could be that there is a disadvantage to mating within the human body, or that mating per se and pathogenesis within the human body are incompatible.
Fourth, and related to the preceding characteristic, opaque-phase cells are far more virulent than white-phase cells in a cutaneous mouse model (23). Is it possible that mating is restricted to the opaque phenotype in order to target the process to a location outside the human body, namely, skin? Is it possible that this restriction removes the process from environments that harbor the human immune system, or targets the process to an environment more prone to multiple strain colonization?
One may also ask why homozygotes do not simply express the opaque phenotype constitutively. Why have they developed a switching system fine-tuned by the deacetylases Hda1p and Rpd3, which maintain frequencies of switching between the two phases of around 10−3 (22, 52)? Obviously there is some advantage for homozygotes to express the white phenotype as well as the opaque phase. That advantage, again, is not immediately obvious.
There is little doubt that these newly discovered dependent relationships between MTL homozygosis, white-opaque switching, and mating in C. albicans have challenged us with a number of truly fascinating and unexpected enigmas and questions related to the basic biology and pathogenesis of C. albicans. There is also little doubt, given the differences between S. cerevisiae and C. albicans, that regulatory mechanisms unique to C. albicans mating will be discovered. The rapid rate of discovery experienced in the past few years should, therefore, be matched, if not outdistanced, by the discoveries that will be made in the next few years. Who would have thought only a few years ago that C. albicans has sex, and that it must switch to do it?
Acknowledgments
We are indebted to T. Srikantha and C. Pujol for valuable discussions and to K. Daniels for help with the figures.
The research from the Soll laboratory on switching and mating reported here was supported by NIH grants AI2392 and DE14219.
REFERENCES
- 1.Anderson, J., R. Mihalik, and D. R. Soll. 1990. Ultrastructure and antigenicity of the unique cell wall pimple of the Candida opaque phenotype. J. Bacteriol. 172:224-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, J. M., and D. R. Soll. 1987. Unique phenotype of opaque cells in the white-opaque transition of Candida albicans. J. Bacteriol. 169:5579-5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Balan, I., A. M. Alarco, and M. Raymond. 1997. The Candida albicans CDR3 gene codes for an opaque-phase ABC transporter. J. Bacteriol. 179:7210-7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blignaut, E., C. Pujol, S. Lockhart, S. Joly, and D. R. Soll. 2002. Ca3 fingerprinting of Candida albicans isolates from HIV+ individuals reveal a new clade in South Africa. J. Clin. Microbiol. 40:826-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calderone, R. A. 2002. Candida and candidiasis. ASM Press, Washington, D.C.
- 6.Chibana, H., B. B. Magee, S. Grindle, Y. Ran, S. Scherer, and P. T. Magee. 1998. A physical map of chromosome 7 of Candida albicans. Genetics 149:1739-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu, W. S., B. B. Magee, and P. T. Magee. 1993. Construction of an SfiI macrorestriction map of the Candida albicans genome. J. Bacteriol. 175:6637-6651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cross, F., L. H. Hartwell, C. Jackson, and J. B. Konopka. 1988. Conjugation in Saccharomyces cerevisiae. Annu. Rev. Cell Biol. 4:429-457. [DOI] [PubMed] [Google Scholar]
- 9.Enger, L., S. Joly, C. Pujol, P. Simonson, M. A. Pfaller, and D. R. Soll. 2001. Cloning and characterization of a complex DNA fingerprinting probe for Candida parapsilosis. J. Clin. Microbiol. 39:658-669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fries, B. C., D. L. Goldman, R. Cherniak, R. Ju, and A. Casadevall. 1999. Phenotypic switching in Cryptococcus neoformans results in changes in cellular morphology and glucuronoxylomannan structure. Infect. Immun. 67:6076-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goldman, D. L., B. C. Fries, S. P. Franzot, L. Montella, and A. Casadevall. 1998. Phenotypic switching in the human pathogenic fungus Cryptococcus neoformans is associated with changes in virulence and pulmonary inflammatory response in rodents. Proc. Natl. Acad. Sci. USA 95:14967-14972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haber, J. 1998. Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet. 32:561-599. [DOI] [PubMed] [Google Scholar]
- 13.Howlett, J. A., and C. A. Squier. 1980. Candida albicans ultrastructure: colonization and invasion of oral epithelium. Infect. Immun. 29:252-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hube, B., M. Monod, D. Schofield, A. Brown, and N. Gow. 1994. Expression of seven members of the gene family encoding aspartyl proteinases in Candida albicans. Mol. Microbiol. 14:87-99. [DOI] [PubMed] [Google Scholar]
- 15.Hull, C. M., and A. D. Johnson. 1999. Identification of a mating type-like locus in the asexual pathogenic yeast Candida albicans. Science 285:1271-1275. [DOI] [PubMed] [Google Scholar]
- 16.Hull, C. M., and J. Heitman. 2002. Fungal mating: Candida albicans flips a switch to get in the mood. Curr. Biol. 12:R78278-R78284. [DOI] [PubMed] [Google Scholar]
- 17.Hull, C. M., R. M. Raisner, and A. D. Johnson. 2000. Evidence for mating of the “asexual” yeast Candida albicans in mammals. Science 289:307-310. [DOI] [PubMed] [Google Scholar]
- 18.Janbon, G., F. Sherman, and E. Rustchenko. 1998. Monosomy of a specific chromosome determines l-sorbose utilization: a novel regulatory mechanism in Candida albicans. Proc. Natl. Acad. Sci. USA 95:5150-5155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson, A. D. 1995. Molecular mechanisms of cell-type determination in budding yeast. Curr. Opin. Genet. Dev. 5:552-558. [DOI] [PubMed] [Google Scholar]
- 20.Keleher, C. A., C. Goutte, and A. D. Johnson. 1988. The yeast cell-type-specific repressor α2 acts cooperatively with a non-cell-type-specific protein. Cell 53:927-936. [DOI] [PubMed] [Google Scholar]
- 21.Keleher, C. A., S. Passmore, and A. D. Johnson. 1989. Yeast repressor α2 binds to its operator cooperatively with yeast protein Mcm1. Mol. Cell. Biol. 9:5228-5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klar, A., T. Srikantha, and D. R. Soll. 2001. A histone deacetylation inhibitor and mutant promote colony-type switching of the human pathogen Candida albicans. Genetics 158:919-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kvaal, C., S. Lachke, T. Srikantha, K. Daniels, J. McCoy, and D. R. Soll. 1999. Misexpression of the opaque phase-specific gene PEP1 (SAP1) in the white phase of Candida albicans confers increased virulence in a mouse model of cutaneous infection. Infect. Immun. 67:6652-6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lachke, S., S. Joly, K. Daniels, and D. R. Soll. 2002. Phenotypic switching and filamentation in Candida glabrata. Microbiology 148:2661-2674. [DOI] [PubMed] [Google Scholar]
- 25.Lachke, S., T. Srikantha, and D. R. Soll. 2003. The regulation of EFG1 in white-opaque switching in Candida albicans involves overlapping promoters. Mol. Microbiol. 48:523-536.. [DOI] [PubMed]
- 26.Lachke, S., T. Srikantha, L. Tsai, K. Daniels, and D. R. Soll. 2000. Phenotypic switching in Candida glabrata involves phase-specific regulation of the metallothionein gene MT-II and the newly discovered hemolysin gene HLP. Infect. Immun. 68:884-895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lan, C.-Y., G. Newport, L. A. Murillo, T. Jones, S. Scherer, R. W. Davis, and N. Agabian. 2002. Metabolic specialization associated with phenotypic switching in Candida albicans. Proc. Natl. Acad. Sci. USA 99:14907-14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lo, H. J., J. R. Kohler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti, and G. R. Fink. 1997. Nonfilamentous Candida albicans mutants and avirulence. Cell 90:939-949. [DOI] [PubMed] [Google Scholar]
- 29.Lockhart, S. R., C. Pujol, K. Daniels, M. Miller, A. Johnson, and D. R. Soll. 2002. In Candida albicans, white-opaque switchers are homozygous for mating type. Genetics 162:737-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lockhart, S. R., K. J. Daniels, R. Zhao, D. Wessels, and D. R. Soll. 2003. The cell biology of mating in Candida albicans. Eukaryot. Cell 2:49-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lockhart, S. R., M. Nguyen, T. Srikantha, and D. R. Soll. 1998. A MADS box protein consensus binding site is necessary and sufficient for activation of the opaque-phase-specific gene OP4 of Candida albicans. J. Bacteriol. 180:6607-6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Magee, B. B., and P. T. Magee. 2000. Induction of mating in Candida albicans by construction of MTLa and MTLα strains. Science 289:310-313. [DOI] [PubMed] [Google Scholar]
- 33.Miller, M. G., and A. D. Johnson. 2002. White-opaque switching in Candida albicans is controlled by the mating type (MTL) locus and allows efficient mating. Cell 110:293-302. [DOI] [PubMed] [Google Scholar]
- 34.Morrow, B., T. Srikantha, and D. R. Soll. 1992. Transcription of the gene for a pepsinogen, PEP1, is regulated by white-opaque switching in Candida albicans. Mol. Cell. Biol. 12:2997-3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morrow, B., T. Srikantha, J. Anderson, and D. R. Soll. 1993. Coordinate regulation of two opaque-specific genes during white-opaque switching in Candida albicans. Infect. Immun. 61:1823-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Odds, F. C. 1988. Candida and candidosis. Baillière Tindall, London, United Kingdom.
- 37.Pomes, R., C. Gil, and C. Nombela. 1985. Genetic analysis of Candida albicans morphological mutants. J. Gen. Microbiol. 131:2107-2113. [DOI] [PubMed] [Google Scholar]
- 38.Pujol, C., S. Joly, S. R. Lockhart, S. Noel, M. Tibayrenc, and D. R. Soll. 1997. Parity among the randomly amplified polymorphic DNA method, multilocus enzyme electrophoresis, and Southern blot hybridization with the moderately repetitive DNA probe Ca3 for fingerprinting Candida albicans. J. Clin. Microbiol. 35:2348-2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pujol, C., M. Pfaller, and D. R. Soll. 2002. Ca3 fingerprinting of C. albicans bloodstream isolates from the United States, Canada, South America, and Europe reveals a European clade. J. Clin. Microbiol. 40:2729-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rikkerink, E. H., B. B. Magee, and P. T. Magee. 1988. Opaque-white phenotype transition: a programmed morphological transition in Candida albicans. J. Bacteriol. 170:895-899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Slutsky, B., J. Buffo, and D. R. Soll. 1985. High frequency switching of colony morphology in Candida albicans. Science 230:666-669. [DOI] [PubMed] [Google Scholar]
- 42.Slutsky, B., M. Staebell, J. Anderson, L. Risen, M. Pfaller, and D. R. Soll. 1987. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J. Bacteriol. 169:189-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sobel, J. D., G. Muller, and H. R. Buckley. 1984. Critical role of germ tube formation in the pathogenesis of Candida vaginitis. Infect. Immun. 44:576-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soll, D. R. 1992. High frequency switching in Candida albicans. Clin. Microbiol. Rev. 5:183-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soll, D. R. Candida albicans. In A. Craig and A. Scherf (ed.), Antigenic variation, in press. Academic Press, London, United Kingdom.
- 46.Soll, D. R., M. Staebell, C. J. Langtimm, M. Pfaller, J. Hicks, and T. V. G. Rao. 1988. Multiple Candida strains in the course of a single systemic infection. J. Clin. Microbiol. 26:1448-1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sprague, G. F., Jr., and J. W. Thorner. 1992. Pheromone response and signal transduction during the mating process of Saccharomyces cerevisiae, p. 657-744. In E.W. Jones, J. R. Pringle, and J. R. Broach (ed.), Molecular and cellular biology of the yeast Saccharomyces: gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Srikantha, T., and D. R. Soll. 1993. A white-specific gene in the white-opaque switching system of Candida albicans. Gene 131:53-60. [DOI] [PubMed] [Google Scholar]
- 49.Srikantha, T., L. Tsai, and D. R. Soll. 1997. The WH11 gene of Candida albicans is regulated in two distinct developmental programs through the same transcription activation sequences. J. Bacteriol. 179:3837-3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Srikantha, T., L. Tsai, K. Daniels, and D. R. Soll. 2000. EFG1 null mutants of Candida albicans switch but cannot express the complete phenotype of white-phase budding cells. J. Bacteriol. 182:1580-1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Srikantha, T., L. Tsai, K. Daniels, L. Enger, K. Highley, and D. R. Soll. 1998. The two-component hybrid kinase regulator CaNIK1 of Candida albicans. Microbiology 144:2715-2729. [DOI] [PubMed] [Google Scholar]
- 52.Srikantha, T., L. K. Tsai, A. Klar, and D. R. Soll. 2001. The histone deacetylases HDA1 and RPD3 play distinct roles in the regulation of high-frequency phenotypic switching in Candida albicans. J. Bacteriol. 183:4614-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.White, T. C., H. Miyasaki, and N. Agabian. 1993. Three distinct secreted aspartyl proteinases in Candida albicans. J. Bacteriol. 175:6126-6135. [DOI] [PMC free article] [PubMed] [Google Scholar]