Abstract
In eukaryotes, RNA polymerase (pol) I exclusively transcribes the large rRNA gene unit (rDNA) and mRNA is synthesized by RNA pol II. The African trypanosome, Trypanosoma brucei, represents an exception to this rule. In this organism, transcription of genes encoding the variant surface glycoprotein (VSG) and the procyclins is resistant to α-amanitin, indicating that it is mediated by RNA pol I, while other protein-coding genes are transcribed by RNA pol II. To obtain firm proof for this concept, we generated a T. brucei cell line which exclusively expresses protein C epitope-tagged RNA pol I. Using an anti-protein C immunoaffinity matrix, we specifically depleted RNA pol I from transcriptionally active cell extracts. The depletion of RNA pol I impaired in vitro transcription initiated at the rDNA promoter, the GPEET procyclin gene promoter, and a VSG gene expression site promoter but did not affect transcription from the spliced leader (SL) RNA gene promoter. Fittingly, induction of RNA interference against the RNA pol I largest subunit in insect-form trypanosomes significantly reduced the relative transcriptional efficiency of rDNA, procyclin genes, and VSG expression sites in vivo whereas that of SL RNA, αβ-tubulin, and heat shock protein 70 genes was not affected. Our studies unequivocally show that T. brucei harbors a multifunctional RNA pol I which, in addition to transcribing rDNA, transcribes procyclin genes and VSG gene expression sites.
African trypanosomes are unicellular eukaryotic parasites causing sleeping sickness in humans and related diseases in livestock. Trypanosomiasis is endemic to large parts of tropical Africa and in generally fatal if left untreated. The African trypanosome, Trypanosoma brucei, has a biphasic life cycle, which alternates between a mammalian host and its transmitting insect vector, the tsetse fly. The bloodstream form of T. brucei, living in the mammalian bloodstream, is covered with a dense surface coat made up of identical glycoproteins known as variant surface glycoproteins (VSG). By undergoing antigenic variation of the VSG coat, the parasite escapes immune attack by the host (3, 4, 56). When the parasite is ingested by a tsetse fly, it differentiates into the procyclic or insect form and its VSG coat is shed and replaced by a coat of procyclin or procyclic acidic repetitive protein. Both surface coats appear to be important for efficient growth of trypanosomes (37; V. Carruthers and G. A. M. Cross, personal communication; E. Vassella, P. Butikofer, J. Jelk, M. Engstler, T. W. Pearson, and I. Roditi, Mol. Parasitol. Meet. XIII, abstr. 13B, 2002).
Transcription of protein-coding genes and mRNA maturation in trypanosomes and other kinetoplastids is characterized by mechanisms that are unusual compared to transcription mechanisms in most other eukaryotes. These mechanisms include polycistronic transcription of clustered genes, trans splicing of the common 5′ capped 39-nucleotide noncoding spliced leader (SL; in trypanosomes, this is also referred to as the miniexon) onto the main coding exon of every mRNA, and posttranscriptional control of the steady-state mRNA level. A potential unique regulatory mechanism of T. brucei is that RNA polymerase (pol) I may transcribe a specific subset of protein-coding genes (10, 28). In other eukaryotes, mRNA is synthesized exclusively by RNA pol II because mRNA capping, an essential step in pre-mRNA maturation, occurs cotranscriptionally and the capping enzyme interacts with the C terminus of the RNA pol II largest subunit (8, 31, 59). In trypanosomes, most individual mRNAs are processed from large precursors by trans splicing and polyadenylation and hence can obtain their caps posttranscriptionally through the addition of the capped SL, derived from the SL RNA. trans splicing thus uncouples capping and RNA pol II-mediated transcription and hence makes it possible for other RNA polymerases to synthesize mRNA.
Evidence that T. brucei utilizes RNA pol I for transcription of a subset of endogenous protein-coding genes was obtained by nuclear run-on experiments. Comparison of the α-amanitin sensitivity of transcription revealed that VSG and procyclin genes were transcribed by an α-amanitin-resistant RNA pol while most other protein-coding genes, such as the hsp70 and αβ-tubulin genes, were transcribed by a conventional α-amanitin-sensitive RNA pol II (12, 21, 38, 39). This observation raised the possibility that VSG gene expression sites (VSG ESs) and the procyclin gene loci are transcribed by an RNA pol I-like enzyme rather than by RNA pol II. Several independent lines of evidence support this notion. First, the rDNA promoter can efficiently direct reporter gene expression in transformed trypanosomes (40, 60). Second, the hypothesis that α-amanitin-resistant transcription from procyclin gene and VSG ES promoters may be mediated by one of the two RNA pol II largest subunit sets present in T. brucei was excluded by a study of RNA pol II mutant cell lines. There are two alleles of RNA pol II (pol II-A and pol II-B). Transcription of procyclin genes, of the region downstream of the VSG ES promoter, and of rRNA genes was still resistant to α-amanitin in RNA pol II-B allele double-knockout cell lines, while transcription of other protein-coding genes remained sensitive to the toxin and presumably was conducted by the RNA pol II-A polymerase (11). Third, procyclin and rDNA promoters have upstream control elements, which are functionally interchangeable (19). Fourth, transcription elongation on procyclin, VSG, and rRNA genes is equally resistant to the addition of α-amanitin and the detergent N-lauroylsarcosine (Sarkosyl) whereas transcription of other protein-coding genes is highly sensitive to these compounds (21, 23, 41). Fifth, in situ hybridization revealed that in bloodstream form trypanosomes, RNA pol I colocalizes with the actively transcribed VSG ES outside of the nucleolus in a discrete nuclear compartment called the expression site body (ESB) (32). Finally, transcription competition experiments indicated that procyclin, VSG, and rDNA promoters interact with common transcription factors in vitro (24). Similarly, introduction of a large number of episomal procyclin promoters into procyclic trypanosomes significantly reduced the transcriptional efficiency derived from the endogenous procyclin, VSG, and rDNA promoters, suggesting that the episomal promoters competed for common transcription factors in vivo (27). In contrast to this large amount of evidence, the requirement for Mg2+ and Mn2+ ions in nuclear run-on assays indicated that the RNA polymerase transcribing the VSG genes is RNA pol II and not RNA pol I (17). To provide direct evidence for RNA pol I-mediated transcription of procyclin genes and VSG ESs, we determined transcriptional efficiencies when the expression level of the RNA pol I largest subunit was affected both in vitro and in vivo.
We demonstrate that depletion of RNA pol I from cell extracts nearly abolished in vitro transcription initiated at rDNA, procyclin gene, and VSG ES promoters, while SL RNA gene promoter transcription remained unaffected. Similarly, down-regulation of the expression level of the RNA pol I largest subunit by RNA interference (RNAi) in procyclic trypanosomes drastically reduced the transcriptional efficiency of the procyclin and rRNA genes, while gene transcription by RNA pol II and RNA pol III was not significantly affected. In sum, these data unequivocally show that RNA pol I transcribes procyclin genes and VSG ESs in T. brucei.
MATERIALS AND METHODS
Trypanosomes.
The procyclic form of T. brucei stock 427-60, originally obtained from R. Brun, was maintained in SDM-79 medium at 25°C (6). Procyclic trypanosome strain 29-13 which harbors the T7 RNA polymerase gene and the tetracycline repressor gene was obtained from the laboratory of G. Cross and was used as the parental strain for transformation using tetracycline-inducible double-stranded RNA (dsRNA) expression constructs (58). For all transformations, procyclic trypanosomes at a cell density of 5 × 106 8 × 106 cells/ml were used. For induction of dsRNA, trypanosomes were cultured in medium containing 1.0 μg of tetracycline per ml. For measuring growth curves, trypanosomes were continuously maintained at concentrations between 2 × 106 and 1.5 × 107 cells/ml. If needed, the volume of each culture was expanded during growth and the total numbers of trypanosomes in expanded cultures were calculated for establishing growth curves.
Description of plasmid constructs.
The pI-ble-KO construct, which was used to delete the RNA pol I largest subunit (TbRPA1) gene, consists of sequences from 5′ to 3′, a ∼1-kb BamHI-Tth111I fragment of the 5′-flanking region of the TbRPA1 coding sequence, the phleomycin resistance gene, and the SalI-XbaI fragment of ∼1.2 kb from the 3′ end of the TbRPA1 gene. The pRPIC-hph construct, which was used to introduce a protein C tag at the end of the TbRPA1 gene, contains two sequence units in the same orientation. From 5′ to 3′, the first unit consists of the 3′-terminal 434 bp of the TbRPA1 coding sequence, the 36-bp long protein C epitope-coding sequence 5′-GAAGATCAGGTGGATCCTCGTCTTATTGATGGGAAA-3′, the stop codon, and 470 bp of the 3′-flanking region of TbRPA1; and the second unit consists of the intergenic region of hsp70 genes 2 and 3 (29), the hygromycin resistance gene (hph), and the βα-tubulin intergenic region (25). The SalI site located in the 3′ coding region of the TbRPA1 gene was used to linearize plasmid pRPCI-hph for transformation. Three TbRPA1 dsRNA-expressing constructs (pIds-2.2, pIds-0.8, and pIds-1.5) contain the 2.2-kb Mlu-HindIII fragment of the 5′ coding region of TbRPA1, the 0.8-kb HindIII-SalI fragment in the middle of the TbRPA1 coding region, and the 1.5-kb -SalI-HindIII fragment of the 3′ coding region of TbRPA1, respectively, which replaced the tubulin sequence located between the XhoI and HindIII sites in the original pZJM construct (57). Transcription of the TbRPA1-derived sequence is under the control of two opposing T7 promoter-tetracycline operators. Constructs pIds-2.2, pIds-0.8, and pIds-1.5 were linearized at the NotI site located at the rDNA derived sequence, and these plasmids are designed for integration into the nontranscribed spacer region of the rDNA locus.
Stable DNA transformation.
Ten- or 20-μg of linearized plasmid was electroporated into procyclic trypanosomes by using a BTX electroporator as previously described (38). At 36 h postelectroporation, hygromycin B (40 μg/ml), phleomycin (2 μg/ml), or G418 (20 μg/ml) was added to the cell cultures to select stably transformed trypanosomes. The individually transformed drug-resistant trypanosomes were cloned by limiting dilution using microtiter dishes and were supplemented with 2 × 106 to 5 × 106 wild-type trypanosomes per ml.
Southern genomic blot analysis.
Nuclear DNA was isolated from parasites. Following digestion with restriction endonucleases, the DNA was separated on a 0.8% agarose gel and transferred to nitrocellulose filters. The blots were hybridized with32P-labeled probes. The final posthybridization wash was performed in 0.1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% sodium dodecyl sulfate (SDS) at 65°C. All of the clonal trypanosome cell lines were analyzed by Southern blotting analysis to confirm that the correct integration or replacement event occurred in each cell line.
Nascent RNA analysis.
Nuclear preparation and nuclear run-on reactions were performed as described previously (38). Hybridizations were performed for 48 h. The final posthybridization wash was performed in 0.1× SSC-0.1% SDS at 65°C. Hybridization signals were quantified using a Molecular Dynamics PhosphorImager. Probes used for nuclear run-on assays are as follows. The HindIII-EcoRI fragment spanning the 5′ end of the 18S rRNA gene (rDNA) was described by Kooter and Borst (21). The αβ-tubulin gene probe (Tubulin), pTb α-β T-1 was described by Thomashow et al. (53). The VSG 118 ES promoter probe (VSG-P) contained a 1.3-kb HindIII-PstI fragment derived from VSG 118 ES promoter probe 1 as described by Gottesdiener et al. (16). The 5S rRNA gene probe was described by Lenardo et al. (30). Mini-exon gene clone pCL103 was described by Laird et. al. (22). The procyclin cDNA was described by Rudenko et al. (39). The 1.8-kb HindIII fragment spanning the hsp70 gene (Hsp) was as previously described (14). Purified inserts from each probe were loaded on the slot blots for hybridization with the synthesized nascent RNA.
RNA isolation and Northern blot analysis.
All RNA samples were isolated by guanidine thiocyanate lysis and purified by centrifugation through CsCl cushions. RNA samples were separated in 1% formaldehyde-agarose gels and transferred to nitrocellulose filters. Northern blots were hybridized with probes. Following hybridization, the filters were washed to a final stringency of 0.1× SSC-0.1% SDS at 65°C.
Trypanosome cell extract preparation and RNA pol I immunodepletion.
Cell extracts were prepared as described previously (23), except that the concentration of dithiothreitol was reduced from 2 to 0.5 mM and no EDTA was added. For immunodepletion of RNA pol I, anti-protein C affinity matrix (Roche) was equilibrated either in calcium ion-containing EC-100 buffer (150 mM sucrose, 20 mM HEPES-KOH [pH 7.7], 100 mM KCl, 20 mM potassium l-glutamate, 3 mM MgCl2, 1 mM CaCl2, 0.5 mM dithiothreitol, 10 μg of leupeptin per ml) or in EGTA-containing EE-100 buffer (same as buffer EC-100 but contains 0.5 mM EGTA instead of 1 mM CaCl2). Then 250 μl of cell extract was mixed with the same volume of buffer EC-100 or EE-100, mixed with 250 μl of pelleted and equilibrated beads, and rotated for 30 min at 4°C. Subsequently, the beads were pelleted by a 1-min centrifugation at 4°C and 6,000 × g. The supernatant was transferred to a second tube and again mixed with 250 μl of pelleted and equilibrated beads. After a second 30-min incubation on the rotator, the beads were pelleted and the supernatant was taken off and immediately used for in vitro transcription assays. For RNA pol I elution, the beads were washed in 12 volumes of buffer EC-100, mixed with 0.5 ml of elution buffer (150 mM sucrose, 20 mM HEPES-KOH [pH 7.7], 100 mM KCl, 20 mM potassium l-glutamate, 5 mM EGTA [pH 8.0], 5 mM EDTA [pH 8.0], 0.5 mM dithiothreitol, 10 μg of leupeptin per ml), and rotated for 15 min at 4°C. The elution step was repeated twice, and the eluate was pooled and concentrated on a Centricon 3 concentrator (Millipore).
Western blot analysis.
For Western blot analysis of TbRPA1 in cell extracts, 1 μl of undiluted cell extract (∼20 μg of protein), 2 μl of immunodepleted extracts, and a corresponding volume of RNA pol I eluate were separated on SDS-5% polyacrylamide gels and blotted onto polyvinylidene difluoride membranes (Roche). For the analysis of TbRPA1 in RNAi cell lines, total-cell extracts from 0.5 × 107 induced and uninduced cells were loaded in each lane. The blots were probed with monoclonal anti-protein C antibody HPC4 (diluted 1:1,000) (Roche), polyclonal anti-TbRPA1 antiserum (diluted 1:2,000) (44), and, in control reactions, polyclonal anti-40K antiserum, which is directed against the U2 snRNP-specific 40K protein of T. brucei (34), or anti-Tb-29 (26). Antibodies bound to polypeptides were detected with a peroxidase-labeled secondary antibody and the BM chemiluminescence blotting substrate (Roche).
In vitro transcription and RNA analysis.
In vitro transcription reactions including template constructs GPEET-trm, Rib-trm, VSG-trm, and SLins19, as well as RNA preparation and primer extension analysis, were carried out as described in detail previously (18, 23, 24). In short, 40-μl transcription reaction mixtures were incubated out for 60 min at 28°C and contained 8 μl of extract and 20 mM potassium l-glutamate, 20 mM KCl, 3 mM MgCl2, 20 mM HEPES-KOH (pH 7.7), 0.5 mM ATP, 0.5 mM GTP, 0.5 mM CTP, 0.5 mM UTP, 20 mM creatine phosphate, 0.48 mg of creatine kinase per ml, 2.5% polyethylene glycol, 0.2 mM EDTA, 0.5 mM EGTA, 4.25 mM dithiothreitol, 10 μg of leupeptin per ml, and 40 μg of template per ml in the form of circular plasmid DNA (35 μg of template per ml, 15 μg of SLins19 per ml). Subsequently, RNA was prepared by the single-step method of Chomczynski and Sacchi (9) and analyzed by primer extension with32P-end-labeled oligonucleotides Tag-PE (23) and SLtag (18) using the Superscript II reverse transcriptase (Invitrogen) as specified by the manufacturer. Primer extension products were separated on denaturing 6% polyacrylamide-50% urea gels and visualized by autoradiography. Densitometry of transcription and immunoblot signals was carried out with the E.A.S.Y Win32 imaging system (Herolab).
RESULTS
Isolation of TbRPA1 from T. brucei strain 427-60.
The T. brucei gene for the largest subunit of RNA pol I (TbRPA1) was previously isolated independently by Smith et al. (49) and Jess et al. (20). DNA sequence analysis revealed that one of the TbRPA1 clones encodes an extra 21 amino acids at the N terminus. To avoid potential errors due to heterogeneity of TbRPA1 among different strains of trypanosomes, we cloned a 9.7-kb EcoRI-XbaI fragment spanning the entire TbRPA1 gene from T. brucei 427-60 (clone pG9-1) (Fig. 1). By pulsed-field gel electrophoresis analysis of chromosome-sized DNA, the two alleles of TbRPA1 were demonstrated to be located at chromosome bands 15 and 16, based on the nomenclature of Gottesdiener et al. (reference 15 and data not shown). All of the TbRPA1-derived sequences used in our study are derived from clone pG9-1.
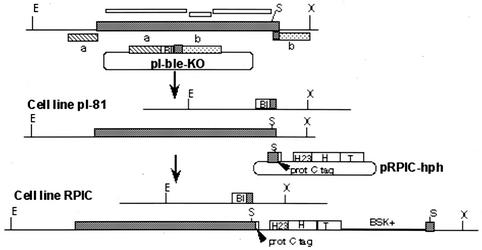
FIG. 1.
Physical maps of the RNA pol I largest-subunit locus in wild-type trypanosomes and the RPIC cell line. (Top) Structure of the RNA pol I largest-subunit (TbRPA1) locus in wild-type trypanosomes and plasmid pI-ble-KO for gene replacement. The large grey box represents the coding region of the TbRPA1 of T. brucei. a and b indicate the 5′ and 3′ ends, respectively, of TbRPA1-derived targeting sequences. The three white bars above the map indicate regions that were cloned into the pZJM construct for expression of dsRNA of TbRPA1. (Middle) Structure of the TbRPA1 locus in cell line pI-81 and plasmid pRPIC-hph used for gene integration. (Bottom) Structure of the TbRPA1 locus in the cell line RPIC. Abbreviations: Bl, phleomycin resistance gene; H, hph gene; T, αβ-tubulin intergenic region (53); H23, intergenic region of hsp70 gene 2 and 3 (14); BSK+, Bluescript SK(+) vector; prot C tag, protein C tag. Restriction enzymes: E, EcoRI; X, XbaI; S, SalI.
Epitope tagging of RNA pol I in a procyclic cell line (RPIC).
One of our strategies to demonstrate that RNA pol I initiates transcription at procyclin gene and VSG ES promoters was to immunodeplete the enzyme from transcriptionally active extracts with antibodies directed against TbRPA1. However, the study of RNA polymerases from various eukaryotic organisms demonstrated that the largest subunits of RNA pol I, pol II, and pol III are homologous to one another, contain eight highly conserved domains denoted a to h, and are immunologically related to each other (reviewed in reference 45). Homology among these largest subunits has made it difficult to raise polyclonal antibodies specific for a particular RNA pol largest subunit. Nevertheless, we obtained a RNA pol I largest-subunit-specific antibody for TbRPA1 by raising a polyclonal antiserum against an internal TbRPA1 peptide sequence. This antiserum specifically detected TbRPA1 in Western blot analysis (see below) but failed to precipitate native RNA pol I from cell extracts to a significant extent (data not shown). To ensure a high-affinity interaction of an antibody with RNA pol I without losing specificity, we decided to epitope tag TbRPA1 instead of raising antiserum against a larger portion of the native polypeptide. As an epitope, we chose the 12-amino-acid human protein C activation peptide region, which binds to a Ca2+-dependent monoclonal antibody with high affinity (51). This Ca2+-dependent interaction has the advantage that bound complexes can be eluted under mild conditions in the presence of a calcium chelator. To generate a procyclic cell line which exclusively expresses epitope-tagged RNA pol I, we modified both alleles of the single-copy TbRPA1 gene. One TbRPA1 allele was deleted and replaced by construct pI-ble-KO containing a phleomycin resistance gene (ble) via homologous recombination (Fig. 1). The resulting cell line was named pI-81. To introduce the protein C epitope-coding sequence directly upstream of the TbRPA1 stop codon, we designed an integration construct (pRPIC-hph), which contained the 3′-terminal part of the TbRPA1 gene with the insertion of the protein C epitope and the hygromycin phosphotransferase gene (hph), as a selectable marker (Fig. 1). pRPIC-hph was linearized inside the TbRPA1 sequence and then introduced into the pI-81 cell line. Through gap repair, the DNA construct inserted into the 3′ end of the remaining TbRPA1 allele and introduced the protein C epitope (Fig. 1). The corresponding cell line was denoted RPIC. By Southern blot analysis, we confirmed that RPIC cells had correctly integrated both constructs, contained solely a tagged allele of TbRPA1, and did not carry the protein C epitope anywhere else in the genome (data not shown).
Immunodepletion of RNA pol I from RPIC cell extracts specifically interferes with rDNA, VSG ES, and procyclin gene transcription in vitro.
Next, we prepared transcriptionally competent cytoplasmic extracts from RPIC cells and, as a control, from wild-type cells. To analyze whether RPIC cells express epitope-tagged TbRPA1, both wild-type and RPIC extracts were separated on an SDS-polyacrylamide gel, blotted, and probed with the polyclonal anti-TbRPA1 immune serum or with the monoclonal anti-protein C epitope antibody (Fig. 2A). As revealed by the anti-TbRPA1 immune serum, TbRPA1 migrated as a polypeptide of approximately 200 kDa, which closely corresponded to the calculated size of 195.9 kDa. The anti-protein C antibody interacted exclusively with the same band but, as expected, only in the RPIC lane (Fig. 2A). Hence, we concluded that RPIC cells expressed protein C epitope-tagged TbRPA1 and that the anti-protein C antibody did not cross-react with endogenous trypanosome proteins.
FIG. 2.
In vitro transcription analysis of RNA pol I-depleted extracts. (A) Immunodetection of TbRPA1. Nondepleted extracts prepared from wild-type (WT) and RPIC cells were separated on an SDS-5% polyacrylamide gel, blotted, and probed with the polyclonal anti-TbRPA1 antibody (left panel) or the monoclonal anti-protein C antibody (right panel). Arrows on the left indicate positions of protein marker bands. (B) RNA pol I depletion from extracts. Flowthrough fractions of extracts subjected to anti-protein C affinity chromatography in the absence (−Ca++) or presence (+Ca++) of calcium ions were analyzed with anti-TbRPA1 antiserum and, as a control, with anti-40K antiserum. In addition, a corresponding amount of eluate from the affinity matrix incubated with RPIC extract in the presence of calcium ions (IP) was loaded onto the gel. (C and D) In vitro transcription analysis of RNA pol I-depleted extract. Standard transcription reactions were carried out with wild-type (C) and RPIC (D) extracts treated with anti-protein C affinity matrix and template constructs GPEET-trm (GPEET), Rib-trm (rDNA), and VSG-trm (VSG). In each reaction, the control template SLins19 (SL) was cotranscribed. In a further control, vector DNA was transcribed in nondepleted cell extract. Transcription signals were obtained by primer extension analysis of RNA prepared from in vitro transcription reactions using32P-end-labeled oligonucleotides GLESS_PE and SLtag. Extension products were separated on 6% polyacrylamide-50% urea gels and visualized by autoradiography. Arrows on the right point to correctly sized products. Marker, MspI-digested pBR322.
Subsequently, we developed a protocol for immunodepletion of RNA pol I from cell extracts using anti-protein C affinity matrix. To demonstrate the specificity, both wild-type and RPIC cell extracts were immunodepleted in the presence of 1 mM CaCl2 and, as a control, in the presence of 0.5 mM EGTA. Western blot analysis of immunodepleted extracts with anti-TbRPA1 antiserum showed that anti-protein C immunoaffinity chromatography in the presence or absence of calcium ions did not affect RNA pol I abundance in extracts of wild-type cells (Fig. 2B, compare lanes 1 and 2). In contrast, addition of CaCl2 to RPIC extract decreased the RNA pol I level by 98% (compare lanes 3 and 4). RNA pol I retained on the immunoaffinity matrix did elute in the presence of EGTA and was not degraded (lane 5). Hence, the anti-protein C antibody recognized its epitope in native RNA pol I and very efficiently bound this enzyme in crude cell extracts. Finally, immunodepleted extracts were tested in transcription reactions. As DNA templates, the previously described gene constructs GPEET-trm, Rib-trm, and VSG-trm, which contain the GPEET procyclin gene promoter, the rDNA promoter, and a VSG ES promoter, respectively, were transcribed (23). As a control, the SL RNA gene template SLins19, which most probably is transcribed by RNA pol II (13, 52), was added to each reaction mixture. To exclude the possibility that immunoaffinity chromatography in the presence of EGTA or calcium chloride had a different effect on transcription efficiency, we tested the wild-type extracts first. As shown in Fig. 2C, EGTA or calcium chloride did not affect transcription in wild-type extracts; both extracts with and without Ca2+ generated transcription signals of similar strength (compare lanes 2, 3, and 4 with lanes 5, 6, and 7, respectively). The analysis of RPIC extracts revealed a different outcome. Transcription of GPEET-trm, Rib-trm, and VSG-trm was dramatically reduced in the RPIC extract depleted of RNA pol I (Fig. 2D, compare lanes 2, 3, and 4 with lanes 5, 6, and 7, respectively). This defect was specific to RNA pol I because transcription of the SLins19 control template remained unaffected. Therefore, we concluded that in our T. brucei in vitro system, RNA pol I, and not a modified, α-amanitin-resistant RNA pol II enzyme, initiates transcription at procyclin gene and VSG ES promoters.
Selective down-regulation of the RNA pol I largest-subunit mRNA by RNAi.
It has been shown that RNAi can effectively down-regulate gene expression in trypanosomes and in many other eukaryotes (1, 33, 54). This method is based on the fact that introduction of dsRNA into cells results in specific degradation of mRNAs containing the same sequence. We determined the transcriptional impact of TbRPA1 down-regulation by the RNAi approach.
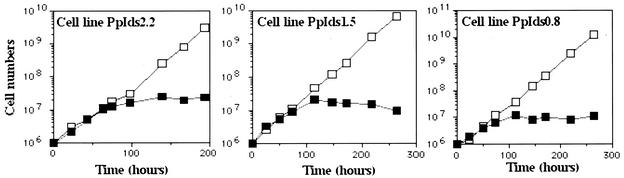
The pZJM vector, provided by P. Englund, was used to express dsRNA of TbRPA1 in trypanosomes. Expression of the dsRNA in pZJM is controlled by two opposing and inducible T7 promoter-tetracycline operators (57). Three constructs, allowing the expression of dsRNAs encoded by different regions of TbRPA1, were generated and referred to as pIds-2.2, pIds-1.5, and pIds-0.8 (Fig. 1; see Materials and Methods for descriptions). Linearized plasmid DNAs were introduced into the procyclic cell line 29-13, which expresses T7 RNA pol and the tetracycline repressor (a gift from Dr. Cross [58]. Clonally transformed cell lines were established and maintained under uninduced conditions. Three corresponding cell lines were referred to as cell lines PpIds2.2, PpIds1.5, and PpIds0.8, respectively. The integration of plasmid DNA at the nontranscribed rDNA spacer region in each cell line was confirmed by Southern blot analysis (data not shown). At 72 h after the induction of TbRPA1 dsRNA by addition of tetracycline to the culture, the growth efficiency of each transformed cell line declined rapidly and, after prolonged induction, eventually led to cell death, as expected for cells lacking RNA pol I (Fig. 3).
FIG. 3.
Effect of the TbRPA1 RNAi on cell growth. The growth curves of trypanosome cell lines PIds2.2, Pids1.5, and Pids0.8 were determined in the absence (open squares) or presence (solid squares) of 1 μg of tetracycline per ml (for the induction of dsRNA expression). Cells were continuously maintained at a log phase. If needed, medium can be added to expand the culture. The total number of cells in each culture was calculated at different time points.
Steady-state TbRPA1 mRNA levels from the three cell lines, before and 24 or 48 h after tetracycline induction of dsRNA expression, were analyzed by Northern blot analysis. Addition of tetracycline alone did not affect TbRPA1 expression, as shown by the control cell line 29-13, which did not contain any of the dsRNA expression plasmid. (Fig. 4, lanes C). In the three cell lines which did contain an RNAi construct, PpIds2.2, PpIds1.5, and PpIds0.8, we observed a mild reduction of the TbRPA1 mRNA level 24 h after TbRPA1 RNAi induction (data not shown). Down-regulation of TbRPA1 mRNA was drastically enhanced 48 h after induction and was specific to TbRPA1 because mRNA levels of the RNA pol II largest subunit remained unaffected (Fig. 4, lanes 1 to 3, − and + tetracycline, respectively). After normalization with the level of tubulin mRNA in each lane, we estimated that 48 h after induction of TbRPA1, RNAi resulted in ∼90% reduction of TbRPA1 mRNA and did not significantly affect the level of RNA pol II mRNA.
FIG. 4.
Northern blot analysis of the effect of the TbRPA1 RNAi on mRNA levels. Approximately equal amounts (∼10 μg) of total RNA from cell lines 29-13 (lanes C), PIds2.2 (lanes 1), Pids1.5 (lanes 2), and Pids0.8 (lanes 3), cultured in the absence (−) or presence (+) of 1 μg of tetracycline per ml for 48 h, were separated in 1% formaldehyde-agarose gels. (Top panel) The blot was hybridized to a TbRPA1 coding region probe. The final posthybridizational wash was performed in 0.1 × SSC-0.1% SDS at 65°C. (Middle panel) Hybridization with the β-tubulin gene probe, indicating that similar amounts of RNA were loaded in each lane. (Bottom panel) Probes on the filter was stripped off and rehybridized with an RNA pol II largest-subunit probe.
Selective reduction of transcriptional efficiency of a subset of genes by the RNA pol I largest subunit RNAi.
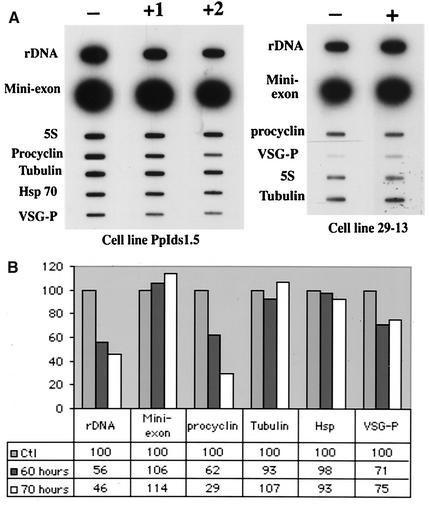
By using nuclear run-on assays, we analyzed the transcriptional efficiencies of different genes before and after the induction of TbRPA1 RNAi in procyclic cell lines. Since we found a dramatic reduction of the TbRPA1 mRNA level at 48 h after induction and observed that cell growth was dramatically reduced at 72 h after induction, we measured the effect of TbRPA1 RNAi on nascent RNA synthesis between 48 and 72 h after induction. Figure 5 shows an analysis of the relative rates of nascent RNA synthesis for various genes before and 60 or 70 h after induction of TbRPA1 RNAi. We found that up to 60 h after induction, TbRPA1 RNAi did not significantly affect the overall transcriptional activity of most genes except that of the rDNA and procyclin genes and VSG ES promoter sequences. However, induction of TbRPA1 RNAi for 70 h or longer significantly reduced the overall transcription efficiency of all genes in isolated nuclei. It is possible that a significant amount of this transcriptional reduction was caused by the death of trypanosome cells within the 70-h period of RNAi induction. To exclude the variation of the number of living cells in each nuclear preparation, we compared and standardized the relative transcriptional efficiencies with that of the RNA pol III-transcribed 5S rRNA gene in the same preparation (Fig. 5B). We found that in comparison to uninduced trypanosomes, induction of TbRPA1 RNAi for 60 h decreased the transcriptional efficiency of rRNA and procyclin genes to 56 and 46%, respectively (Fig. 5B, rDNA and procyclin). The reduction was even more prominent, namely, to 46 and 29%, respectively, when RNAi was induced for 70 h (Fig. 5B). In our procyclic cell lines, TbRPA1 RNAi was less efficient in affecting VSG 118 ES promoter sequence transcription. Nevertheless, a reduction of approximately 30% was reproducibly detected in all three TbRPA1 RNAi cell lines. Importantly, and in contrast to these results, the relative transcriptional efficiencies of the SL RNA gene and of the RNA pol II-transcribed αβ-tubulin and hsp70 genes were not significantly affected by the induction of TbRPA1 RNAi (Fig. 5). Hence, the transcriptional efficiencies of these genes and of the 5S rRNA gene remained constant in all experiments and excluded the possibility of a nonspecific transcriptional effect. Similarly, the analysis of nuclei isolated from cell line 29-13 before and 70 h after the addition of tetracycline showed that the treatment of tetracycline alone did not affect transcription efficiency in general (Fig. 5A, right panel). For each cell line, duplicated experiments were performed and reproducible results were obtained. Based on the results of nuclear run-on analysis, we concluded that induction of TbRPA1 RNAi for a certain period specifically interfered with the in vivo transcription of rDNA, procyclin genes, and nucleotide sequences downstream of a VSG ES promoter while it had no effect on the transcription of genes controlled by RNA pol II and III.
FIG. 5.
Effect of the TbRPA1 RNAi on nascent RNA synthesis in procyclic form trypanosomes. (A) Nuclear run-on analysis. Nuclei were isolated from the PpIds1.5 cell line (left panel) and the 29-13 cell line (right panel) before (lanes −) and 60 h (lane +1) or 70 h (lane +2 and + on the right hand panel) after the addition of 1 μg of tetracycline per ml to induce dsRNA expression. Newly synthesized nascent RNA was labeled by the incorporation of [α-32P]UTP and was hybridized with slot blots containing the following probes: T. brucei 18S rRNA gene (rDNA), the mini-exon clone CL103 (Mini-exon), the 5S rRNA probe (5S), the procyclin coding region clone CPT4 (Procyclin), the coding region of the αβ-tubulin gene (Tubulin), a 1.8-kb HindIII fragment from the coding region of the hsp70 gene (Hsp), and the VSG 118 ES promoter sequence (VSG-P). (B) Quantification of the relative transciptional efficiency of different genes. The radioactivity bound on the nitrocellulose filters from the left panel of panel A was quantified with a PhosphorImager. The transcriptional efficiency of each gene relative to that of the 5S RNA gene was determined for each nuclear preparation. The relative transcriptional efficiency of all genes in uninduced trypanosomes was arbitrarily set to 100.
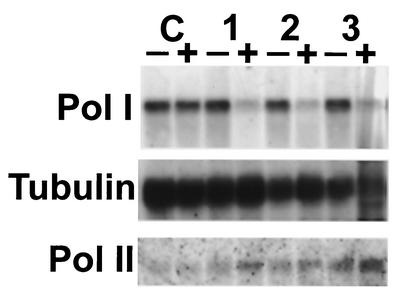
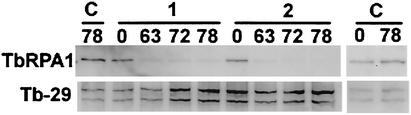
To confirm that the reduction of the rRNA, procyclin, and VSG gene transcription by TbRPA1 RNAi is a result of the down-regulation of TbRPA1 protein expression, we compared the TbRPA1 protein level after 63, 72, and 78 h of TbRPA1 RNAi induction to that in uninduced cells by Western blot analysis (Fig. 6). The control hybridization with antibodies recognizing the Tb-29 proteins demonstrated the relative amounts of protein loaded in each lane (Fig. 6, bottom panels) (26). We calculated the relative amount of TbRPA1 protein in each lane after adjusting the amount of sample loaded in each lane. It appeared that 63 h of TbRPA1 RNAi induction reduced the TbRPA1 protein level to ∼15% of that in uninduced cells and that 72 or 78 h of RNAi induction reduced the TbRPA1 protein level to ∼5 to 7% of that in uninduced cells in cell lines PpIds0.8 and PpIds1.5 (Fig. 6, lanes 1 and 2, respectively). Addition of tetracycline alone did not affect the level of TbRPA1 protein, as measured using cell line 29-13 (Fig. 6, lanes C). We did not analyze the PpIds2.2 cell line, because the tetracycline-irresponsive population was increased in the PpIds2.2 cell line at the time that we performed the protein analysis.
FIG. 6.
Effect of the TbRPA1 RNAi on the TbRP1 protein level in procyclic form trypanosomes. Total protein lysates (∼5 × 106 trypanosomes), derived from cell line 29-13 (lanes C), cell line PpIds0.8 (lanes 1), and cell line PpIds1.5 (lanes 2) incubated in the absence (lanes 0) or presence of tetracycline for 63 h (lanes 63), 72 h (lanes 72), and 78 h (lanes 78) were size separated in SDS-5% polyacrylamide gels and electrophoretically transferred to polyvinylidene difluoride membranes. The blots were probed with anti-TbRPA1 (top panel [TbRPA1]). The same blots were later probed with anti-Tb-29 (bottom panel [Tb-29] [26]) to demonstrate that the relative amounts of protein were loaded in each lane.
DISCUSSION
Our studies show that T. brucei utilizes RNA pol I for transcription of rRNA genes, procyclin genes, and VSG promoter region sequences in procyclic form trypanosomes. Immunodepletion of RNA pol I from procyclic cell extracts specifically abolishes transcription initiated at rDNA, VSG ES, and procyclin gene promoters, in vitro. Attempts to reconstitute the RNA pol I activity in the RNA pol I-depleted cell extracts failed due to our failure to elute sufficient amounts of active RNA pol I from the affinity matrix. In vivo, down-regulation of the expression of the RNA pol I largest subunit slectively reduced the transcriptional efficiency of rDNA, procyclin genes, and VSG ES sequences. However, the extent of transcriptional down-regulation by RNA pol I RNAi was different on the rDNA, procyclin genes, and VSG ES sequences. This effect could be partly attributed to the fact that RNA pol I may interact with different subsets of transcription factors or a different higher-order structure when it transcribes rRNA, VSG, or procyclin genes.
A major difference between rRNA production and procyclin and VSG gene expression is that rRNA is constitutively expressed and not trans spliced whereas procyclin and VSG genes are differentially expressed in procyclic and bloodstream forms, respectively, and the synthesis of their mature mRNAs is dependent on an efficient trans-splicing machinery. In eukaryotes, RNA pol I-mediated transcription of rDNA is confined to the nucleolus, which in general lacks RNA-splicing factors such as spliceosomal U small nuclear ribonucleoproteins (snRNPs) (47, 48, 50). T. brucei appears to be no exception to this rule because polyclonal antibodies raised either against a set of proteins common to spliceosomal U snRNPs or against the U2 snRNP-specific 40K protein did not stain the nucleolus of trypanosomes (34). This observation suggested that the nucleolus of T. brucei may not be able to support trans splicing or mRNA processing in general. Consistent with this idea, the active VSG ES colocalizes with RNA pol I in the ESB outside the nucleolus (32). In procyclics, such a compartment was not found, and the location of transcription driven by the procyclin gene promoter is still unclear. In situ hybridization experiments by one group revealed that RNA of the neomycin resistance gene driven by the procyclin and rRNA promoters was mapped either to the nucleolus or to the nucleolar periphery (11, 40). In contrast, experiments by another research group showed that most of the nuclear localization of Hph and chloramphenicol acetyltransferase RNAs derived from a procyclin transcription unit did not overlap with the nucleolus (7). Hence, it remains to be clarified whether native procyclin gene transcription takes place in the nucleolus.
Another aspect of RNA pol I transcription and RNA processing is that the RNA-processing machinery in T. brucei may interact with RNA pol II but not with RNA pol I. In higher eukaryotes, RNA processing enzyme complexes such as the spliceosome and the polyadenylation complexes interact with the C terminus of the RNA pol II largest subunit, which is characterized by the repeated heptapeptide Tyr-Ser-Pro-Thr-Ser-Pro-Ser (reviewed in reference 36). The homologous protein domain in T. brucei lacks the repeated heptapeptide motif, although it is rich in amino acid residues that compose the repeat. The C terminus of TbRPA1 is divergent from the C terminus of the RNA pol II largest subunit, and it is not clear whether RNA pol I is as effective as RNA pol II in recruiting RNA-processing complexes to its pre-mRNA in T. brucei. Interestingly, Vanhamme et al. found that pre-mRNA derived from inactive VSG ESs is poorly processed, supporting the notion that RNA pol I may be unable to efficiently interact with the RNA-processing machinery (55). On the other hand, RNA pol I mediated mRNA synthesis in a subnuclear compartment such as the ESB, which may be allowing efficient maturation of RNA pol I-derived pre-mRNA.
Interestingly, the C terminus of TbRPA1 appears to have a greater functional relevance in the bloodstream form than in the procyclic form. We have successfully constructed procyclic cell lines expressing exclusively RNA pol I carrying either the protein C tag or the TY tag (M. G.-S. Lee et al., unpublished data) at the TbRPA1 C terminus. Apparently, the C-terminal tag did not affect the function of TbRPA1 in procyclics. However, our repeated attempts to establish the corresponding bloodstream form cell lines failed (data not shown). When using bloodstream form cells, We succeeded in deleting one native TbRPA1 allele or in producing cell lines which contained one native TbRPA1 allele and one C-terminally tagged TbRPA1 allele (either the protein C tag or the TY tag). However, we were not able to manipulate both alleles in the same cell.
The essential multifunctional role of the T. brucei RNA pol I may depend on transcription factors or factor domains which are unique to the parasite. Several observations support the hypothesis that the efficiency of T. brucei RNA pol I transcription complexes varies depending on the genes transcribed, on the parasite life cycle stage, or on the chromosomal locations. First, the segregation of RNA pol I molecules into two different subnuclear compartments, the nucleolus and the ESB, in bloodstream forms may require separate determinants. Second, structural differences of rDNA, procyclin gene, and VSG ES promoters exist. For example, the VSG promoter does not have definable promoter elements upstream of position −67 relative to the transcription initiation site, whereas the other two promoters extend roughly to position −250 (5, 35, 46). Furthermore, the most distal rDNA promoter element has a high sequence similarity to SL RNA gene promoter elements and binds essential SL RNA gene transcription factors in vitro. In contrast, the corresponding region of the procyclin gene promoter neither shares this sequence homology nor binds to those factors (A. Günzl et al., unpublished data). Third, the different sensitivities of RNA pol I-mediated transcription of different genes to manganese ions may argue that different RNA pol I complexes may exist within a certain life cycle stage. It is also possible that due to different conformational needs in promoter recognition, specific ion concentrations may be required for optimal transcription of a specific promoter. Bloodstream form nuclear run-on assays revealed that transcription elongation on a VSG ES, in contrast to that on rDNA, is resistant to elevated Mn2+ concentrations (17). Similarly, in procyclic extracts, procyclin gene promoter transcription exhibited the same level of resistance whereas VSG ES and rDNA promoter transcription did not (23). Fourth, procyclin and VSG gene expression is regulated in part at the transcriptional level. In bloodstream forms, the promoter of the active VSG ES is up-regulated about 20-fold (42). Although this fact may be attributed to the ESB, it does not exclude the existence of VSG-specific trans activators. Biebinger et al. showed that the activity of the procyclin gene promoter was fivefold higher in procyclic form cells than in bloodstream form cells independent of the chromosomal integration site (2). These data were supported by the finding that in the procyclic cell extract, procyclin gene promoter transcription was about fourfold stronger than rDNA and VSG ES promoter transcription (23). Fifth, chromosomal location differentially affects the activity of VSG ES and rDNA promoters. In bloodstream form trypanosomes, the α-amanitin-resistant transcription driven by both VSG and rRNA promoters is repressed at an RNA pol II-transcribed locus (60; M. G.-S. Lee, unpublished data). However, the rRNA promoter can be functionally active at an RNA pol II locus in procyclic trypanosomes (10). The differential activity of the rRNA gene promoter in different life cycle stages of T. brucei may suggest that the function or structure of RNA pol I exhibits unique properties in each stage of the parasite.
In conclusion, we have shown that RNA pol I of T. brucei transcribes, in addition to rDNA, the procyclin genes and the VSG ESs. As discussed, there is strong evidence that specific determinants or factors exist for the transcriptional control of the VSG and procyclin genes. Identification and characterization of the corresponding polypeptides appears to be a promising objective of future studies.
Acknowledgments
We thank L. H. T. Van der Ploeg for critical reading of the manuscript, P. Englund for providing the pZJM plasmid, and G. A. M. Cross for providing the procyclic 29-13 cell line; we also thank H. I. Wang for technical support.
This work was supported by a grant from the German Research Foundation (DFG) to A.G. (Gu 371) and an NIH grant AI28953 to G.S.M.L., who is a Burroughs Wellcome Fund New Investigator in molecular parasitology.
REFERENCES
- 1.Bass, B. L. 2000. Double-stranded RNA as a template for gene silencing. Cell 101:235-238. [DOI] [PubMed] [Google Scholar]
- 2.Biebinger, S., S. Rettenmaier, J. Flaspohler, C. Hartmann, J. Pena-Diaz, L. E. Wirtz, H.-R. Hotz, D. Barry, and C. Clayton. 1996. The PARP promoter of Trypanosoma brucei is developmentally regulated in a chromosomal context. Nucleic Acids Res. 24:1202-1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borst, P. 2002. Antigenic variation and allelic exclusion. Cell 109:5-8. [DOI] [PubMed] [Google Scholar]
- 4.Borst, P., and S. Ulbert. 2001. Control of VSG gene expression sites. Mol. Biochem. Parasitol. 114:17-27. [DOI] [PubMed] [Google Scholar]
- 5.Brown, S. D., J. Huang, and L. H. T. Van der Ploeg. 1992. The promoter for the procyclic acidic repetitive protein (PARP) genes of Trypanosoma brucei shares features with RNA polymerase I promoters. Mol. Cell. Biol. 12:2644-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brun, R., and M. Schönenberger. 1979. Cultivation and in vitro cloning of procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 36:289-292. [PubMed] [Google Scholar]
- 7.Chaves, I., J. Zomerdijk, M. A. Dirks, R. W. Dirks, A. K. Raap, and P. Borst. 1998. Subnuclear localization of the active variant surface glycoprotein gene expression site in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 95:12328-12333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho, E. J., T. Takagi, C. R. Moore, and S. Buratowski. 1997. mRNA capping enzyme is recruited to the transcription complex by phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 11:3319-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 10.Chung, H. M., G.-S. M. Lee, and L. H. T. Van der Ploeg. 1993. RNA polymerase I-mediated protein-coding gene expression in Trypanosoma brucei. Parasitol. Today 8:414-418. [DOI] [PubMed] [Google Scholar]
- 11.Chung, H. M., G.-S. M. Lee, P. Dietrich, J. Huang, and L. H. T. Van der Ploeg. 1993. Disruption of large subunit RNA polymerase II genes in Trypanosoma brucei. Mol. Cell. Biol. 13:3734-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clayton, C. E., J. P. Fueri, J. E. Itzhaki, V. Bellofatto, D. R. Sherman, G. S. Wisdom, S. Vijayasarathy, and M. R. Mowatt. 1990. Transcription of the procyclic acidic repetitive protein genes of Trypanosoma brucei. Mol. Cell. Biol. 10:3036-3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gilinger, G., and V. Bellofatto. 2001. Trypanosome spliced leader RNA genes contain the first identified RNA polymerase II gene promoter in these organisms. Nucleic Acids Res. 29:1556-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glass, D. J., R. I. Polvere, and L. H. T. Van der Ploeg. 1986. Conserved sequences and transcription of the hsp70 gene family in Trypanosoma brucei. Mol. Cell. Biol. 6:4657-4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottesdiener, K., J. Garcia-Anoveros, G.-S. M. Lee, and L. H. T. Van der Ploeg. 1990. Chromosome organization of the protozoan Trypanosoma brucei. Mol. Cell. Biol. 10:6079-6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gottesdiener, K., H-M. Chung, S. Brown, G-S. M. Lee, and L. H. T. Van der Ploeg. 1991. Characterization of VSG gene expression site promoters and promoter-associated DNA rearrangement events. Mol. Cell. Biol. 11:2467-2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Grondal, E. J. M., R. Evers, K. Ksubek, and A. W. C. A. Cornelissen. 1989. Characterization of the RNA polymerases of Trypanosoma brucei: trypanosomal mRNAs are composed of transcripts derived from both by RNA pol II and III. EMBO J. 8:3383-3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Günzl, A., E. Ullu, M. Dörner, S. P. Fragoso, K. F. Hoffmann, J. D. Milner, Y. Morita, E. K. Nguu, S. Vanacova, S. Wünsch, A. O. Dare, H. Kwon, and C. Tschudi. 1997. Transcription of the trypanosoma brucei spliced leader RNA gene is dependent only on the presence of upstream regulatory elements. Mol. Biochem. Parasitol. 85:67-76. [DOI] [PubMed] [Google Scholar]
- 19.Janz, L., and C. Clayton. 1994. The PARP and rRNA promoters of Trypanosoma brucei are composed of dissimiliar sequence elements that are functionally interchangeable. Mol. Cell. Biol. 14:5804-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jess, W., A. Hammer, and A. W. C. A. Cornelissen. 1989. Complete sequence of the gene encoding the largest subunit of RNA polymerase I of Trypanosoma brucei. FEBS Lett. 249:123-128. [DOI] [PubMed] [Google Scholar]
- 21.Kooter, J. M., and P. Borst. 1984. Alpha-amanitin-insensitive transcription of variant surface glycoprotein genes provides further evidence for discontinuous transcription in trypanosomes. Nucleic Acids Res. 12:9457-9472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laird, P., J. M. Kooter, N. Loosbroek, and P. Borst. 1985. Mature mRNAs of Trypanosoma brucei possess a 5′ cap acquired by discontinuous RNA synthesis. Nucleic Acids Res. 13:4253-4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laufer, G., G. Schaaf, S. Bollgönn, and A. Günzl. 1999. In vitro analysis of alpha-amanitin-resistant transcription from the rRNA, procyclic acidic repetitive protein, and variant surface glycoprotein gene promoters in Trypanosoma brucei. Mol. Cell. Biol. 19:5466-5473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laufer, G., and A. Günzl. 2001. In-vitro competition analysis of procyclin gene and variant surface glycoprotein gene expression site transcription in Trypanosoma brucei. Mol. Biochem. Parasitol. 113:55-65. [DOI] [PubMed] [Google Scholar]
- 25.Lee, G.-S. M., and L. H. T. Van der Ploeg. 1991. The hygromycin B-resistance coding gene as a selectable marker for stable transformation of Trypanosoma brucei. Gene 105:255-257. [DOI] [PubMed] [Google Scholar]
- 26.Lee, G.-S. M., D. Russell, P. A. D'Alesandro, and L. H. T. Van der Ploeg. 1994. Identification of membrane-associated proteins in Trypanosoma brucei, encoding an internal, EARLRAEE amino acid repeat. J. Biol. Chem. 269:8408-8415. [PubMed] [Google Scholar]
- 27.Lee, G.-S. M. 1995. A foreign transcription unit in the inactivated VSG gene expression site of the procyclic form of Trypanosoma brucei and formation of large episomes in stably transformed trypanosomes. Mol. Biochem. Parasitol. 69:223-238. [DOI] [PubMed] [Google Scholar]
- 28.Lee, G.-S. M., and L. H. T. Van der Ploeg. 1997. Transcription of protein-coding genes expression in trypanosomes by RNA polymerase I. Annu. Rev. Microbiol. 51:463-89. [DOI] [PubMed] [Google Scholar]
- 29.Lee, M.G.M. 1996. An RNA polymerase II promoter in the hsp70 locus of Trypanosoma brucei. Mol. Cell. Biol. 16:1220-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lenardo, M. J., D. M. Dorfman, and J. E. Donelson. 1985. Characterization of the Trypanosoma brucei 5S ribosomal RNA gene and transcript: the 5S rRNA is a spliced leader-independent species. Gene 35:131-141. [DOI] [PubMed] [Google Scholar]
- 31.McCracken, S., N. Fong, E. Rosonina, K. Yankulov, G. Brothers, D. Siderovski, A. Hessel, S. Foster, S. Shuman, and D. L. Bentley. 1997. 5′-Capping enzymes are targeted to pre-mRNA by binding to the phosphorylated carboxy-terminal domain of RNA polymerase II. Genes Dev. 11:3306-3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Navarro, M., and K. Gull. 2001. A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature 414:759-763. [DOI] [PubMed] [Google Scholar]
- 33.Ngo, H., C. Tschudi, K. Gull, and E. Ullu. 1998. Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 95:14687-14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palfi, Z., and A. Bindereif. 1992. Immunological characterization and intracellular localization of trans-spliceosomal small nuclear ribonucleoproteins in Trypanosoma brucei. J. Biol. Chem. 267:20159-20163. [PubMed] [Google Scholar]
- 35.Pham, V. P., C. C. Qi, and K. M. Gottesdiener. 1996. A detailed mutational analysis of the VSG gene expression site promoter. Mol. Biochem. Parasitol. 75:241-254. [DOI] [PubMed] [Google Scholar]
- 36.Proudfoot, N. J., A. Furger, and M. J. Dye. 2002. Integrating mRNA processing with transcription. Cell 108:501-512. [DOI] [PubMed] [Google Scholar]
- 37.Roditi, I., and M. Liniger 2002. Dressed for success: the surface coats of insect-borne protozoan parasites. Trends Microbiol. 10:128-134. [DOI] [PubMed] [Google Scholar]
- 38.Rudenko, G., D. Bishop, K. Gottesdiener, and L. H. T. Van der Ploeg. 1989. Alpha-amanitin resistant transcription of protein coding genes in insect and bloodstream form Trypanosoma brucei. EMBO J. 13:4259-4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rudenko, G., S. Le Blancq, J. Smith, G-S. M. Lee, A. Rattray, and L. H. T. Van der Ploeg. 1990. Procyclic acidic repetitive protein (PARP) genes located in an unusually small α-amanitin-resistant transcription unit: PARP promoter activity assayed by transient DNA transfection of Trypanosoma brucei. Mol. Cell. Biol. 10:3492-3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rudenko, G., H. M. Chung, V. P. Pham, and L. H. T. Van der Ploeg. 1991. RNA polymerase I can mediate expression of CAT and neo protein-coding genes in the parasitic protozoan Trypanosoma brucei. EMBO J. 10:3387-3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rudenko, G., G.-S. M. Lee, and L. H. T. Van der Ploeg. 1992. The PARP and VSG genes of Trypanosoma brucei do not resemble RNA polymerase II transcription units in sensitivity to Sarkosyl in nuclear run-on assays. Nucleic Acids Res. 20:303-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rudenko, G., P. A. Blundell, M. C. Taylor, R. Kieft, and P. Borst. 1994. VSG gene expression site control in insect from Trypanosoma brucei. EMBO J. 13:5470-5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ruepp, S., A. Furger, U. Kurath, C. K. Renggli, A. Hemphill, R. Brun, and I. Roditi. 1997. Survival of Trypanosoma brucei in the tsetse fly is enhanced by the expression of specific forms of procyclin. J. Cell Biol. 137:1369-1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schimanski, B., B. Klumpp, G. Laufer, R. J. Marhöfer, P. Selzer, and A. Günzl. 2003. The second largest subunit of Trypanosoma brucei's multifunctional RNA polymerase I has a unique N-terminal extension domain. Mol. Biochem. Parasitol. 126:193-200. [DOI] [PubMed] [Google Scholar]
- 45.Sentenac, A., M. Riva, P. Thuriaux, J.-M. Buhler, I. Treich, C. Carles, M. Werner, A. Ruet, J. Huet, C. Mann, N. Chiannilkulchai, S. Settler, and S. Mariotte. 1992. Yeast RNA polymerase subunits and genes, p. 27-54. In S. L. McKnight and K. R. Yamamoto (ed.), Transcriptional regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 46.Sherman, D. R., L. Janz, M. Hug, and C. Clayton. 1991. Anatomy of the PARP gene promoter of Trypanosoma brucei. EMBO J. 10:3379-3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shaw, P. J. 1995. The nucleolus. Annu. Rev. Cell Dev. Biol. 11:93-121. [DOI] [PubMed] [Google Scholar]
- 48.Sheer, U., and D. Weisenberger. 1994. The nucleolus. Curr. Opin. Cell Biol. 6:354-359. [DOI] [PubMed] [Google Scholar]
- 49.Smith, J. L., J. R. Levin, and N. Agabian. 1989. Molecular characterization of the Trypanosoma brucei RNA polymerase I and III largest subunit genes. J. Biol. Chem. 264:18091-18099. [PubMed] [Google Scholar]
- 50.Sollner-Webb, B., and E. B. Mougey. 1991. News from the nucleolus: rRNA gene expression. Trends Biochem. Sci. 16:58-62. [DOI] [PubMed] [Google Scholar]
- 51.Stearns, D. J., S. Kurosawa, P.J. Sims, N. L. Esmon, and C. T. Esmon. 1988. The interaction of a Ca2+-dependent monoclonal antibody with the protein C activation peptide region. Evidence for obligatory Ca2+ binding to both antigen and antibody. J. Biol. Chem. 263:826-832. [PubMed] [Google Scholar]
- 52.Sturm, N. R., M. C. Yu, and D. A. Campbell. 1999. Transcription termination and 3′ End processing of the spliced leader RNA in kinetoplastids. Mol. Cell. Biol. 19:1595-1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomashow, L. S., M. Milhausen, W. J. Rutter, and N. Agabian. 1983. Tubulin genes are tandemly linked and clustered in the genome of Trypanosoma brucei. Cell 32:35-43. [DOI] [PubMed] [Google Scholar]
- 54.Ullu, E., A. Djikeng, H. Shi, and C. Tschudi. 2002. RNA interference: advances and questions. Phil. Trans. R. Soc. Lond. Ser. B 357:65-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vanhamme, L., P. Poelvoorde, A. Pays, P. Tebabi, H. V. Xong, and E. Pays. 2000. Differential RNA elongation controls the variant surface glycoprotein gene expression sites of Trypanosoma brucei. Mol. Microbiol. 36:328-340. [DOI] [PubMed] [Google Scholar]
- 56.Vanhamme, L., L. Lecordier, and E. Pays. 2001. Control and function of the bloodstream variant surface glycoprotein expression sites in Trypanosoma brucei. Int. J. Parasitol. 31:522-530. [DOI] [PubMed] [Google Scholar]
- 57.Wang, Z., J. C. Morris, M. E. Drew, and P. T. Englund. 2000. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 275:40174-40179. [DOI] [PubMed] [Google Scholar]
- 58.Wirtz, E., S. Leal, C. Ochatt, and G. A. Cross. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89-101. [DOI] [PubMed] [Google Scholar]
- 59.Xue, Z., E. Maldonado, R. Pillutla, H. Cho, D. Reinberg, and A. J. Shatkin. 1997. Mammalian capping enzyme complements mutant Saccharomyces cerevisiae lacking mRNA guanylyltransferase and selectively binds the elongating form of RNA polymerase II. Proc. Natl. Acad. Sci. USA 94:12898-12903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zomerdijk, J. C. B. M., R. Kieft, and P. Borst. 1991. Efficient production of functional mRNA mediated by RNA polymerase I in Trypanosoma brucei. Nature 353:772-775. [DOI] [PubMed] [Google Scholar]