Abstract
The actin filament network at the leading edge of motile cells relies on localized branching by Arp2/3 complex from “mother” filaments growing near the plasma membrane. The nucleotide bound to the mother filaments (ATP, ADP and phosphate, or ADP) may influence the branch dynamics. To determine the effect of the nucleotide bound to the subunits of the mother filament on the formation and stability of branches, we compared the time courses of actin polymerization in bulk samples measured using the fluorescence of pyrene actin with observations of single filaments by total internal reflection fluorescence microscopy. Although the branch nucleation rate in bulk samples was nearly the same regardless of the nucleotide on the mother filaments, we observed fewer branches by microscopy on ADP-bound filaments than on ADP-Pi-bound filaments. Observation of branches in the microscope depends on their binding to the slide. Since the probability that a branch binds to the slide is directly related to its lifetime, we used counts of branches to infer their rates of dissociation from mother filaments. We conclude that the nucleotide on the mother filament does not affect the initial branching event but that branches are an order of magnitude more stable on the sides of new ATP- or ADP-Pi filaments than on ADP-actin filaments.
INTRODUCTION
At the leading edge of motile cells, a host of molecules participates in the continuous assembly and disassembly of a cytoskeleton capable of producing protrusive forces. Actin filaments form the scaffold on which all of the other molecules act. By attaching to the sides of these filaments, Arp2/3 complex contributes to the formation of a dense network of short branched filaments (1–3). Within one to two micrometers of the leading edge, the network is remodeled into long, unbranched filaments (4) capable of providing the traction to sustain the forces necessary for global cell motility (5).
Timing is important in the assembly and disassembly of these filaments to maintain distinct filament networks within the dynamic cytoskeleton. Actin filaments contain their own internal timer with the subunits hydrolyzing bound ATP within seconds followed by the release of the γ-phosphate within minutes. Proteins utilizing the filament's nucleotide-bound state could aid in timing the disassembly or rearrangement of the network. For example, ADF/cofilin increases the rate of phosphate dissociation from ADP-Pi filaments and then binds with higher affinity, promotes dissociation of branches, and severs ADP-actin filaments (6,7).
ATP hydrolysis and phosphate dissociation might also influence the ability of filaments to bind Arp2/3 complex and support the formation of branches, but the published evidence on this matter is inconsistent. Experiments on branch formation in bulk samples using the fluorescence of pyrene actin to measure filament nucleation implicated the nucleotide bound to the Arp2/3 complex and not nucleotide bound to the mother filament (8,9). Microscopic observations of filaments stabilized with rhodamine-phalloidin or filaments on glass slides showed more branches on newly polymerized segments of filaments than older filaments (7,10,11), but these studies lacked quantitative information on whether ATP hydrolysis or phosphate release influence observed branching. One study concluded that only the tip of the barbed end is available for branching (12), which seems unlikely given microscopic observations of branching from the sides of filaments (10,13).
Here we take a new approach to investigate how the nucleotide bound to mother filaments influences their ability to support branching. We compared the number of branches observed by total internal reflection fluorescence (TIRF) microscopy with the number of filament ends formed in bulk samples where polymerization was measured by the fluorescence of pyrene-labeled actin. Branches formed at similar rates in bulk samples whether they contained aging actin filaments or exclusively ADP-Pi-bound actin filaments. On the other hand, we observed fewer branches microscopically on older filaments with bound ADP than on new ATP-actin filaments or ADP-actin filaments saturated with phosphate. The number of branches observed on filaments assembled from ATP-actin declined with time, with two exponential rates on highly adhesive surfaces and one exponential rate on less adhesive surfaces. We conclude from our experiments on branching in bulk samples that the nucleotide bound to the mother filament does not influence branch formation, but our microscopic observations show that ATP hydrolysis and γ-phosphate dissociation increase the rate of branch dissociation ∼10-fold as mother filaments age.
MATERIALS AND METHODS
Proteins
Actin was prepared from an acetone powder of rabbit skeletal muscle (14), gel filtered on a column of Sephacryl S300, stored in buffer G (2 mM Tris, pH 8.0, 0.2 mM ATP, 0.1 mM CaCl2, 0.5 mM dithiothreitol (DTT)) at 4°C, and used within 3 weeks of preparation. Actin was dialyzed for 24 h versus buffer G and centrifuged at 100,000 × g for 2 h immediately before use. Mg-ADP actin monomers were prepared by treating Mg-ATP actin with soluble hexokinase and glucose (15) and used within 4 h.
Arp2/3 complex was prepared from bovine thymus using a modification of a previously described protocol (16). Two frozen bovine thymuses were broken into small chunks and blended in 100 ml homogenization buffer (10 mM Tris, pH 8.0, 1 mM DTT, 1 mM MgCl2, 50 mM KCl, 2 mM EGTA, 5% glycerol, 4 Complete EDTA-free protease inhibitor tablets (Roche Diagnostics, Mannheim, Germany)/L, and 1 mM phenylmethylsulfonyl fluoride). The homogenate was centrifuged at 2600 rpm for 30 min. Lipids were removed from the surface. The supernatant and pellet were further centrifuged at 6000 rpm for 15 min. The supernatant was filtered through cheesecloth and centrifuged at 100,000 × g for 1 h. The supernatant was passed over a fast-flow Q-Sepharose (Amersham Pharmacia Biotech, Uppsala, Sweden) column in Q-buffer (10 mM Tris, pH 8.0, 1 mM DTT, 1 mM MgCl2, 50 mM KCl, 0.2 mM ATP, 1 mM EGTA, 2% glycerol) with two protease inhibitor tablets without EDTA/L. The flow-through was subject to precipitation with a 35% followed by a 55% saturated ammonium sulfate solution. The pellet from the second step was dialyzed into Q-buffer without KCl overnight.
The protein was further purified on a fast-flow Q-Sepharose column in Q-buffer (10 mM Tris, pH 8.0, 1 mM DTT, 1 mM MgCl2, 15 mM KCl, 0.2 mM ATP, 1 mM EGTA, 2% glycerol) after some dilution to ensure that the initial conductivity of the solution matched a 15 mM KCl solution. A linear gradient of salt between 15 mM and 300 mM KCl was used to elute the proteins. Fractions with actin-nucleating activity were further purified by binding to human GST (glutathione S-transferase C-terminal fragment)-WASp (Wiskott-Aldrich syndrome protein)-VCA (verprolin homology - connecting - acidic motifs) on a glutathione-Sepharose column and eluted with Q-buffer containing 300 mM KCl. A final step of purification over a Source S column (10 mM 1,4-piperazinediethanesulfonic acid (PIPES), pH 6.8, 25–150 mM KCl, 0.2 mM EGTA, 0.2 mM MgCl2, 1 mM DTT) yielded pure Arp2/3 complex. The complex was dialyzed into ATP polymerization buffer (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 10 mM imidazole, pH 7.0, 0.2 mM ATP, 1 mM DTT, 5% glycerol) and frozen as small aliquots in liquid nitrogen. Aliquots were thawed for each experiment.
GST-WASp-VCA was expressed in Escherichia coli BL21 cells and purified by affinity chromatography on glutathione-Sepharose and ion exchange chromatography on Source Q-Sepharose and stored at −80°C. Protein concentrations were determined by absorbance using the following extinction coefficients: actin (ɛ290 = 25,974 M−1cm−1), Arp2/3 complex (ɛ290 = 139,000 M−1cm−1), GST-WASp-VCA (ɛ280 = 46,200 M−1cm−1).
Protein labeling
We labeled actin on lysine with Alexa Fluor 488 succinimidyl ester (Molecular Probes, Eugene, OR) in a protocol similar to Kellogg et al. (17). Actin filaments were dialyzed against labeling buffer (50 mM PIPES, pH 6.8, 50 mM KCl, 0.2 mM CaCl2, 0.2 mM ATP) for 4 h to remove DTT and to adjust the pH. We incubated actin filaments in labeling buffer with a 20-fold molar excess of dye overnight at 4°C. Filaments were pelleted at 100,000 × g for 2 h, resuspended in buffer G, and dialyzed for 48 h versus buffer G (1 L, 3 changes). The actin monomers were centrifuged at 100,000 × g for 2 h to remove remaining oligomers. The top 70% of the supernatant was gel filtered on a column of Sephacryl S-300 equilibrated with buffer G. The final label concentration was calculated using ɛ495 = 71,000 M−1cm−1 (Molecular Probes product information, MP10168). The final protein concentration was determined assuming a 0.11 fractional contribution to the A290 signal from the label. The ratio of label per molecule was <0.8 in all preparations. The rate of elongation was on average 10.7 ± 2.0 subunits/(μM × s) with 15% or 30% of the actin monomers labeled on lysine residues with Alexa Fluor 488. This value matches the elongation rate of unlabeled actin (18), so that labeling actin with Alexa 488 at these concentrations does not affect actin filament elongation as much as labeling actin on cysteine 374 with rhodamine (10) or Oregon green (19).
Total internal reflection fluorescence microscopy
Glass flow cells were prepared from cleaned glass slides and 24 × 40 mm No. 2 glass coverslips using stretched Parafilm as spacers to create a chamber 20–50-μm deep and 5-mm wide (20). In most cases, chambers were prepared in a high humidity environment on a grounded metal plate to minimize static. Chambers were flushed first with 20–200 nM N-ethylmaleimide (NEM)-inactivated skeletal muscle myosin (21) in high salt buffer (500 mM KCl, 1 mM EGTA, 1 mM MgCl2, 10 mM imidazole) for 30 s at 22°C. Chambers were then flushed sequentially with high salt buffer and KMEI buffer (50 mM KCl, 1mM EGTA, 1 mM MgCl2; 10 mM imidazole, PH 7.0). Bovine serum albumin in KMEI buffer was used to block the slides for 2 min, and imaging buffer was introduced into the chamber. The imaging buffer contained 1 mM EGTA, 1 mM MgCl2, 10 mM imidazole, 100 mM DTT, 20 μg/ml catalase, 100 μg/ml glucose oxidase, 3 mg/ml glucose, and 0.5% methylcellulose (2000 cps at 2%) (10,19) with one of three salt solutions, each with the same conductivity at room temperature: i), 50 mM KCl; ii), 25 mM potassium phosphate, pH 7.0 (15.37 mM H2KPO4 and 9.63 mM HK2PO4) with 19.9 mM KCl; or iii), 25 mM potassium sulfate with 3.5 mM KCl. The potassium sulfate introduces a similar bivalent anion to control for the presence of the phosphate ion in solution. In agreement with previous results, sulfate did not affect measurable polymerization parameters (22).
For TIRF microscopy we used an Olympus (Tokyo, Japan) IX70 inverted microscope with the excitation beam entering through a prism from the top of the slide (10,19). The excitation wavelength was 490 nm. Filaments were visible with 10% or more of the subunits labeled with Alexa Fluor. Generally 30% labeled actin was used in experiments. Initial “seed” filaments are grown for several minutes on the slide before flowing in a mixture of Arp2/3 complex, GST-WASp-VCA, and actin monomer. Images were taken every 5–10 s with a Hamamatsu (Hamamatsu City, Japan) CCD camera ORCA-ER charge-coupled device camera. Humidifiers kept the ambient relative humidity above 35%.
Measurement of adhesion of filaments to slides
The adhesion of filaments to slides varied between experiments and influenced observed branch densities. We determined the adhesiveness of each slide independently using the motion of filaments on the slide. First, we corrected the images for global stage drift by subtracting the center of mass positions (xcm, ycm) of all the measured filaments at a given time from the raw coordinates of the filament segments. We measured the changes in corrected position of each segment of each actin filament (Xij(tk+1), Yij(tk+1)) relative to the previous time position (tk). Here k is the ordinal number of the image, i is the ordinal number of the filament, and j is the ordinal number of the segment. Reaction 1 defines the seed mobility, and the total number of filaments (nf) was constant through the movie. Each filament contained a number of equal length segments (ms) that depended on the length of the filament (Li) at the time (tk). A total of pt time points are separated by time Δt. Higher mobility implies fewer attachment points.
Mother filament age determination
The use of full filament histories to determine age was necessary for quantitative results because filament growth could be interrupted by slide affects, breakage, or annealing. We recorded a full time-lapse series of the growth of at least 20 filaments for each experiment and traced their growth through time. Each complete experiment included two series of images: one series documented the growth of the seed filaments, and the other series provided information after the addition of the Arp2/3 complex and activators. Each image in the series contained an absolute time stamp used to determine time differences. We measured filaments grown from an original mother filament as well as filaments nucleated later, but not filaments that dropped into the plane of focus fully formed with no history. Our measurement technique used a combination of a custom automated line detection given a starting point and manual correction as necessary. Each measured filament in an image was reduced to a series of (x,y) coordinates of five-pixel (∼530 nm) filament segments with smaller end segments for a total of m segments that varied with filament length. The total filament length, L, at the time of each image is the sum of the lengths of all the segments.
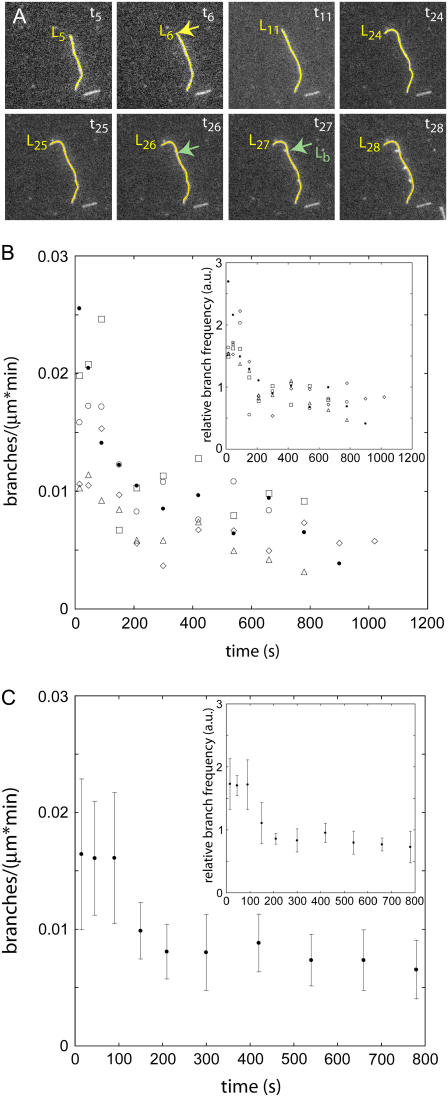
Fig. 1 A illustrates how we determined the time that each branch began and the age of that segment of the mother filament at that point in time. When defining the time that each branch began, we first looked late in the time-lapse sequence when the branch was over 1-μm long and clearly visible growing from the mother filament, as it is in Fig. 1, frame 28. We then tracked the branch backward through time until the first indication of the branch was detectable as a bright spot. Knowing that branches grow at six subunits (16 nm) per second, we assumed that the branch started in the previous frame, 10 s before the frame with the bright spot. We used the subscript k to denote an image number in the time-lapse series of images taken of the slide surface. In the time between tk−1 and tk each filament grew from length Lk−1 to length Lk. Thus, the segment of filament between Lk−1 and Lk was newly created at time (tk + tk−1)/2. The age of this segment at any later time was found by subtracting creation time from the current image time. For example, the age of the “mother filament” at the branch point, Lb, that occurred at time t26 was age = t26 − t6 where t6 was the time the length segment at Lb was first created. A linear interpolation was used for points falling between the discrete segment positions or points on the seeds that were formed before the first slice of the seed movie. We divided the number of branches that originated on segments of a given age by the length of segments of that age to obtain the branch count per unit length for each age.
FIGURE 1.
Actin branch formation by Arp2/3 complex observed by fluorescence microscopy on aging mother filaments. Comparison of five separate experiments demonstrates the variability in the observations. (A) Time series of fluorescence micrographs of a branching actin filament. The images in this series were taken 10 s apart. Conditions: 0.6 μM Alexa 488-ATP-actin monomers, 200 nM WASp-VCA, 5 nM Arp2/3 complex in TIRF buffer without phosphate. The mother filament is traced in yellow with a length marker at the barbed end. A branch point is labeled with a green arrow in the frame in which it becomes visible and in the frame immediately before it appears. The length to the branch point from the pointed end is labeled (Lb). The segment of the mother filament that serves as the substrate for the branch labeled in frame t26 (the number refers to the frame number) was first created in the frame labeled t6 (Lb = L6). The difference of these two times yields the age of the mother filament at the time of branching, age = t26 − t6. (B) The observed branch density per minute (branches per micrometer of filament in 60 s) on segments of filaments with ages ranging from 0 to 1100 s. The symbols represent data from five similarly prepared slides. (Inset) These five data sets were normalized by dividing individual branch densities per minute in a given age segment by the total branch density per minute over all ages and times to correct for variations in protein concentrations. (C) Average branch densities per minute observed as a function of the age of the mother filament segment for the five experiments in B. The error bars are ∼30% mean ± 1 SD. (Inset) Normalized mean branch densities per minute have lower standard deviations of 15%.
Uncertainties
The following sources contribute to uncertainties in these experiments: length measurement error, global variations in concentrations, and differences in adhesive effects of the substrate. To ascertain the magnitude of the uncertainties in the experiments, we repeated experiments with a single set of conditions on the same day. We traced and tallied branches on 20 filaments in each experiment. Some scatter existed between experiments, but the trends in observed branch frequency with age were consistent (Fig. 1 B). To correct for variations in concentrations we divided the observed branch frequencies on filaments of different ages by the total frequency of observed branches on all filaments in the field (inset, Fig. 1 B). We used this type of normalization to compare the observed branch frequencies along the filaments looking for age dependence but not to draw quantitative comparisons between different experiments. Without this normalization standard deviations between experiments were 30% of the mean branch frequency (Fig. 1 C). With this normalization the standard deviations were 15% of the mean (Fig. 1 C, inset). We used these percentage errors to display all data unless otherwise stated.
RESULTS
Branch formation in bulk samples
We used the time course of polymerization of bulk samples with pyrenyl-labeled actin to measure new end formation by ATP actin monomers with Arp2/3 complex, GST-WASp-VCA, and actin filament seeds. We calculated the concentration of growing barbed ends from the instantaneous polymerization rate, the elongation rate constant, and the concentration of ATP-actin at every point in the experiment. This method measures the concentration of all growing barbed ends whether the filament is a branch or free in solution. We grew actin filament seeds by polymerizing ATP-actin monomers for 1 h in the presence or absence of phosphate. In the absence of phosphate most of the subunits in these seeds had bound ADP, whereas in the presence of phosphate the seeds were primarily ADP-Pi bound. We performed the nucleation experiments with buffers that allowed for free aging (no phosphate) or that maintained the homogeneous nucleotide (with phosphate).
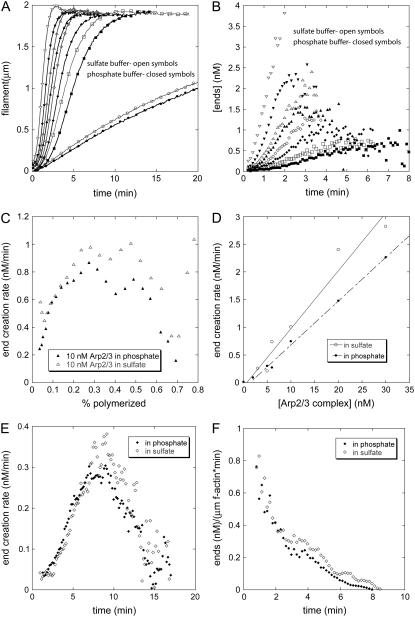
In reactions of ATP-actin monomers and actin filament seeds the polymerization rate increased with time, slightly less so with saturating concentrations of phosphate than controls with sulfate (Fig. 2 A). From the time courses of polymerization, we derived the concentration of ends over time (Fig. 2 B). The concentration of ends increased with the concentration of Arp2/3 complex and was consistently higher in sulfate than in phosphate. The slopes in Fig. 2 B yield end creation rates, shown in Fig. 2 C for 10 nM Arp2/3 complex. The end creation rate increased during the first third of the polymerization reaction because the concentration of polymerized actin increased over time, providing more substrate for branching. The end creation rates were the same in sulfate and in phosphate until 30% of the actin was polymerized, after which the rate was faster in sulfate. At the point where half of the actin was polymerized, the end creation rate was directly proportional to the concentration of Arp2/3 complex (Fig. 2 D) and higher in sulfate than phosphate. The results were similar using a lower concentration of ATP-actin monomers (Fig. 2 E). The end creation rates fell late in all of these reactions (Fig. 2, C, E, and F) owing to the depletion of actin monomers. Arp2/3 complex requires filaments to nucleate efficiently, indicating that branch formation is the primary mechanism for creating filament ends in these experiments. The initial end creation rates show that ATP-actin monomers and Arp2/3 complex formed branches at the same rate on mother filaments saturated with ADP-Pi and on a mixture of ADP and ageing ATP actin filaments.
FIGURE 2.
Calculation of nucleation rates from the time course of polymerization of bulk samples of actin monomers, Arp2/3 complex, WASp-VCA, and actin filament seeds. The polymer concentration was measured from the fluorescence of pyrene-actin. (A–D) Conditions: 2 μM Mg-ATP-actin, 200 nM GST-WASp-VCA, 0–20 nM active Arp2/3 complex, 5 nM actin filament seeds (30 min old, formed from ATP actin monomers in the same buffer used for the nucleation reaction) in polymerization buffer with (open symbols) 25 mM sulfate or (solid symbols) 25 mM phosphate. (A) Time courses of polymerization in phosphate or sulfate buffer with variable concentrations of active Arp2/3 complex: (circles) 0 nM; (squares) 2 nM; (diamonds) 5 nM; (triangles) 10 nM; and (inverted triangles) 20 nM. This stock solution of Arp2/3 complex was 25% active based on the concentration of ends produced with 5 nM of complex. (B) Calculated concentration of barbed ends created by Arp2/3 complex over time in the samples in A. Ends provided by the seeds were subtracted. (C) End creation rate with 2.5 nM active Arp2/3 complex depending on the fraction of the total actin polymerized. (D) Dependence of the end creation rate on the concentration of active Arp2/3 complex with 25 mM sulfate (open circles) or phosphate (solid circles). These rates were calculated at the point where 50% of the actin monomers were polymerized. (E and F) Rates of barbed end formation under the conditions used in TIRF experiments (Figs. 1, 3, and 4). Conditions: 0.6 μM actin monomer, 5 nM Arp2/3 complex (55% active), 200 nM GST-WASp-VCA, seeds grown from ATP-bound monomers for 10 min. Symbols: open diamonds 25 mM sulfate; filled diamonds 25 mM phosphate buffer. (E) End creation rate as a function of time for conditions matching the TIRF experiments formed in 25 mM sulfate (open diamonds) or phosphate (solid diamonds). (F) Rate of ends formed per micrometer of mother filament as a function of time after initiating polymerization in 25 mM phosphate or 25 mM sulfate.
Branch formation observed by microscopy
We used TIRF microscopy to visualize individual branches on the sides of mother filaments on glass slides coated with NEM-myosin. This method records those branches that grow parallel to the microscope slide within the evanescent field. Branches that grew long enough on the surface coated with NEM-myosin to be distinguished from the mother filament showed no tendency to detach from the mother filament or move out of the evanescent field. This implies that filaments recorded here were stabilized on the slide surface. In no case was a fully formed filament seen to couple to the side of a mother filament to form a branch.
The key variable in these experiments was the nucleotide state of the mother filament at the time a branch became visible. Mother filaments were grown from actin monomers with bound ATP, ADP, or ADP-Pi. The ADP and ADP-Pi mother filaments were stable over time in their respective buffers. The ATP mother filaments aged as they hydrolyzed the bound ATP and dissociated the γ-phosphate. To document the age of each segment of each mother filament, we took time-lapse images of filaments growing for up to 10 min with only actin monomers in polymerization buffer. After adding Arp2/3 complex, fresh actin monomers, and GST-WASp-VCA to activate Arp2/3 complex, we acquired more images for a further 5–10 min. We used the recorded history of each filament to extrapolate back the age of each segment at the time that each branch began.
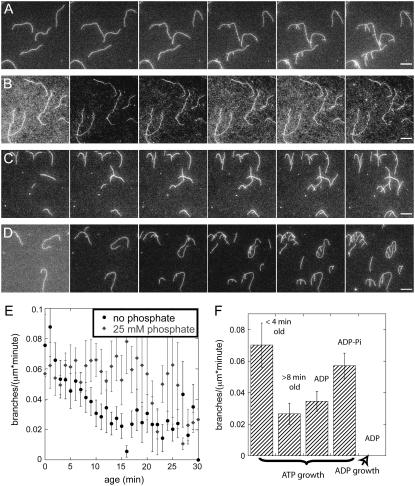
We tested four combinations of actin monomers and filaments to determine how the nucleotide bound to monomers and filaments influenced the observation of branches by TIRF microscopy. For quantitative analysis of the experimental conditions, we combined several experiments and averaged the branch frequencies per micrometer. Using a total of 2 cm actin filament and ∼1000 branching events, we determined the average branch frequency per micrometer as a function of age (Fig. 3 E). The error bars in Fig. 3 E represent the standard deviation of the mean and are largest on the oldest filament where the sample size is small. We observed no branches on ADP-bound mother filaments after the introduction of Arp2/3 complex, ADP actin monomers, and activators in ADP buffer (Fig. 3 B). The absence of a hydrolyzable nucleotide bound to Arp2/3 complex may play a role in this result (7,8,15,22). We observed branches at a low frequency on ADP-bound mother filaments when we polymerized ATP-actin monomers with Arp2/3 complex and an activator in ATP buffer (Fig. 3 C).
FIGURE 3.
Effect of nucleotide on the branch density observed by fluorescence microscopy. Conditions: 0.6 μM actin monomer, 5 nM Arp2/3 complex (55% active), 200 nM GST-VCA in TIRF buffer with 25 mM sulfate or phosphate. (A–D) Time series of fluorescence micrographs at 60-s intervals in different buffer conditions. (A) ATP-actin monomers in ATP/sulfate buffer with mother filaments polymerized from ATP-actin monomers starting 10 min before the first frame. ATP branches grew from aging mother filaments. (B) ADP-actin monomers in ADP/sulfate buffer with mother filaments polymerized from ADP-actin monomers starting 15 min before the first frame. No ADP branches grew from ADP-actin mother filaments. (C) ATP-actin monomers in ATP/sulfate buffer with mother filaments polymerized from ADP-actin monomers starting 15 min before the first frame. ATP branches grew from ADP mother filaments. (D) ATP-actin monomers in ATP/phosphate buffer with mother filaments polymerized from ATP-actin monomers in phosphate starting 10 min before the first frame. ATP branches grew from ADP-Pi mother filaments. (E) Average observed branch frequency per micron as a function of the age of the mother filament. Mean ± 1 SD. (Solid circles) Branching by ATP-actin monomers in ATP/sulfate buffer from aging mother filaments polymerized from ATP-actin monomers. (Shaded symbols) Branching by ATP-actin monomers in ATP/phosphate buffer from mother filaments polymerized from ATP-actin monomers in phosphate buffer. (F) Summary of the average branch densities per minute observed under various conditions: ATP-actin monomers on aging ATP-actin filaments <4 min old or >8 min old; ATP-actin monomers on ADP-filaments; ATP-actin monomers on ADP-Pi filaments; and ADP-actin on ADP filaments. Averaging was performed over 14–18 experiments containing a total of 2 cm of actin and 1000 events for each condition. Scale bars are 5 μm.
The number of branches observed with ATP-actin monomers, Arp2/3 complex, and an activator in ATP buffer depended on the age of each segment of mother filament polymerized from ATP-actin (Figs. 1, B and C, and 3 E). After averaging branch frequencies from all of our slide conditions, we observed 2–3 times more branches on young segments of filaments (<4 min old) than on old segments of filaments (>8 min old) (Fig. 3, E and F). The observed frequency of new branches on old segments of filaments plateaued after 10 min at a minimum value similar to the frequency of branches on ADP-bound mother filaments without phosphate. Under these conditions, the frequency of observed branches decayed with the age of each mother filament segment with a half time of ∼6.3 min (Fig. 3 E, solid circles).
When 25 mM phosphate was present with ATP-actin monomers, Arp2/3 complex, and an activator in ATP buffer, branches appeared at the same frequency on all parts of mother filaments regardless of their age (Fig. 3, D and E, shaded diamonds). This phosphate concentration largely saturates the γ-phosphate site of ADP-actin filaments, forming ADP-Pi-bound filaments (22). Within error, we observed branches on the phosphate-bound mother filaments at the same frequency as on segments of mother filaments grown from ATP-actin in the absence of phosphate that were <4 min old. Given the standard deviations in Fig. 3 F, we can conclude with 99% certainty that the average number of branches observed on young filaments and ADP-Pi-bound filaments is statistically larger than the average number of branches on old filaments and ADP-bound filaments.
These direct observations by fluorescence microscopy differed in two ways from our measurements on bulk samples with the same concentrations of proteins. For this comparison we calculated the branch density per micrometer-minute of polymerized actin in bulk samples from the end creation rate, assuming that that all new filaments were created as branches in the presence of Arp2/3 complex and an activator (Fig. 1 F). First, the number of branches observed by microscopy depended on the nucleotide bound to the mother filament but not in bulk samples. Second, we calculated the rate of barbed end formation in bulk samples by averaging the data over the first 3 min and obtained 0.5 ends/(μm×min). This value was more than fivefold higher than observed by microscopy, even on ATP-actin filaments that were <3-min old or on ADP-Pi filaments.
Influence of the slide surface on the observation of branches
The adhesiveness of the slide influenced the number of branches observed but not the main points in previous sections. The ability to observe branches depends on first stabilizing the branch. In our experiments, the factor that allows us to stabilize the branches is the slide surface itself, and the ability of the surface to hold new branches can vary according to the number of filament adhesion points available.
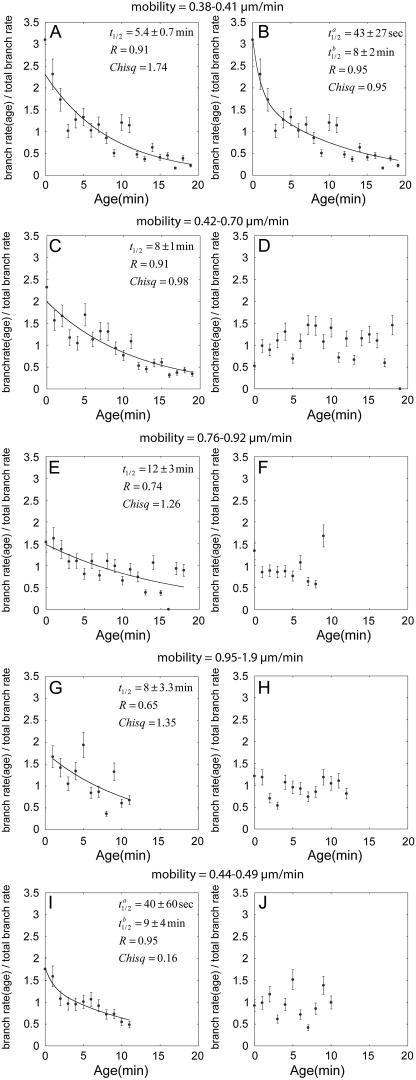
To demonstrate this effect of the surface, we normalized the observations from over 40 experiments, each with 10 and 20 filaments, by dividing the observed branch density per minute for a given age by the total branch density per minute over all the filaments in the experiment. This normalization eliminated effects of differing protein concentrations while allowing a comparison of the age dependence of the branching on slides with differing adhesion strengths. We used the lateral motion of filaments on the slide (“mobility”) to gauge adhesion. This mobility was inversely related to the density of nonspecific adhesions to the glass (that varied between stocks of slides) and of specific tethers by NEM-myosin (that varied with the total amount of myosin used to coat the slide). We grouped the experimental data according to the motilities and averaged over 4–7 experiments.
The normalized number of branches observed decayed with the age of the mother filament assembled from ATP-actin in sulfate buffer (Fig. 4, A–C, E, G, and I) but was independent of the age of the mother filament in the presence of 25 mM phosphate (Fig. 4, D, F, H, and J). The decay in the number of branches observed on the age of ATP-mother filaments was better fit by a double exponential than a single exponential when the filaments adhered strongly to the substrate (Fig. 4, A and B). However, with fewer attachment points, the fast exponential was not apparent, leaving only a single exponential with a decay constant of between 8 and 12 min (Fig. 4, C, E, and G). Although the slow exponential was expected from the results in Figs. 1 and 3, the decay of <1 min is new and indicative of a previously undetected age dependence of the branch stability.
FIGURE 4.
Normalized branch density per minute observed microscopically as a function of the age of the mother filament with different nucleotide composition and surface adhesiveness. Dividing the individual branch density per minute at each age segment by the total branch density per minute for each experiment provided the normalization. Adhesiveness is inversely related to the mobility of the filaments on the surface, noted in μm/min above each graph. General conditions: 0.6 μM Mg-ATP actin, 5 nM total Arp2/3 complex (50% active), and 200 nM GST-VCA. (A–C, E, G, and I) 25 mM sulfate buffer. (A and B) Highly adhesive low mobility (0.38–0.41 μm/min) substrates in sulfate buffer. The decline in the observed branch density per minute with age of the mother filament fits a double exponential (B, t1/2 = 1 min, t1/2 = 12 min, R = 0.95) much better than a single exponential (A, t1/2 = 8.8 min, R = 0.91). (C, E, and G). When the adhesiveness of the substrate was lower, the observed branch density per minute declined with the age of the mother filament as a single exponential noted on each graph. (I) With a higher rate of filament elongation using 1 μM actin monomer, the decline in branch densities per minute fits two exponentials (t1/2 = 1 min, t1/2 = 14 min, R = 0.94) even on a slide of moderate adhesiveness. (D, F, H, and J) 25 mM phosphate buffer ionically balanced with KCl. Observed branch density per minute was independent of the age of the mother filament on all substrates with both concentrations of actin monomers.
We also observed the fast exponential on less adhesive slides, provided that we used a high concentration of actin monomers to increase the rate of elongation (Fig. 4 I). The ability to capture the fast decay at high adhesion or fast polymerization indicates that highly unstable branches can only be captured when adhesion density is strong or branch growth is fast enough to span more disperse adhesion points.
DISCUSSION
By comparing actin filament branching rates observed in bulk samples with those found by TIRF microscopy, we learned that the nucleotide bound to mother filaments governs the stability of branches but not the rate that Arp2/3 complex forms branches. Branches dissociate more rapidly from ADP-actin mother filaments than from ADP-Pi mother filaments. Dissociation of branches has no effect on the time course of polymerization of bulk samples, explaining why the nucleotide state of mother filaments does not influence their ability to promote branching in bulk samples. However, the rate of dissociation strongly affects the probability that a branch will be captured by the slide in real time microscopic assays or by phalloidin in others assays (7,11,23). The discussion reviews the reactions and the interpretation of the two types of assays, followed by our calculations of branch dissociation rates.
Reactions observed in bulk samples using fluorimeter assays
Fluorimeter assays of bulk samples and microscopy of single filaments measure different aspects of the branching reactions. Because spontaneous nucleation was very slow under the conditions used here and because nucleation by Arp2/3 complex requires binding to a preexisting filament, most of the new filament ends created in bulk samples were formed as branches. In a simplified branching model, a VCA activator molecule brings together an actin monomer (G-actin) and an Arp2/3 complex on the side of an existing filament to produce a branch:
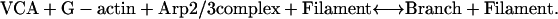 |
(1) |
In reaction 1, filaments act catalytically to form branches. Each filament and each branch has a free barbed end capable of elongation. New barbed ends, whether on a free filament or on an attached branch, are indistinguishable in bulk assays. A branch can dissociate to form an independent filament with the same barbed end, releasing VCA and Arp2/3 complex to recycle:
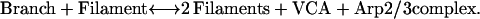 |
(2) |
The reaction described by reaction 2 has little or no effect on the number of ends measured in bulk polymerization assays, assuming that the polymerization reaction proceeds quickly enough that the newly released Arp2/3 complex and VCA do not have time to start a new branch. Late in the polymerization reaction, the fluorimeter assay detected slightly fewer ends in phosphate than in the sulfate control. One possible explanation for this observation is that Arp2/3 complex dissociates more slowly from phosphate-bound mother filaments, slowing the recycling of Arp2/3 complex.
This work and previous experiments with phalloidin (7,11,23) show that branches dissociate in seconds to minutes. On the other hand, the reverse of reaction 2, the assembly of a branch from two free filaments, is extremely slow and was not observed with the low concentration of 5 nM Arp2/3 complex used in this study.
Branch products measured microscopically
Microscopic assays detect only branches associated with their mother filament. Since we never observe branches dissociating, all of the observed branches must be stabilized by interactions with myosin or other adhesion points on the slide. In this reaction, a branch (Br) interacts with a stabilizing influence (st) on the slide to form a stabilized branch (stBr):
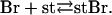 |
(3) |
The probability that a branch is stabilized before it dissociates (and is lost to observation) depends on the length of the branch (and therefore its rate of elongation) and the adhesiveness of the slide. Given a uniform elongation rate and uniformly distributed adhesion sites, the probability that a branch is captured is inversely related to the rate that it dissociates.
Branches that grow vertically out of the evanescent field or dissociate before binding to the slide are lost to direct observation but are recorded in bulk assays. That is, bulk assays measure Br + filament, whereas microscopic assays measure only stBr. Therefore we can use measurements of branches formed in bulk samples and microscopic counts of stabilized branches to infer rates of branch dissociation from mother filaments.
Comparison of branching from ADP- and ADP-Pi mother filaments
In bulk samples Arp2/3 complex and VCA formed new filament ends, presumed to be branches under the conditions employed, at about the same rate on mother filaments that age freely with or without saturating phosphate, whereas saturating phosphate increased the number of branches observed by TIRF microscopy on ADP filaments independent of their age. The larger number of branches that we observed by microscopy on filaments saturated with phosphate was previously missed in a study that used only 2 mM phosphate (11), which did not fully saturate the filaments (22). However, even with saturating phosphate the number of branches observed per micrometer of filament was less than in bulk samples. Our interpretation of this difference is that many filaments dissociate from the mother filaments before being stabilized for microscopic observation and branches dissociate faster from ADP filaments than ADP-Pi filaments.
Saturating concentrations of phosphate may associate with the nucleotide in the Arp2/3 complex itself and affect the stability of branches. Within error, the number of branches observed microscopically on new filaments is the same as the branch density on filaments in saturating phosphate buffer. Since the branch density observed microscopically is proportional to the branch stability, any effect of phosphate-bound Arp2/3 complex on branch stability is subtle.
Branching on aging ATP-actin filaments
The gap between the number of branches formed in bulk samples and the number observed microscopically widens as ATP-actin mother filaments age. We observed the maximum of branches on segments of filaments assembled from ATP-actin <1-min old, provided that the surface was highly adhesive (Fig. 4 B) or we used 1 μM actin monomers (Fig. 4 I) so that the branches grew about twice as fast as in our standard conditions. Since the bulk assays show that all segments of filaments are equally supportive of branch formation, our interpretation is that branches start dissociating immediately after their formation, not that the ends favor branching as proposed previously (12). The difference in the lifetime of these transient branches that occurs between new filament and filament over 1-min old may have been lost to observation in earlier experiments (7,11) due to slow binding of rhodamine-phalloidin to actin filaments.
ATP hydrolysis and phosphate dissociation from mother filaments, daughter filaments, or Arp2/3 complex may contribute to this fast loss of stability during the first minute after polymerization of ATP-actin. On these young filaments, more than 80% of the subunits have bound ADP-Pi, so the branches lose stability before phosphate dissociates from all of the subunits in the mother filaments. Perhaps phosphate dissociation from just one of the three mother filament subunits that bind each Arp2/3 complex (24) destabilizes branches.
After the first minute, the stability of the branches declines further with a half time on the order of 8 min as mother filaments age. Three lines of evidence suggest that this decline in the number of branches observed as a function of age is related to ATP hydrolysis and phosphate release. First, the decline in branches has about the same half time as γ-phosphate dissociation (6,25). Second, a concentration of phosphate that saturates the γ-phosphate site on ADP-actin filaments prevents the decay in branch density observed by microscopy on filaments freshly polymerized from ATP-actin (Fig. 4). Third, phalloidin, which inhibits γ-phosphate dissociation (26), and the phosphate analog BeF3 also stabilize branches (11).
Branch dissociation rates
We calculated the relative rates of branch dissociation from mother filaments with different nucleotide states and put lower limits on the rate constants. We assumed that branches dissociate unless captured by the slide and that slides capture branches randomly at a rate proportional to the length of the branch (and therefore directly proportional to the elongation rate) and to the density of adhesion points. We assumed that branch dissociation is a competing first order reaction and subtracted the dissociation rate from the stabilization rate to get the net capture rate. Of these quantities, only the branch dissociation rate was variable on a single slide.
Using the two decay constants in observed branches from Fig. 4 B, we calculated that the rate constant for branch dissociation from ADP-bound mother filaments (kD) is almost 10 times higher than the rate constant for branch dissociation from new ATP-actin filaments (kn = 0.1 kD). Additionally, the rate constant for branch dissociation from mixed ADP/ADP-Pi filament (kpi) was approximately half that of pure ADP filament (kpi = 0.5 kD). We could distinguish branches from mother filaments easily after they had reached a length of 1 μm, which took 60 s with 0.6 μM actin in our experiments. Branches that we saw did not dissociate and were well adhered to the substrate, so kn ∼ 0.02 s−1 was the lower limit for the branch dissociation rates from the newest segments of ATP-actin mother filaments. Using the relative rates of capture from the different segments of aging filaments, we calculated that kD ∼ 0.2 s−1 and kpi ∼ 0.04 s−1.
Implications for cells
In motile cells highly branched actin networks are confined to the first 1–2 μm of the leading edge. The branched network exists for a short time, on the order of a few seconds, before remodeling into longer unbranched filaments, disassembling, and recycling (1,4). The time course of debranching observed in this work may contribute to the remodeling. Especially intriguing is the observation of transient branches that dissociate from filaments with >80% of the subunits in the ADP-Pi state. With the aid of accessory proteins such as cofilin to accelerate phosphate release, branches could easily be limited to the first micrometer of a leading edge and exist for mere seconds.
References
- 1.Pollard, T. D., and G. G. Borisy. 2003. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 112:453–465. [DOI] [PubMed] [Google Scholar]
- 2.Machesky, L. M. 1997. Cell motility: complex dynamics at the leading edge. Curr. Biol. 7:R164–R167. [DOI] [PubMed] [Google Scholar]
- 3.Mullins, R. D., and M. D. Welch. 2002. Cellular control of actin nucleation. Annu. Rev. Cell Dev. Biol. 18:247–288. [DOI] [PubMed] [Google Scholar]
- 4.Svitkina, T. M., and G. G. Borisy. 1999. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J. Cell Biol. 145:1009–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flanagan, L. A., J. Chou, H. Falet, R. Neujahr, J. H. Hartwig, and T. P. Stossel. 2001. Filamin A, the Arp2/3 complex, and the morphology and function of cortical actin filaments in human melanoma cells. J. Cell Biol. 155:511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blanchoin, L., and T. D. Pollard. 1999. Mechanism of interaction of Acanthamoeba actophorin (ADF/cofilin) with actin filaments. J. Biol. Chem. 274:15538–15546. [DOI] [PubMed] [Google Scholar]
- 7.Blanchoin, L., T. D. Pollard, and R. D. Mullins. 2000. Interaction of ADF/cofilin, Arp2/3 complex, capping protein and profilin in remodeling of branched actin filament networks. Curr. Biol. 10:1273–1282. [DOI] [PubMed] [Google Scholar]
- 8.Le Clainche, C., D. Pantaloni, and M.-F. Carlier. 2003. ATP hydrolysis on actin-related protein 2/3 complex causes debranching of dendritic actin arrays. Proc. Natl. Acad. Sci. USA. 100:6337–6342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dayel, M. J., R. D. Holleran, and D. M. Mullins. 2001. Arp2/3 complex requires hydrolyzable ATP for nucleation of new actin filaments. Proc. Natl. Acad. Sci. USA. 98:14871–14876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amann, K. J., and T. D. Pollard. 2001. Direct real-time observation of actin filament branching mediated by Arp2/3 complex using total internal reflection microscopy. Proc. Natl. Acad. Sci. USA. 98:15009–15013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ichetovkin, I., W. Grant, and J. Condeelis. 2002. Cofilin produces newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex. Curr. Biol. 12:79–84. [DOI] [PubMed] [Google Scholar]
- 12.Pantaloni, D., R. Boujemaa, D. Didry, P. Gounon, and M.-F. Carlier. 2000. The Arp2/3 complex branches filament barbed ends: functional antagonism with capping proteins. Nat. Cell Biol. 2:385–391. [DOI] [PubMed] [Google Scholar]
- 13.Fujiwara, I., S. Suetsugu, S. Uemura, T. Takenawa, and S. Ishiwata. 2002. Visualization and force measurement of branching by Arp2/3 complex and N-WASP in actin filament. Biochem. Biophys. Res. Commun. 293:1550–1555. [DOI] [PubMed] [Google Scholar]
- 14.Spudich, J. A., and S. Watt. 1971. The regulation of rabbit skeletal muscle contraction. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246:4866–4871. [PubMed] [Google Scholar]
- 15.Pollard, T. D. 1984. Polymerization of ADP-actin. J. Cell Biol. 99:769–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Higgs, H. N., L. Blanchoin, and T. D. Pollard. 1999. Influence of the Wiskott-Aldrich syndrome protein (WASp) C terminus and Arp2/3 complex on actin polymerization. Biochemistry. 38:15212–15222. [DOI] [PubMed] [Google Scholar]
- 17.Kellogg, D., T. Mitchison, and B. M. Alberts. 1988. Behavior of microtubules and actin filaments in living Drosophila embryos. Development. 103:675–685. [DOI] [PubMed] [Google Scholar]
- 18.Pollard, T. D., P. C.-H. Tseng, D. L. Rimm, D. P. Bichell, R. C. Williams, and J. Sinard. 1986. Characterization of alpha-actinin from Acanthamoeba. Cell Motil. Cytoskeleton. 6:649–661. [DOI] [PubMed] [Google Scholar]
- 19.Kuhn, J. R., and T. Pollard. 2005. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys. J. 88:1387–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kron, S. J., and J. A. Spudich. 1986. Fluorescent actin filaments move on myosin fixed to a glass surface. Proc. Natl. Acad. Sci. USA. 83:6272–6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pemrick, S., and A. Weber. 1998. Mechanism of inhibition of relaxation by N-ethylmaleimide treatment of myosin. Biochemistry. 15:5193–5198. [DOI] [PubMed] [Google Scholar]
- 22.Carlier, M.-F., and D. Pantaloni. 1988. Binding of phosphate to F-ADP-actin and role of F-ADP-Pi-actin in ATP-actin polymerization. J. Biol. Chem. 263:817–825. [PubMed] [Google Scholar]
- 23.Blanchoin, L., K. J. Amann, H. N. Higgs, J. B. Marchand, D. A. Kaiser, and T. D. Pollard. 2000. Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASp/Scar proteins. Nature. 404:1007–1011. [DOI] [PubMed] [Google Scholar]
- 24.Volkmann, N., K. J. Amann, S. Stoilova-McPhie, C. Egile, D. C. Winter, L. Hazelwood, J. E. Heuser, R. Li, T. D. Pollard, and D. Hanein. 2001. Structure of Arp2/3 complex in its activated state and in actin filament branch junctions. Science. 293:2456–2459. [DOI] [PubMed] [Google Scholar]
- 25.Melki, R., S. Fievez, and M.-F. Carlier. 1996. Continuous monitoring of Pi release following nucleotide hydrolysis in actin or tubulin assembly using 2-amino-6-mecapto-7-methylpurine ribonucleoside and purine-nucleoside phosphorylase as an enzyme-linked assay. Biochemistry. 35:12038–12045. [DOI] [PubMed] [Google Scholar]
- 26.Dancker, P., and L. Hess. 1990. Phalloidin reduces the release of inorganic phosphate during actin polymerization. Biochim. Biophys. Acta. 1035:197–200. [DOI] [PubMed] [Google Scholar]