Abstract
Trichoderma virens is a mycoparasitic fungus used in biocontrol of soilborne plant pathogens. It inhibits or kills plant-pathogenic fungi through production of antifungal antibiotics and parasitism of hyphae and sclerotia. Conidiation, or the production of asexual spores, an inducible process triggered by light or nutrient stress, is an important trait in survival and also development of formulation products. In many fungi, signaling pathways, including mitogen-activated protein kinase (MAPK) cascades, have been implicated in parasitism of host plants as well as in the production of asexual spores. Here, we have studied the role of a MAPK gene, that for TmkA, in conidiation and antagonistic properties of a biocontrol strain of T. virens. Through single- and double-crossover recombination, we obtained three tmkA loss-of-function mutants. The TmkA transcript was not detectable in these mutants. The mutants conidiated in the dark, although photoinduction was normal and the light sensitivities of the wild type and the mutant were the same. The mutants had, overall, normal colony morphology, but their radial growth rate was reduced by about 16%, with no decrease in biomass production. Against Rhizoctonia solani hyphae, the knockout mutants exhibited mycoparasitic coiling and lysis of host hyphae similar to that of the wild type. The mutants, however, were less effective in colonizing the sclerotia of R. solani. On Sclerotium rolfsii, the MAPK loss-of-function mutants had reduced antagonistic properties in confrontation assays and failed to parasitize the sclerotia. TmkA-dependent and -independent pathways are thus involved in antagonism against different hosts. Finally, in contrast to the case for other filamentous fungi studied so far, signaling through a MAPK represses, rather than induces, asexual sporulation.
Members of the genus Trichoderma attack and parasitize plant pathogens and are used as commercial biofungicides. Trichoderma virens (Gliocladium virens) can parasitize many soilborne pathogens, including Sclerotium rolfsii, Rhizoctonia solani, and Pythium spp., and also produces antifungal metabolites. Trichoderma has been reported to form specialized structures upon contact with the host; in particular, the mycoparasite coils around the host hyphae (7, 9). Furthermore, induction of genes encoding hydrolytic enzymes and the secretion of these enzymes are involved in the mycoparasitic interaction (1, 5, 14, 21, 38). Both coiling and enzyme secretion are induced responses, presumably triggered by molecules released from the host or located on its surface, as suggested by biomimetic experiments in which Trichoderma coils around lectin-coated nylon fibers (11). The other major developmental pathway in Trichoderma is conidiation. Blue light or nutrient stresses induce conidiation of Trichoderma, providing a model for photobiological studies (3, 10). Thus, mycoparasitic and reproductive (asexual) pathways are both triggered by external signals. Cellular signaling pathways are beginning to be understood in genetic model systems such as Neurospora and Aspergillus and in some of the plant pathogens such as Magnaporthe and Ustilago, as well as in Candida and Cryptococcus, which cause human diseases (17). Relatively little is known about the role of conserved eukaryotic signaling pathways in the antagonistic properties of mycoparasitic fungi used in plant disease biocontrol. A G protein belonging to the fungal Gi class is involved in conidiation and the mycoparasitic coiling response of Trichoderma atroviride (31). Loss of function (mutants obtained by antisense expression) of the Tga1 Gα subunit gene resulted in intense sporulation and loss of coiling and the ability to overgrow a host, R. solani. The severely reduced growth of the mutants, however, may be a major reason for the loss of antagonism.
The mitogen-activated protein kinase (MAPK) protein phosphorylation cascade transduces a variety of signals in eukaryotes (30), including yeasts, where there are five MAPK genes transmitting signals for mating, filamentous growth, cell integrity, response to osmotic stress, and ascospore formation (8). Genes encoding MAPK homologs are essential for development and pathogenicity of filamentous fungi (36). In the rice blast fungus Magnaporthe grisea, for example, three MAPK genes have been isolated. Pmk1, the prototype of the YERK1 (yeast and fungal extracellular signal-related kinase) class (16), is dispensable for growth and sporulation while required for appressorium differentiation and invasive growth (35). Fusarium oxysporum Fmk1 is required for invasive growth on tomato and for transcription of a pectate lyase gene but is dispensable for vegetative growth and sporulation (6). Colletotrichum lagenarium Cmk1 mutants are defective in conidiation and pathogenicity (34). The gray mold fungus Botrytis cinerea (39) requires the Pmk1 homolog Bmp1 for penetration and maceration of plant tissues; mutants were nonpathogenic on carnation flowers and tomato leaves. Cochliobolus heterostrophus Chk1 mutants lack appressoria and are impaired in mating, conidiation, and the ability to infect host leaves (18). The Claviceps purpurea (ergot) MAPK homolog Cmk1 is required for infection of the ovarian tissues of rye (25). Deletion of Ustilago maydis Kpp2 (Ubc3) affected cell fusion, induction of pheromone-responsive genes, and pathogenicity (24, 29). MAPKK Fuz7 mutants (2) show a more drastic loss of virulence, so that there may be redundant MAPKs downstream of Fuz7. Thus, a variety of pathogens, employing diverse modes of infection, all require a MAPK belonging to the YERK1 class defined by rice blast Pmk1 and yeast Fus3 and Kss1.
Mycoparasitism is a fungus-fungus interaction and thus represents a different model of host-pathogen interactions. Conserved eukaryotic signaling elements are nevertheless likely to be important, as in plant-fungus interactions. Likewise, the signals for the induction of conidiation could be transduced through these elements. Both mycoparasitism (involved in the process of biocontrol) and conidiation (involved in the process of formulation) are important phenomena so far as application of Trichoderma spp. as commercial biofungicides is concerned. We have initiated a program to study the role of the signal transduction process in the biocontrol fungus T. virens, with a view to manipulate these pathways to improve the biocontrol potential of Trichoderma spp. In this study, we isolated a MAPK homolog belonging to the YERK1 class, TmkA, from a biocontrol strain of T. virens; obtained loss-of-function mutants by homologous integration; and studied the role of the gene in antagonistic properties and conidiation.
MATERIALS AND METHODS
Fungal strains and culture conditions.
T. virens IMI 304061 was isolated from soil of Pantnagar, India, by using S. rolfsii as bait (26) and was subsequently characterized as a mycoparasite on the sclerotia of S. rolfsii and R. solani and on the hyphae of R. solani (28). S. rolfsii was isolated from ginger rhizome (27), R. solani (ITCC 4110) was collected from the Indian Type Culture Collection, New Delhi, India. Routinely, the fungi were maintained on potato dextrose agar (PDA) at room temperature and stored as glycerol stocks at −80°C for long-term storage.
Library construction (cDNA and cosmid), sequencing, and primer synthesis.
PCR amplification, Southern hybridization, plasmid extraction, screening of cDNA and cosmid library, and ligation, etc., were performed by standard protocols (32) or according to the manufacturers' protocols when kits were used. For reverse transcriptase PCR (RT-PCR), 3 μg of total RNA was treated with 1 Kunitz unit of RNase-free DNase (Qiagen) for 20 min at 37°C in New England Biolabs restriction enzyme buffer 4 (total volume, 10 μl), and then incubated at 65°C for 15 min. One microliter of the treated RNA was used as template for RT-PCRs with avian myeloblastosis virus reverse transcriptase and Tfl DNA polymerase (Access RT-PCR; Promega). The primers were MKORFf (CCAGCCCGACCATCATGTCTC) and MKORFr (CGCATAATCTCTTGGTAAATCAGTTG) for tmkA and hphfor (GAGGGCGAAGAATCTCGTGC) and hphrev (CACTGACGGTGTCGTCCATC) for the hygromycin phosphotransferase gene (hph). RT incubation was for 1 h at 48°C, followed by RT inactivation and template denaturation for 2 min at 94°C. Amplification was 40 cycles; for tmkA primers the annealing temperature was 60°C, and for hph primers it was 55°C.
For construction of a cDNA library, the fungus was grown in 100 ml of PDA (Difco) in a 250-ml flask at room temperature (24 to 25°C) for 48 h at 150 rpm in an orbital shaker and was left without shaking for 2 h before RNA extraction. Total RNA was extracted as described previously (12). mRNA was isolated by using a PolyAttract mRNA isolation kit (Promega). Five micrograms of mRNA was used for the synthesis of cDNA, using a Stratagene ZAP Express cDNA synthesis kit (the vector used was UniZap XR-Amp). After ligation, 1 μl of the 5-μl ligation mix was used for packaging in Stratagene's Gigapack III Gold packaging extract. The titer determination was done by infecting XL1-Blue MRF′ cells and counting the PFU. The library had a total 4 × 106 PFU. For determining the size of the insert, single clones from the primary library were excised by using XL1-Blue MRF′ cells and Ex-Assist helper phage, and the excised phagemids were transfected to XLOLR cells. The plasmid was extracted, digested with EcoRI and XhoI, and gel separated, and the average insert size was calculated to be slightly more than 1 kb. The primary library was amplified in XL1-Blue MRF′ cells and stored in 7% dimethyl sulfoxide at −80°C. The titer in the amplified library was 106 PFU/μl, and total volume was about 250 ml.
For making the cosmid library, high-molecular-weight (>200-kb) genomic DNA was isolated from T. virens mycelia and partially digested with MboI. Ten micrograms of the partially digested DNA was dephosphorylated with calf intestinal alkaline phosphatase. Approximately 5 μg of the cosmid vector pAN26 (procured from the Fungal Genetics Stock Center), which has two cos sites, was first digested with HindIII, dephosphorylated in a 200-μl volume (as described above), and again digested with BamHI. A 2.5-μg quantity of partially digested and dephosphorylated genomic DNA was ligated to 1 μg of the vector DNA at 16°C, in a 20-μl total volume, overnight. Four microliters of the ligation mix was packaged into Gigapack III XL packaging extract (Stratagene). The titer determination was done by transfecting a part of the library into XL1-Blue MRF′ cells, and the titer was found to be about 150,000 per packaging. The average insert size was more than 40 kb. The library was amplified and stored at −80°C.
Biolistic and protoplast transformation.
T. virens was transformed by using both particle gun bombardment (biolistic transformation) and polyethylene glycol-mediated transformation of the protoplast. Biolistic transformation was done with tungsten M17 particles (Bio-Rad) at 650 or 900 lb/in2 as described previously (19). Protoplast transformation was done essentially as described previously (20). The protoplasts were released from germinating conidia by 2 to 2.5 h of digestion at 30°C with gentle shaking (70 rpm, rotary shaker) in an enzyme mixture which consisted of 0.2 g of β-d-glucanase (InterSpex, San Mateo, Calif.), 0.4 g of Driselase (InterSpex), and approximately 2 mg of chitinase (Sigma C6137); stirred for 5 min at room temperature in 70 ml of 0.7 M NaCl; centrifuged for 10 min at 10,000 rpm (Sorvall SS34), and sterile filtered (0.22-μm-pore-size filter). Protoplasts were counted, and mycelial fragments were removed by filtering through three layers of sterile gauze and washed in STC (sorbitol, 1.2 M; Tris [pH 7.5], 10 mM; CaCl2, 50 mM) before transformation of about 108 protoplasts with 20 to 30 μg of plasmid DNA or linearized fragments for double-crossover integration, as described previously (20). Selection was on 200 mg of hygromycin B (Calbiochem) per liter (final concentration). Routinely, the transformants were maintained on PDA with 50 mg of hygromycin B per liter.
Growth rate, confrontation assay, coiling, and sclerotial parasitism.
The growth rate of the mutants vis-à-vis the wild type was determined by inoculating 5-mm-diameter mycelial disks of the fungi at the center of PDA plates and recording the colony diameter every 24 h. For biomass production, the fungi were grown in 100 ml of potato dextrose broth at room temperature with constant shaking at 150 rpm for 48 h, and the dry weight was recorded. The confrontation assay to assess the ability of T. virens to overgrow S. rolfsii and R. solani was done as described previously (28). The ability of wild-type T. virens and mutants to coil around R. solani hyphae was studied on 1.5% agar plates, where the interacting fungi were inoculated opposite each other and the interaction was studied under a microscope at the zone of interaction after staining with cotton blue in lactophenol. The ability to parasitize the sclerotia of R. solani and S. rolfsii in soil was assessed by using a soil plate assay as described previously (28) except that autoclaved soil was used in order to avoid interference from the native Trichoderma population and 109 conidia were added to each plate.
Photoinduction.
Mycelial colonies were grown and photoinduced as described previously (31). Cultures were inoculated in 9-cm-diameter petri dishes containing 3 ml of potato dextrose-yeast-casein hydrolysate medium, at the center of an 8-cm-diameter disk of Whatman no. 50 hardened filter paper overlying Whatman no. 1 filter paper. At the indicated times, the cultures were exposed to 480 μmol of blue light m−2 and were fixed in ethanol 24 h later. Photoinduced conidiation was measured essentially as described previously for T. atroviride (10) except that cultures were not fixed in ethanol; conidia were suspended in 5 ml of water by using a cell spreader, and the optical density (scatter) was measured at 600 nm and calibrated by counting in a hemocytometer. The optical density of a similar extraction of a colony with no conidia was subtracted to correct for background due to mycelial fragments. Curves were fit to an exponential model with the SIMPLEX algorithm (4) and plotted with Microsoft Excel.
Nucleotide sequence accession number.
The tmkA gene sequence has been deposited in the GenBank database under accession number AY141978.
RESULTS
Isolation of TmkA.
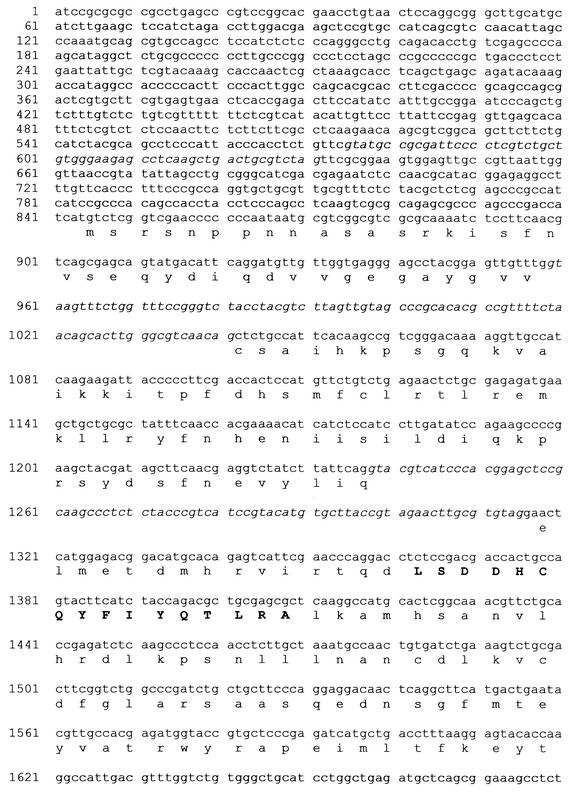
Degenerate primers designed according to conserved MAPK signatures (35) were used to obtain a 383-bp product, which was cloned and used to screen cDNA and cosmid libraries. A full-length cDNA clone named pD111 and a 3,243-bp BamHI fragment from a 45-kb cosmid clone (pMPCS2) were obtained. The organization of the gene and its features are shown in Fig. 1. The tmkA gene is very similar to its close fungal homologs of the YERK1 class. For example, another member of the genus, T. atroviride, has a member of this class (Tmk1 gene; accession number AF452096) showing 91% nucleotide homology to tmkA, after removing the introns from both sequences before comparison. At the amino acid level, the identity is 98.6%. Several of the few differences between Tmk1 and TmkA are located in the N terminus: MSRSNPPNN in T. virens and MSRSTAPN in T. atroviride. A consensus signature (noted in Fig. 1) for the YERK1 subfamily (16) is identical to that of C. heterostrophus Chk1 and differs by only one amino acid from that of M. grisea Pmk1. The 5′ untranslated region contains an intron 268 bp upstream of the ATG. The coding sequence is interrupted by two introns. These are conserved in the T. atroviride Tmk1 gene. Despite the 91% homology of the coding sequence, the third intron in the T. atroviride sequence is absent from the tmkA gene. In the upstream region, there are no obvious canonical TATA or CAAT boxes, as is common in fungal genes, although there is a TATA-like sequence located 174 bp upstream of the ATG. A putative polyadenylation signal, AATACA, is found 26 bp upstream of the polyadenylation site (Fig. 1).
FIG. 1.
Features of tmkA. The conserved signature of the YERK1 fungal MAPK class (16) is indicated in boldface in the amino acid sequence. Introns, located by sequencing of cDNA, are shown in italic. The polyadenylation site is indicated by an arrow, and a putative polyadenylation signal is underlined.
Isolation of TmkA loss-of-function mutants.
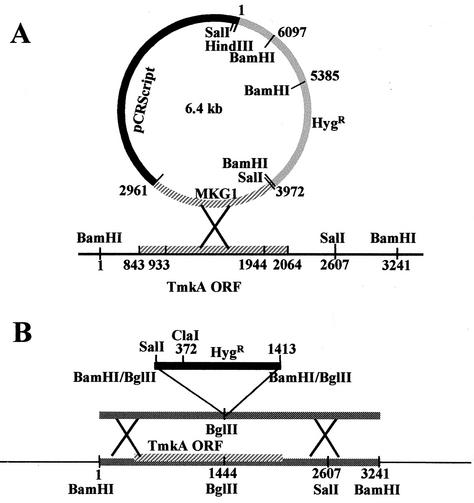
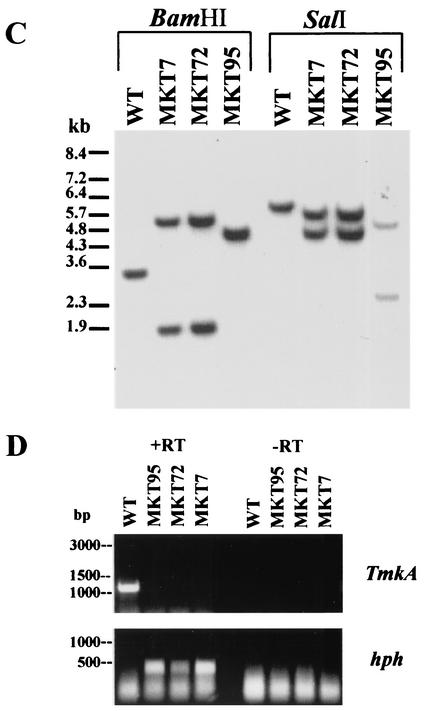
To determine the function of this gene, loss-of-function mutants were obtained by homologous integration through single-crossover and double-crossover events (Fig. 2). The single-crossover construct was made by amplifying and cloning a 1,011-bp PCR product (pMKG1) from the genomic DNA by using primer pair MKS (GGTGAGGGAGCCTACG) and MKAS (GGCTCGAGGTAGGGGTGC), which amplifies the coding region including two introns and excludes 30 amino acids from the 5′ end and 40 amino acids from the 3′ end. The product was ligated to the hygromycin resistance cassette (TrpC promoter, hph, and TrpC terminator). The double-crossover construct was made by inserting a BamHI fragment (TrpC promoter and hph) into the BglII site of the 3,243-bp fragment from the cosmid clone; this places the marker gene inside the coding region with a 1,446-bp left flank and 1,797-bp right flank. We obtained three transformants having homologous integration events, out of about 100 transformants screened, from each of single-crossover biolistic, single-crossover protoplast, and double-crossover protoplast transformations. Mutant MKT7 was obtained by single-crossover recombination through biolistic transformation, MKT72 was obtained through single-crossover protoplast transformation, and MKT95 was obtained through double-crossover protoplast transformation. Transformants were purified by single-spore isolation, and the homologous recombination was confirmed by Southern hybridization with the 1,011-bp MKG fragment as a probe (Fig. 2C). The TmkA transcript did not give a clear signal by Northern hybridization but was detected by RT-PCR in the wild type and was absent from all three mutants (Fig. 2D). The hygromycin phosphotransferase transcript was detected in the mutants but not in the wild type. All of the ectopic transformants were unstable, losing the selective marker if not grown on hygromycin. Likewise, we have observed previously (P. K. Mukherjee and B. A. Horwitz, unpublished data) that intensely fluorescent transformants carrying gfp introduced by transformation with circular DNA lose their fluorescence upon growth on nonselective medium. Presumably it is lost by a reversal of the single-crossover integration event. All of the homologous recombinants, in contrast, remained stable even after repeated subculture on PDA without hygromycin. Most of the ectopic transformants, however, were stable, retaining resistance and continuing to grow if maintained on hygromycin.
FIG. 2.
Disruption of tmkA by homologous integration. (A) Strategy for single-crossover integration. Map positions are labeled in base pairs, starting with the first BamHI site. (B) Strategy for double-crossover integration. Map positions are labeled in base pairs, starting with the first BamHI site. (C) Southern analysis of transformants. Genomic DNA was digested with the indicated enzymes, and the blot was probed with the fragment labeled MKG1 in panel A. WT, wild type. (D) RT-PCR analysis of transformants. No products were detected when RT was omitted from the reactions (right lanes).
Growth, morphology and conidiation.
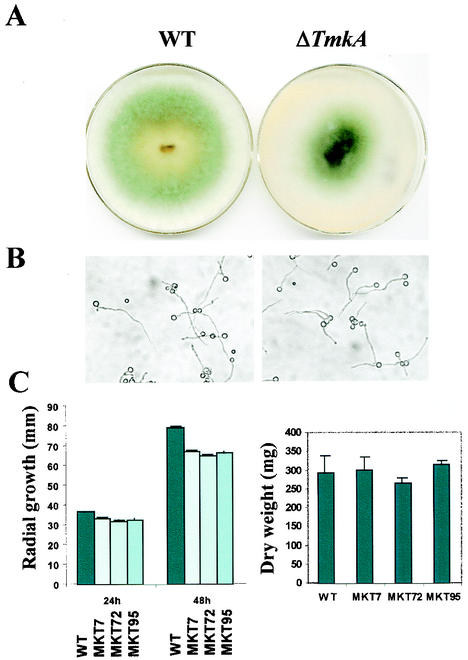
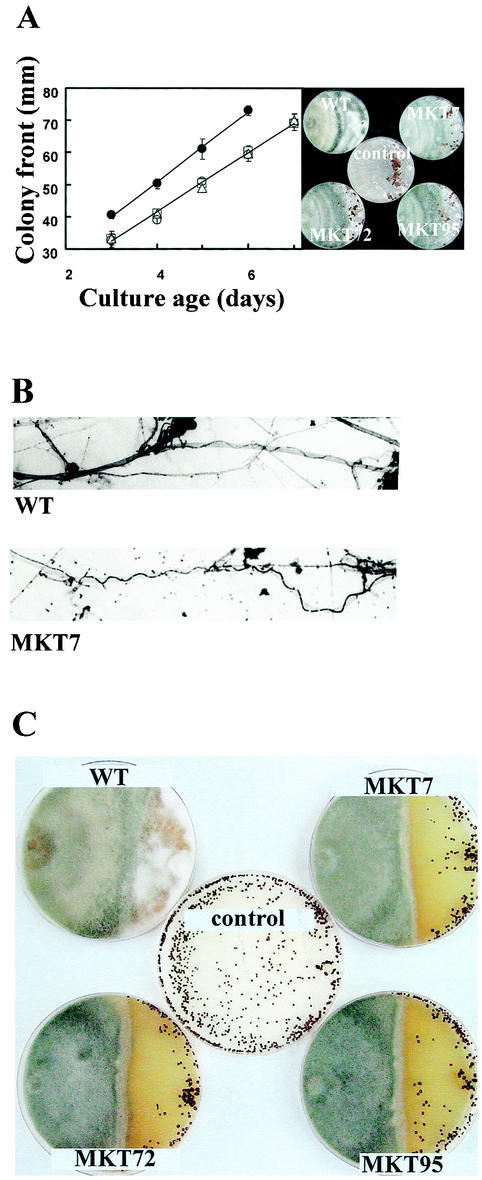
The radial growth rate of the tmkA mutant lines was reduced by about 16%. Colonies of the mutants began producing dense growth and sporulating at the center, unlike the wild type, which begins to sporulate towards the colony periphery (Fig. 3). Conidia of the mutants appeared to be normal, and there was no significant delay or decrease in germination (Fig. 3). Due to early aerial growth, the colony surfaces of the mutants were convex, while wild-type colonies were concave. The rates of increase in biomass in liquid shake culture were identical, within variability, in the wild type and mutants (Fig. 3).
FIG. 3.
Development, colony morphology, and growth of the wild type (WT) and tmkA mutants. (A) Colonies were grown for 3 days on PDA medium under continuous light at 30°C. (B) Appearance of germinating conidia at 8 h after inoculation on glass slides in liquid potato dextrose medium. (C) Growth rate (left panel), as measured by colony radius at 24 and 48 h, and biomass (right panel), measured after 48 h of growth in shake culture. Error bars indicate standard errors.
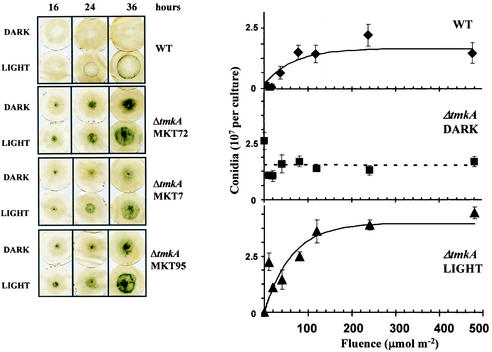
In total darkness, wild-type colonies showed, at most, a very faint ring of conidiophore induction at about 24 h of growth from the inoculum. A brief exposure to blue light induced conidiation in a ring located where the colony perimeter was at the time of photoinduction. In contrast, colonies of the mutants began to sporulate almost concurrently with their growth (Fig. 4). In addition to their sporulation in the dark, the tmkA mutants were able to respond to light, forming a defined ring where the perimeter was located at the time of photoinduction. This light-induced ring is clearly visible against the background of diffuse light-independent sporulation (Fig. 4). The extent of light-independent conidiation was not affected by the fluence of the light pulse, as expected, since its induction took place before the colony received light (Fig. 4). The sensitivities to light of the mutants and the wild type were identical within the variability, although the extent of conidiation was about twofold higher in the mutant (Fig. 4).
FIG. 4.
Photoinduction of conidiation. The wild type (WT) and three tmkA loss-of-function mutants were grown in total darkness. At the indicated times, the cultures were exposed to 480 μmol of blue light m−2 and fixed in ethanol 24 h later (left panels). Fluence response curves were obtained for photoinduction for the wild type and the tmkA mutant MKT95 (right panels). For the mutant, the central region and light-induced ring were analyzed separately; each region was cut from the filter paper-supported cultures before spore extraction. The amounts of light required to reach 63% (1 − 1/e) saturation, calculated from the curve fits, are k = 66.2 and 62.0 μmol m−2 for the wild type and MKT95, respectively. In the middle panel, the sporulation in the central region of tmkA mutant colonies (see photographs at the left) was monitored. Error bars indicate standard errors from five replicate cultures.
Antagonistic properties. (i) Confrontation and coiling.
When T. virens, which is a hyperparasite on R. solani (28), was coinoculated with this fungus by placing starters opposite to each other on PDA plates, both the wild type and tmkA mutants could steadily grow over the R. solani colony. Overgrowth is visible on photographs of confrontation plates by both the green color (Trichoderma sporulation) and the disappearance of the normal morphology of the host colony (Fig. 5A). The progression of overgrowth can be seen in the right panel of Fig. 5A, in which the mutants can be seen overgrowing the host, although with some delay compared to the wild type. The wild type completely overgrew the R. solani colony in 6 days, although the mutants took 24 to 36 h more (Fig. 5A). The wild type and the mutants could both colonize the sclerotia in confrontation plates, although the extent of growth on the sclerotia was more with the wild type than with the mutants (Fig. 5B). On water agar plates, all of the mutants as well as the wild type coiled around and lysed R. solani hyphae in 4 days after coinoculation, and we could not see any difference in the extents of coiling (Fig. 5B).
FIG. 5.
Confrontation assays of T. virens and host plant-pathogenic fungi. The host was inoculated at the right side of the plate. Controls, host only. WT, wild type. (A) R. solani versus T. virens. Error bars indicate standard errors for three replicates. Right, typical confrontation plates. Dark structures are the sclerotia of the host. (B) Mycoparasitic coiling of wild-type T. virens and a tmkA mutant on R. solani hyphae 5 days after inoculation. The large-diameter hyphae belong tothe host, while the thinner, dark-stained ones are the mycoparasite, which grows in a wavy or coiling pattern in contact with the host. (C) S. rolfsii versus T. virens, 14 days after inoculation. The dark, roughly spherical structures are the sclerotia of the host. Note that sclerotia are not visible in the wild-type T. virens-S. rolfsii interaction.
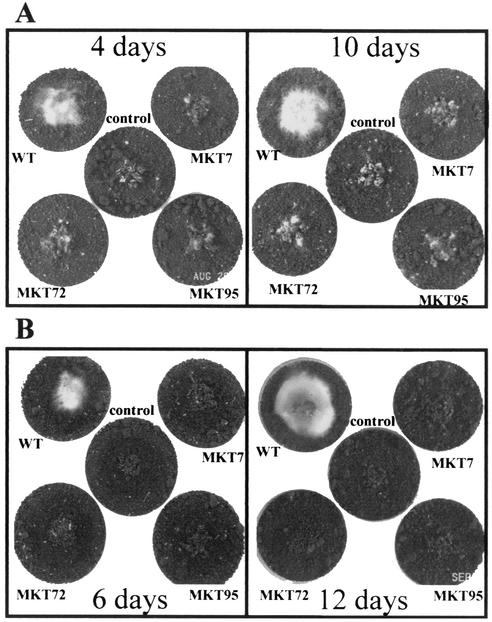
In confrontation assays, T. virens does not coil around S. rolfsii hyphae, but it can grow over and lyse aged mycelia as well as the sclerotia of S. rolfsii (28). In the present study, T. virens could not grow over the S. rolfsii colony until 9 days, after which T. virens growth over the S. rolfsii colony could be seen. After 12 days, there was profuse growth of wild-type T. virens on the S. rolfsii colony, in particular on the areas where sclerotia were formed (Fig. 5C). However, even 21 days after coinoculation, this characteristic dense growth of T. virens on the mycelia and sclerotia of S. rolfsii was not seen with the mutants, although a thin film of Trichoderma growth on the S. rolfsii colony could be seen upon close observation of the plates shown in Fig. 5B. When the sclerotia of R. solani or S. rolfsii were seeded on soil containing the conidia of T. virens, the wild type readily colonized the sclerotia of the pathogens. Growth of wild-type T. virens was visible on R. solani sclerotia 2 days after inoculation, and profuse growth was visible in 3 days. Although the mutants also showed some growth on the sclerotia, they failed to completely colonize the sclerotia even after prolonged incubation (Fig. 6A). The wild-type T. virens started to colonize the sclerotia of S. rolfsii 4 days after inoculation, and by day 6, it produced extensive growth on the sclerotia. Although some superficial growth of the mutants could be seen on the sclerotia (perhaps on sclerotial exudates), the mutants were ineffective in parasitizing the sclerotia of S. rolfsii (Fig. 6B). The sclerotia colonized by the wild type were reduced to a pulpy mass in 6 days, but they remained intact, hard, and viable in plates inoculated with the mutants even after 12 days of incubation.
FIG. 6.
Parasitism of sclerotia by T. virens. The sclerotia were put in the middle of a soil plate (see Materials and Methods) after mixing conidia into the soil. Center plates, sclerotia only, with no Trichoderma conidia. Plates were photographed after the indicated number of days of incubation. (A) Growth on R. solani sclerotia. (B) Growth on S. rolfsii sclerotia.
(ii) Cellulase and chitinase production.
Both the wild type and mutants could utilize chitin and cellulose as sole sources of carbon and could completely clear the medium. However, while the wild-type isolate cleared the chitin medium in 3 days, the mutants took 5 days to do so. On cellulose, the wild type could completely clear in 4 days, while it took 5 days for the mutants to clarify the suspension (data not shown). The growth (biomass) of the mutants on chitin and cellulose was lower by 13 and 15%, respectively, than that of the wild type.
DISCUSSION
In other filamentous fungi studied so far, MAPK genes are either dispensable for conidiation or required for conidiation (for a summary of phenotypes, see reference 36). TmkA of T. virens, in contrast, seems to function to repress conidiation in the dark; loss of TmkA allows T. virens to conidiate in the dark. Conidiation is otherwise inducible by light or nutrient stresses in the genus Trichoderma. The TmkA MAPK pathway may carry a constitutive signal for vegetative hyphal proliferation, as has been postulated for some fungal heterotrimeric G proteins. In Aspergillus, a G-protein (with a Gα subunit belonging to the fungal Gi class) signal transduction pathway controls the balance between vegetative growth and sporulation and secondary product biosynthesis (33). Loss of the Neurospora G-protein α subunit Gna3 results in constitutive sporulation (13), while activation of Gna1 promotes hyphal proliferation (37). In T. atroviride, loss of the Tga1 Gα subunit caused very slow growth accompanied by intense sporulation (31). Different G-protein α subunit genes apparently take over the function of suppressing conidiation in different species. Here, we have shown that a MAPK, TmkA, has this function in T. virens, except that its deletion does not cause any drastic change in colony morphology and growth rate. In Neurospora, loss of a MAPKKK homologous to yeast STE11 promotes conidiation, although it causes additional developmental and growth defects (15). One could thus infer that there is a particular Neurospora MAPK, acting downstream from the MAPKKK Nrc-2, deletion of which would also promote conidiation as we have found here. We predict that mutants with mutations in that MAPK gene would sporulate constitutively but grow normally. TmkA does not carry the signal for photoinduction, as light still induces conidiation in the mutants, and the sensitivity to light is not altered by loss of TmkA (Fig. 4). An additional signal repressing conidiation is probably removed by light, or light may determine the time at which conidiation is initiated. The reduction in radial growth by 16%, without a reduction in biomass (Fig. 3), may result from diversion of resources to sporulation during growth on solid medium. This reduction in growth is not drastic and certainly does not compare with the very restricted growth of T. atroviride mutants lacking the Gαi homolog (31). Since TmkA negatively regulates conidiation and is involved in biocontrol, overexpression (or a constitutively active TmkA allele) might improve biocontrol potential through prolonging vegetative growth. Prolonging vegetative growth would increase the time of active interaction with the host. At the same time, this might improve antagonistic potential through enhanced sclerotial parasitism. It is likely, though, that manipulation of more than one gene will be needed to optimize biocontrol strains.
The reduction of virulence in MAPK loss-of-function mutants of fungal pathogens of plants and animals (17, 36) prompted us to study mycoparasitism of T. virens tmkA mutants. The mutants could overgrow R. solani and also showed hyperparasitic coiling around the hyphae of R. solani similar to that of the wild type (Fig. 5B). The host hyphae appear to be lysed, as they no longer stained with cotton blue; the rapid progression of overgrowth also indicates that the mutants can efficiently obtain their nutrients from the host. TmkA thus is not involved in hyphal parasitism of R. solani. This is in contrast to the case for all other plant pathogens studied, where the homologous MAPK is involved in penetration and/or invasive growth on host plants. The somewhat slower growth of the MAPK mutants in the confrontation assay could result from the slower radial growth rate of the mutants (Fig. 3). T. virens is not a mycoparasite on rapidly growing S. rolfsii hyphae but can overgrow an aging colony and parasitize the sclerotia (28). In the present study wild-type T. virens also could eventually overgrow the colony of S. rolfsii, with prolific growth on the sclerotia, whereas the MAPK mutants failed to produce dense growth and colonize the sclerotia in the confrontation assay, although they formed a thin growth on the S. rolfsii colony. This cannot be due simply to the somewhat decreased growth, since the mutants were able to overgrow R. solani. The tmkA mutants have therefore lost their biocontrol potential (“virulence”) in a host-specific manner.
Parasitism of sclerotia was proposed earlier as the principal mechanism of biocontrol of R. solani and S. rolfsii by T. virens, and a soil plate assay was proposed to assess the biocontrol potential of Trichoderma spp. on these pathogens (28). Using this technique, we have studied the role of TmkA in the mycoparasitism of sclerotia of R. solani and S. rolfsii by T. virens. The results clearly indicated the involvement of TmkA in this process, as the tmkA mutants were either less effective or ineffective in colonization of these resting structures in soil. Both in the confrontation assay and in the soil plate assay, tmkA mutants were less effective in parasitism of sclerotia of R. solani and were ineffective against the sclerotia of S. rolfsii. It is thus apparent that the mutants would be less effective or attenuated in biocontrol potential against these pathogens. It is interesting that the tmkA loss-of-function mutants are as effective as the wild type in mycoparasitic coiling around R. solani hyphae. It therefore appears that the YERK1 MAPK TmkA may not be involved in recognition of the host hyphae leading to mycoparasitic coiling and lysis; rather, it might be involved in activation of transcription factors for genes encoding one or more enzymes responsible for the degradation of the host.
Production of hydrolytic enzymes is part of the mycoparasitic process; Trichoderma spp. kill the host fungi through the production of cell wall-lysing enzymes such as cellulases, chitinases, and β-d-glucanases (see, for example, reference 14). A study of the hydrolytic enzymes (14) of T. virens in the wild type and tmkA mutants may uncover a particular enzyme or set of enzymes that is (i) induced downstream of TmkA and (ii) important for colonization of S. rolfsii and R. solani, particularly sclerotia. Pectinolytic enzymes related to virulence are regulated by MAPK pathways in yeast (22) and Fusarium (6).
In Trichoderma, conidiation is inducible, and the fungus does not sporulate in the dark provided that the nutrient supply is sufficient. We predict that there is a conidiation-repressing regulatory factor which is a MAPK target. Phosphorylation by MAPK can provide negative regulation (see, for example reference 23). The role of this regulatory factor might have diverged from that in foliar plant pathogens, where members of the YERK1 class have not been found to suppress conidiation. Another possibility is that different transcription factors will be found to be unique to different types of fungal developmental cycles yet activated by the same conserved MAPKs. Suppression of conidiation by TmkA suggests that this pathway has a basal level of activation in T. virens grown in light or darkness, transducing an as-yet-unidentified signal.
Acknowledgments
A visit by P.K.M. to the lab of B.K.H. in 1999 to 2000 was made possible by a BOYSCAST Visiting Scientist fellowship from the Department of Science and Technology, Government of India. This study was supported in part by grant 233/00-2 from the Israel Academy of Sciences and by internal funding from the Department of Atomic Energy, Government of India.
We thank the head of the Nuclear Agriculture and Biotechnology Division, Bhabha Atomic Research Centre, Mumbai, India, for his encouragement and support.
REFERENCES
- 1.Baek, J. M., C. R. Howell, and C. M. Kenerley. 1999. The role of an extracellular chitinase from Trichoderma virens Gv29-8 in the biocontrol of Rhizoctonia solani. Curr. Genet. 35:41-50. [DOI] [PubMed] [Google Scholar]
- 2.Banuett, F., and I. Herskowitz. 1994. Identification of fuz7, a Ustilago maydis MEK/MAPKK homolog required for a-locus-dependent and -independent steps in the fungal life cycle. Genes Dev. 8:1367-1378. [DOI] [PubMed] [Google Scholar]
- 3.Betina, V. 1984. Photoinduced conidiation in Trichoderma viride. Int. J. Microbiol. 2:55-68. [Google Scholar]
- 4.Caceci, M. S., and W. P. Cacheris. 1984. Fitting curves to data. Byte May 1984:340-362. [Google Scholar]
- 5.Cortes, C., A. Gutierrez, V. Olmedo, J. Inbar, I. Chet, and A. Herrera-Estrella. 1998. The expression of genes involved in parasitism by Trichoderma is triggered by a diffusible factor. Mol. Gen. Genet. 260:218-225. [DOI] [PubMed] [Google Scholar]
- 6.Di Pietro, A., F. I. Garcia-MacEira, E. Meglecz, and M. I. Roncero. 2001. A MAP kinase of the vascular wilt fungus Fusarium oxysporum is essential for root penetration and pathogenesis. Mol. Microbiol. 39:1140-1152. [PubMed] [Google Scholar]
- 7.Elad, Y., I. Chet, P. Boyle, and Y. Henis. 1983. Parasitism of Trichoderma spp. on Rhizoctonia solani and Sclerotium rolfsii—scanning electron microscopy and fluorescence microscopy. Phytopathology 73:85-88. [Google Scholar]
- 8.Gustin, M. C., J. Albertyn, M. Alexander, and K. Davenport. 1998. Mitogen-activated protein kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62:1264-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herrera-Estrella, A., and I. Chet. 1998. Biocontrol of bacteria and phytopathogenic fungi, p. 263-282. In A. Altman (ed.), Agricultural bio/technology. Marcel Dekker, Inc., New York, N.Y.
- 10.Horwitz, B. A., J. Gressel, S. Malkin, and B. L. Epel. 1985. Modified cryptochrome in vivo absorption in dim photosporulation mutants of Trichoderma. Proc. Natl. Acad. Sci. USA 82:2736-2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Inbar, J., and I. Chet. 1992. Biomimics of fungal cell-cell recognition by use of lectin-coated nylon fibers. J. Bacteriol. 174:1055-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones, J., P. Dunsmuir, and J. Bedbrook. 1985. High-level expression of induced chimeric genes in regenerated transformed plants. EMBO J. 4:2411-2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kays, A. M., P. S. Rowley, R. A. Baasiri, and K. A. Borkovich. 2000. Regulation of conidiation and adenylyl cyclase levels by the G alpha protein GNA-3 in Neurospora crassa. Mol. Cell. Biol. 20:7693-7705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim, D. J., J. M. Baek, P. Uribe, C. M. Kenerley, and D. R. Cook. 2002. Cloning and characterization of multiple glycosyl hydrolase genes from Trichoderma virens. Curr. Genet. 40:374-384. [DOI] [PubMed] [Google Scholar]
- 15.Kothe, G. O., and S. J. Free. 1998. The isolation and characterization of nrc-1 and nrc-2, two genes encoding protein kinases that control growth and development in Neurospora crassa. Genetics 149:117-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kultz, D. 1998. Phylogenetic and functional classification of mitogen- and stress-activated protein kinases. J. Mol. Evol. 46:571-588. [DOI] [PubMed] [Google Scholar]
- 17.Lengeler, K. B., R. C. Davidson, C. D'Souza, T. Harashima, W. C. Shen, P. Wang, X. Pan, M. Waugh, and J. Heitman. 2000. Signal transduction cascades regulating fungal development and virulence. Microbiol. Mol. Biol. Rev. 64:746-785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lev, S., A. Sharon, R. Hadar, H. Ma, and B. A. Horwitz. 1999. A mitogen-activated protein kinase of the corn leaf pathogen Cochliobolus heterostrophus is involved in conidiation, appressorium formation, and pathogenicity: diverse roles for mitogen-activated protein kinase homologs in foliar pathogens. Proc. Natl. Acad. Sci. USA 96:13542-13547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lorito, M., C. K. Hayes, A. Di Pietro, and G. E. Harman. 1993. Biolistic transformation of Trichoderma harzianum and Gliocladium virens using plasmid and genomic DNA. Curr. Genet. 24:349-356. [DOI] [PubMed] [Google Scholar]
- 20.Lu, S., L. Lyngholm, G. Yang, C. Bronson, O. C. Yoder, and B. G. Turgeon. 1994. Tagged mutations at the Tox1 locus of Cochliobolus heterostrophus by restriction enzyme-mediated integration. Proc. Natl. Acad. Sci. USA 91:12649-12653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mach, R. L., C. K. Peterbauer, K. Payer, S. Jaksits, S. L. Woo, S. Zeilinger, C. M. Kullnig, M. Lorito, and C. P. Kubicek. 1999. Expression of two major chitinase genes of Trichoderma atroviride (T. harzianum P1) is triggered by different regulatory signals. Appl. Environ. Microbiol. 65:1858-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madhani, H., T. Galitski, E. Lander, and G. Fink. 1999. Effectors of a developmental mitogen-activated protein kinase cascade revealed by expression signatures of signaling mutants. Proc. Natl. Acad. Sci. USA 96:12530-12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuoka, K., N. Kiyokawa, T. Taguchi, J. Matsui, T. Suzuki, K. Mimori, H. Nakajima, H. Takenouchi, T. Weiran, Y. U. Katagiri, and J. Fujimoto. 2002. Rum1, an inhibitor of cyclin-dependent kinase in fission yeast, is negatively regulated by mitogen-activated protein kinase-mediated phosphorylation at Ser and Thr residues. Eur. J. Biochem. 269:3511-3521. [DOI] [PubMed] [Google Scholar]
- 24.Mayorga, M. E., and S. E. Gold. 1999. A MAP kinase encoded by the ubc3 gene of Ustilago maydis is required for filamentous growth and full virulence. Mol. Microbiol. 34:485-497. [DOI] [PubMed] [Google Scholar]
- 25.Mey, G., B. Oeser, M. H. Lebrun, and P. Tudzynski. 2002. The biotrophic, non-appressorium-forming grass pathogen Claviceps purpurea needs a Fus3/Pmk1 homologous mitogen-activated protein kinase for colonization of rye ovarian tissue. Mol. Plant-Microbe Interact. 15:303-312. [DOI] [PubMed] [Google Scholar]
- 26.Mukherjee, P. K., S. M. Shreshtha, and A. N. Mukhopadhyay. 1993. Baiting with Sclerotium rolfsii for selective isolation of Gliocladium virens from natural soil. Biocontrol Sci. Technol. 3:101-104. [Google Scholar]
- 27.Mukherjee, P. K., P. Thomas, and K. Raghu. 1995. Shelf-life enhancement of fresh ginger rhizomes at ambient temperatures by combination of gamma-irradiation, biocontrol and closed polyethylene bag storage. Ann. Appl. Biol. 127:375-384. [Google Scholar]
- 28.Mukherjee, P. K., A. N. Mukhopadhyay, D. K. Sarmah, and S. M. Shreshtha. 1995. Comparative antagonistic properties of Gliocladium virens and Trichoderma harzianum on Sclerotium rolfsii and Rhizoctonia solani—its relevance to understanding the mechanisms of biocontrol. J. Phytopathol. 143:275-279. [Google Scholar]
- 29.Muller, P., C. Aichinger, M. Feldbrugge, and R. Kahmann. 1999. The MAP kinase kpp2 regulates mating and pathogenic development in Ustilago maydis. Mol. Microbiol. 34:1007-1017. [DOI] [PubMed] [Google Scholar]
- 30.Nishida, E., and Y. Gotoh. 1993. The MAP kinase cascade is essential for diverse signal transduction pathways. Trends Biochem. Sci. 18:128-131. [DOI] [PubMed] [Google Scholar]
- 31.Rocha-Ramirez, V., C. Omero, I. Chet, B. A. Horwitz, and A. Herrera-Estrella. 2002. Trichoderma atroviride G-protein alpha-subunit gene tga1 is involved in mycoparasitic coiling and conidiation. Eukaryot. Cell 1:594-605. [DOI] [PMC free article] [PubMed]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 33.Shimizu, K., and N. P. Keller. 2001. Genetic involvement of a cAMP-dependent protein kinase in a G protein signaling pathway regulating morphological and chemical transitions in Aspergillus nidulans. Genetics 157:591-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takano, Y., T. Kikuchi, Y. Kubo, J. Hamer, K. Mise, and I. Furusawa. 2000. The Colletotrichum lagenarium MAP kinase gene CMK1 regulates diverse aspects of fungal pathogenesis. Mol. Plant-Microbe Interact. 13:373-383. [DOI] [PubMed] [Google Scholar]
- 35.Xu, J., and J. Hamer. 1996. MAP kinase and cAMP signaling regulate infection structure formation and pathogenic growth in the rice blast fungus Magnaporthe grisea. Genes Dev 10:2696-2706. [DOI] [PubMed] [Google Scholar]
- 36.Xu, J. R. 2000. MAP kinases in fungal pathogens. Fungal Genet. Biol. 31:137-152. [DOI] [PubMed] [Google Scholar]
- 37.Yang, Q., and K. A. Borkovich. 1999. Mutational activation of a Galphai causes uncontrolled proliferation of aerial hyphae and increased sensitivity to heat and oxidative stress in Neurospora crassa. Genetics 151:107-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeilinger, S., C. Galhaup, K. Payer, S. L. Woo, R. L. Mach, C. Fekete, M. Lorito, and C. P. Kubicek. 1999. Chitinase gene expression during mycoparasitic interaction of Trichoderma harzianum with its host. Fungal Genet. Biol. 26:131-140. [DOI] [PubMed] [Google Scholar]
- 39.Zheng, L., M. Campbell, J. Murphy, S. Lam, and J. R. Xu. 2000. The BMP1 gene is essential for pathogenicity in the gray mold fungus Botrytis cinerea. Mol. Plant-Microbe Interact. 13:724-732. [DOI] [PubMed] [Google Scholar]