Abstract
Cysteine proteases are known to be important pathogenicity factors of the protozoan parasite Entamoeba histolytica. So far, a total of eight genes coding for cysteine proteases have been identified in E. histolytica, two of which are absent in the closely related nonpathogenic species E. dispar. However, present knowledge is restricted to enzymes expressed during in vitro cultivation of the parasite, which might represent only a subset of the entire repertoire. Taking advantage of the current E. histolytica genome-sequencing efforts, we analyzed databases containing more than 99% of all ameba gene sequences for the presence of cysteine protease genes. A total of 20 full-length genes was identified (including all eight genes previously reported), which show 10 to 86% sequence identity. The various genes obviously originated from two separate ancestors since they form two distinct clades. Despite cathepsin B-like substrate specificities, all of the ameba polypeptides are structurally related to cathepsin L-like enzymes. None of the previously described enzymes but 7 of the 12 newly identified proteins are unique compared to cathepsins of higher eukaryotes in that they are predicted to have transmembrane or glycosylphosphatidylinositol anchor attachment domains. Southern blot analysis revealed that orthologous sequences for all of the newly identified proteases are present in E. dispar. Interestingly, the majority of the various cysteine protease genes are not expressed in E. histolytica or E. dispar trophozoites during in vitro cultivation. Therefore, it is likely that at least some of these enzymes are required for infection of the human host and/or for completion of the parasite life cycle.
Cysteine proteases (EC 3.4.22) of the papain superfamily occur in a wide range of organisms including bacteria, plants, invertebrates, and vertebrates (7). In mammals, they are well documented as intracellular enzymes involved in protein turnover within lysosomes. In addition, extracellular cysteine proteases have been implicated in various physiological and pathophysiological processes, including tumor invasion and metastasis (33, 43). Cysteine proteases are important virulence factors of various infectious agents and the main proteolytic enzymes in many protozoon parasites (22, 29). The protozoan Entamoeba histolytica, the causative agent of human amoebiasis, is characterized by its great capacity to destroy host tissue, leading to potentially life-threatening diseases such as ulcerative colitis or liver abscess. Convincing evidence exists that cysteine proteases are essential for E. histolytica-induced pathology. Treatment of the ameba with sublethal doses of a specific cysteine protease inhibitor or the addition of laminin, which blocks the substrate-binding pocket of cysteine proteases, greatly reduces its ability to produce liver abscesses in laboratory animals (18, 37). Likewise, liver abscess formation was totally blocked and significantly less gut inflammation and damage to the intestinal permeability barrier were observed when ameba genetically engineered to produce low levels of cysteine protease activity were used to infect the animals (1, 2, 47).
Until now, a total of eight genes coding for cysteine proteases in E. histolytica have been identified (ehcp1 to ehcp7 and ehcp112) (8, 12, 26, 38, 39) (EhCP7 accession number, AJ409105). Three of the corresponding proteins, namely, EhCP1, EhCP2, and EhCP5, have been purified (15, 30, 32) and were found to account for approximately 90% of the total cysteine protease activity present in lysates of axenically cultured E. histolytica trophozoites (8). Accordingly, Northern blot analysis indicated high-level expression of ehcp1, ehcp2, and ehcp5, low-level expression of ehcp3, and no expression of ehcp4 and ehcp6 (8). So far, the expression pattern of ehcp7 and ehcp112 has not been investigated. Interestingly, in contrast to all other cysteine proteases, functional genes corresponding to ehcp1 and ehcp5 are absent in Entamoeba dispar, a closely related but nonpathogenic Entamoeba species. By comparing the E. histolytica and E. dispar genomic loci containing the gene for CP5, it was found that the position of cp5 within the genomic context is conserved between the two organisms. However, the gene is highly degenerated in E. dispar, since it contains numerous nucleotide exchanges, insertions, and deletions, resulting in multiple stop codons within the cp5 reading frame (46). With respect to E. histolytica pathogenicity, EhCP5 appears to be of special importance, since it is the only cysteine protease known so far that is present on the ameba surface (15). However, the mechanism of surface association remains to be determined, since the molecule does not contain any known surface attachment moiety.
Our current knowledge of the activity of cysteine proteases in E. histolytica is based on analysis of cultured ameba trophozoites. However, culture medium represents an artificial environment which is not comparable to the situation found during infection of the human host. The various ehcp genes known so far have been cloned by screening E. histolytica cDNA or genomic libraries by using (i) antibodies or N-terminal sequence information of purified ameba enzymes, (ii) PCR-amplified fragments obtained with degenerate primers from regions highly conserved between cysteine protease genes of various organisms, or (iii) already cloned ameba ehcp genes as cross-hybridizing probes. The last approach identified only two additional E. histolytica sequences, since the various ehcp genes have only 40 to 80% sequence identity (8). All of the ameba cp genes encode mature cysteine proteases with calculated molecular masses between 24 and 35 kDa. However, earlier studies using substrate gel electrophoresis suggested that E. histolytica contains a considerable number of cysteine proteases ranging from 16 to more than 100 kDa, some of which were considered to be associated with the ameba membrane (3, 19, 24, 27, 30, 35). This may imply that only a subset of E. histolytica cysteine proteases has been identified so far.
The recent progress in sequencing the E. histolytica genome already allows the identification of nearly all ameba genes, since the amount of sequence information deposited in the two available databases is considered to cover more than 99% of the entire E. histolytica genome.
Here we report the identification and analysis of 12 novel genomic sequences encoding cysteine protease in E. histolytica, of which only 4 are expressed in cultured trophozoites.
MATERIALS AND METHODS
Sequence identification.
Homology searches were performed using the E. histolytica databases of The Institute for Genomic Research (http://www.tigr.org) and The Wellcome Trust Sanger Institute (http://www.sanger.ac.uk). The eight known members of the E. histolytica cysteine protease family were used to query the databases, and additionally identified cysteine protease genes were then used for further analysis of the databases.
The sequencing effort of The Institute for Genomic Research is part of the International Entamoeba Genome Sequencing Project, and the funding for this project is being provided by the National Institute of Allergy and Infectious Diseases. The Wellcome Trust has funded the Sanger Institute Pathogen Sequencing Unit, in collaboration with Graham Clark at the London School of Hygiene and Tropical Medicine, to undertake whole-genome shotgun sequencing of E. histolytica.
Signal sequences and their cleavage sites were identified using the SignalP program (http://www.cbs.dtu.dk/services/SignalP). The prediction of the glycosylphosphatidylinositol (GPI) anchor and cleavage site was made by using DGPI (http://dgpi1.pathbot.com). The prediction of transmembrane helices in proteins was made by using TMHMM (http://www.cbs.dtu.dk/services/TMHMM-2.0/). For the prediction of protein-sorting signals and protein localization sites in cells, PSORTII software was used (http://psort.nibb.ac.jp/). The NetNGly WWW server was used to predict the N-glycosylation sites within the proteases (http://www.cbs.dtu.dk/services/NetNGlyc/) (R. Gupta, E. Jung, and S. Brunak, unpublished data). Homology searches were done using BLAST (http://www.ncbi.nlm.nih.gov/blast/). Multiple alignments were performed by CLUSTAL W (http://www2.ebi.ac.uk/clustalw/) (40). The phylogenetic tree was generated using the alignment performed by CLUSTAL W and the Phylip program package with the neigborhood-joining approach (http://evolution.genetics.washington.edu/phylip.html) (11).
Northern blot analysis.
RNA was isolated from E. histolytica (HM-1:IMSS) and E. dispar (SAW 760) trophozoites by using Trizol reagent (Invitrogen). Since a large number of probes had to be compared, Northern blot analysis was performed in the following way. Two agarose gels, each containing a large slot of about 10 cm, were used. Each slot was loaded with 200 μg of total E. histolytica (gel 1) or E. dispar (gel 2) RNA. After RNA separation, the gels were blotted onto nylon membranes, and subsequently the membranes were cutted into 20 strips. One E. histolytica RNA-containing strip and one E. dispar RNA-containing strip were hybridized in parallel. Each pair of strips was sequentially hybridized with the respective ehcp probe as well as with ehcp9 and an E. histolytica actin gene probe. Hybridization was performed in 0.5 M Na2HPO4-7% sodium dodecyl sulfate-1 mM EDTA (pH 7.2) at 55°C. The blots were washed in 40 mM Na2HPO4-1% sodium dodecyl sulfate (pH 7.2) at 55°C. The following oligonucleotides were used for PCR amplification to generate the various hybridization probes: EhCP8S (5′-GAGA GGTACC ATG TTT GGT TTA CTC TT T GTTACT C), EhCP8AS (5′-GAGA GGATCC TTA AAG ATA TTG TGC ACC TGT T), EhCP9S (5′-GAGA GGTACC ATG TT T GCA GT T AT T CT G TTA GG), EhCP9AS (5′-GAGA GGATCC TTA AAT CTC TTT TAC CCC AAC), EhCP10S (5′-GAGA GGTACC ATG TAT CGA AAT AAC CTT TTT TTT CTT), EhCP10AS (5′-GAGA GGATCC TTA TCC CCA TTC ATT TCC ATA AGA), EhCP11S (5′-GAGA GGATCC ATG ATA GGA GTT GTT TTA GTT TTG), EhCP11AS (5′-GAGA GGATCC TTA GAT GAA GAA AAT CAT TAA AAA GAC), EhCP12S (5′-GAGA GGTACC ATG AGC CAT TTG ATT ATT ATT GTT), EhCP12AS (5′-GAGA GGATCC TCA ATT AAA AAT TAT TGA GCG ATG), EhCP13S (5′-GAGA GGTACC ATG TCA ATT TTG TTT ATC ATT ACT TTT CTG), EhCP13AS (5′-GAGA GGATCC CTA CAT TTC ACC ATC ATC ACC TCC AA), EhCP14S (5′-GAGA GGTACC ATG ATA GAA AAA TTT AAT GCC A), EhCP14AS (5′-GAGA GGATCC TTA GAA AAG CAC CAT AAG ACA TG), EhCP15S (5′-GGG TTA CTT CCA AAG AAA AG), EhCP15AS (5′-TTG TTG TGG TAC TAT GAA GTT), EhCP16S (5′-CAA AAA CAT AAT ATT AAA ATA CAA AAT ATT AAT G), EhCP16AS (5′-GAGA GGATCC TTA TAA TAA TGC ATT TAA TAA TAA TG), EhCP17S (5′-CAA CAC CTT CAG AAA TGT TA), EhCP17AS (5′-AGC TTC ATA TGG ATA CCT TAT TC), EhCP18S (5′-GAT AAA CTT CCC ATG GAA TAT GG), EhCP18AS (5′-TCC ACC ACA ACA TCT TCT AAC G), EhCP19S (5′-GAGA GGTACC ATG TTT TTG TTC CTC GTA TTT C), and EhCP19AS (5′-GAGA GGATCC TTA TTT AAG TCT TTC TAT AGA).
Southern blot analysis.
DNA was isolated from E. histolytica (HM-1:IMSS) and E. dispar (SAW 760) trophozoites by using DNAEasy (InVitrogen). For Southern blot analysis, agarose gels were loaded with 20 μg of restriction enzyme-digested DNA. The transfer was performed as described for the Northern blot analysis.
RESULTS AND DISCUSSION
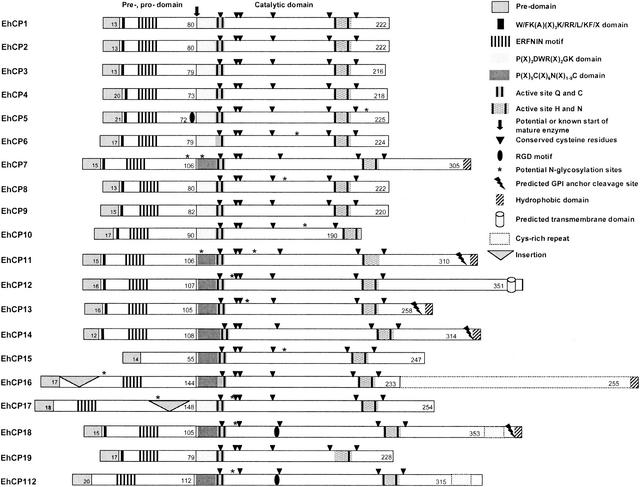
Comprehensive BLAST search analysis of the two currently available E. histolytica genome databases identified a total of 20 different genes with conceptual open reading frames for cysteine proteases of the papain superfamily. These comprise all 8 ehcp genes previously identified (ehcp1 to ehcp7 and ehcp112) as well as 12 so far undescribed ehcp genes, designated ehcp8 to ehcp19. Since the two databases are considered to contain >99% of all E. histolytica genes, it is likely that these 20 ehcp sequences represent the entire repertoire of cysteine protease genes from this protozoan parasite. The various DNA-derived amino acid sequences could be attributed to cysteine proteases since they revealed significant similarity to this class of enzymes from other organisms. In addition, like other cysteine proteases, all of the ameba polypeptides consist of a hydrophobic presequence of 12 to 20 residues, a prodomain of 55 to 148 residues, and a catalytic domain (mature enzyme) of 190 to 488 residues. All cysteine protease-specific signature sequences, the various active-site residues, and six invariant cysteine residues known to be involved in disulfide bridge formation to stabilize the tertiary structure of the enzyme were conserved within the catalytic domain of the various ameba molecules (Fig. 1).
FIG. 1.
Structural organization of 20 E. histolytica cysteine proteases. Numbers indicate the number of amino acid residues forming the predomains, prodomains, or catalytic domains.
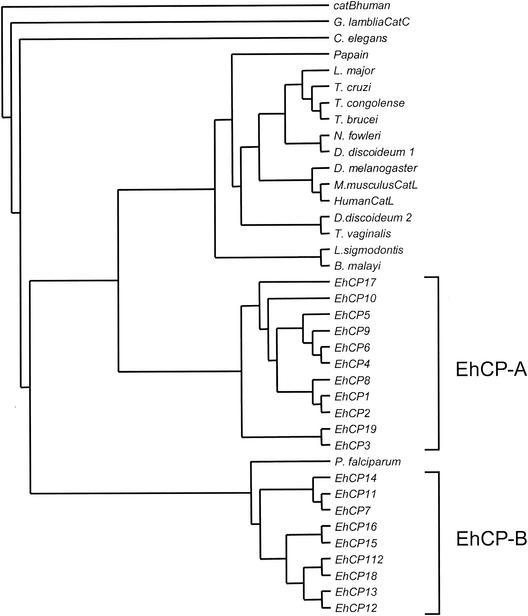
The relatively large number of cp genes within the E. histolytica genome appears to be not simply the result of recent gene amplifications, since pairwise alignments show that the various enzymes have only 10 to 86% sequence identity. In addition, the open reading frames of two of the genes, ehcp14 and ehcp16, are interrupted by short introns of 77 and 59 bp, respectively, both of which contain the ameba-specific 5′ and 3′ splice consensus sequences (45). Construction of phylogenetic trees indicated that the various enzymes form two distinct clades (Fig. 2), suggesting that the 20 ameba cysteine protease genes evolved from two separate ancestors at a very early stage of parasite evolution. Accordingly, the various ameba molecules were grouped into two subfamilies, designated EhCP-A and EhCP-B. In general, proteases belonging to subfamily EhCP-A have shorter prodomains and shorter catalytic domains. The prodomains comprise 72 to 90 residues in subfamily EhCP-A versus 105 to 144 in EhCP-B, and the catalytic domains comprise 190 to 254 residues in EhCP-A versus 230 to 353 in EhCP-B. However, there are two exceptions. EhCP15, which has the shortest prodomain of all ameba enzymes (55 residues), belongs to subfamily EhCP-B, whereas EhCP17, which has the longest prodomain (148 residues), belongs to subfamily EhCP-A. In addition, the two subfamilies can be distinguished on the basis of two sequence characteristics that are located within the N-terminal part of the catalytic domain. Members of EhCP-A contain a conserved Asp-Trp-Arg (DWR) motif at position +6 to +8, and the active-site cysteine is located around position +25. In contrast, members of subfamily EhCP-B do not contain the DWR motif but have a conserved cysteine residue at position +8, and the active-site cysteine is located between positions +30 and +38 (Fig. 3).
FIG. 2.
Phylogenetic tree of all 20 E. histolytica cysteine proteases and representative family members of other organisms. The tree was generated using sequence alignments of the region between the active-site Cys and the active-site His as described in Materials and Methods. Human cathepsin B was used as an outgroup. Two distinct groups are clustered within the E. histolytica protease family (EhCP-A and EhCP-B). The accession numbers for the various proteases are as follows: EhCP1, Q01957; EhCP2, Q01958; EhCP3, CAA60673; EhCP4, CAA62833; EhCP5, CAA62835; EhCP6, CAA62835; EhCP7, CAC34069; EhCP8, AY156066; EhCP9, AY156067; EhCP10, AY156068; EhCP11, AY156096; EhCP12, AY156070; EhCP13, AY156071; EhCP14, AY156072; EhCP15, AY156073; EhCP16, Ay156074; EhCP17, AY156075; EhCP18, AY156076; EhCP19, AY156077; EhCP112, AAF04255; Brugia malayi, AAK16513; human cathepsin B, KHHUB; Dictyostelium discoideum 1, KHDO; Dictyostelium discoideum 2, ACC47482; Drosophila melanogaster, Q95029; Giardia lamblia cathepsin C (CatC), AAK97078; human cathepsin L, KHHUL; Leishmania mexicana, CAA44094; Litomosoides sigmodontis, AAK16515; Mus musculus, NP034114; Naegleria fowleri, AAB01769; Onchocerca volvulus, AAK16514; papain, P00784, Plasmodium falciparum, A45624; Schistosoma mansoni, CAA83538; Trypanosoma brucei, S07051; Trypanosoma congolense, S37048; Trypanosoma cruzi, P25779; and Trichomonas vaginalis, S41427.
FIG. 3.
Comparison between the N termini of the various E. histolytica cysteine protease catalytic domains. Conserved amino acid residues (>50% identity) are printed in bold type, and the active-site residues Gln and Cys are marked by asterisks. For optimal alignment, gaps were introduced into the sequences.
Based on primary sequence and substrate specificity, cysteine proteases can be divided into distinct subclasses such as cathepsin B- or cathepsin L-like enzymes. Sequence alignments revealed the highest similarity of all the ameba polypeptides to members of the cathepsin L-like subclass, irrespective of whether they belong to subfamily EhCP-A or EhCP-B. This was further supported by the finding that 19 of the 20 ameba molecules contain an ERFNIN motif positionally conserved within the pro-region (Fig. 4). This motif, Glu-(X)3-Arg-(X)2-Ile/Val-Phe-(X)2-Asn-(X)3-Ile-(X)3-Asn, and its position within the pro-region are known to be specific for cathepsin L-like proteases (16). However, despite their sequence similarity to cathepsin L-like enzymes, ameba cysteine proteases have previously been shown to be able to degrade Z-Arg-Arg (13, 15, 30, 31), a synthetic peptide that is specifically cleaved by cathepsin B-like enzymes. The substrate specificity of cysteine proteases at the S2 pocket has been ascribed to the chemical properties of the residues corresponding to residue 205 of the papain sequence. Cathepsin B proteases usually have acidic groups (Asp or Glu) at this position, whereas mammalian cathepsin L proteases have an Ala residue at this position (4). Consistent with their substrate specificity, none of the amebic cysteine proteases contain an Ala residue at the postulated S2 pocket site but seven of them contain an Asp residue at this position. A similar observation has been made for cruzipain, a cysteine protease of the protozoan parasite Trypanosoma cruzi. This enzyme shows an overall cathepsin L-like primary structure but has a cathepsin B-like substrate specificity and contains a Glu residue at the postulated S2 pocket site (29).
FIG. 4.
Alignment of the ERFNIN motifs located within the prodomains of the various E. histolytica cysteine proteases. Conserved amino acid residues are printed in bold type.
Four active-site residues, namely, Gln, Cys, His, and Asn, are known to form the catalytic center of cysteine proteases. They can be easily identified on the basis of their location within the primary sequence of the molecules and the fact that they are surrounded by a stretch of well-conserved residues. All ameba cysteine proteases contain the active-site cysteine (Fig. 3), and 16 of 20 have all four residues conserved. Exceptions are EhCP6, EhCP11, EhCP16, and EhCP19, in which one of the residues is substituted (Fig. 3 and 5). Whether this influences the enzyme activity or whether it is due to sequence inaccuracies remains to be determined. However, the latter is less likely, since the respective substitutions were predicted from sequences in both E. histolytica genome databases.
FIG. 5.
Comparison of the E. histolytica cysteine protease sequences spanning the region around active-site residues His and Asn (both marked by asterisks). Conserved amino acid residues are printed in bold type. For optimal alignment, gaps were introduced into the sequences.
Cysteine proteases are located on the surface of a number of cell types. Surface-associated cysteine protease activity has been linked to the invasive properties of tumor cells (20, 34). Likewise, previous studies have suggested that E. histolytica also contains surface-bound cysteine proteases (3, 12, 36). However, the enzymes identified so far do not contain any sequence moiety typical of membrane proteins. In contrast, 7 of the 12 newly identified ameba enzymes, which all belong to the EhCP-B subfamily, have hydrophobic sequences near or directly at the C terminus, which are predicted to constitute transmembrane regions or to be further processed to allow attachment via a GPI anchor. To our knowledge, this property appears to be unique, since it has not been described for any cysteine protease found in higher eukaryotes.
In addition to membrane attachment moieties, EhCP16 contains a C-terminal insertion. This insertion is extraordinary rich in cysteine residues (18%) and has a repetitive structure with the sequence C-(X)5-C-(X2)-C-(X)6-C-(X)10-C-(X)3-C-(X)3-C-(X)2)6-C-(X)6-C-(X)2-C-(X)10-C (Fig. 1). This repeat has significant sequence similarity to furin and PACE 4A, proteins belonging to a family of Ca2+-dependent serine proteases (pro-protein convertases), which have homology to the endoproteases subtilisin (bacteria) and kexin (yeast) (44). These enzymes are responsible for the conversion of precursors of peptide hormones, neuropeptides, and many other proteins into their biologically active forms (6, 23). Furin is expressed in all tissues and cell lines examined so far and is usually localized within the trans-Golgi (23). For PACE 4, it has been postulated that it plays a functional role in the activation of membrane-type metalloproteases and consequently in tumor cell invasion and tumor progression (5, 23). The cysteine-rich repeat close to the C terminus of the protein is considered to play a role in intracellular routing or membrane association (4). In addition to EhCP16, EhCP112 and EhCP18 contain the N-terminal part of the cysteine-rich repeat, C-(X)5-C-(X)2-C-(X)6-C-(X)2-C-(X)11-C, close to the C terminus of the proteins (Fig. 1).
Many of the amebic cysteine proteases contain at least one potential acceptor site for N-linked glycosylation. So far, glycosylation of ameba enzymes has not been determined. It is known for cathepsin L that glycosylation supports protein stability and correct intracellular targeting (10). For some mammalian proteases, glycosylation of the prodomain and subsequent binding to the mannose-6-phosphate receptor is necessary for targeting the molecules to the lysosomal pathway (17). However, only two of the E. histolytica proteases contain putative N- glycosylation sites within the prodomain (Fig. 1). In addition to the mannose-6-phosphate-dependent pathway, a mannose-6-phosphate-independent pathway has been described for cathepsin D and cathepsin L (21, 48). The mechanism is based on the recognition of membrane-associated receptors by the prodomain, which is also known for cruzipain and which has been attributed to the presence of the prodomain sequence Trp/Phe-Lys-(Ala)-X-X-X-Lys/Arg-Arg/Leu/Val-Phe/Tyr (14). Interestingly, this motif is present in Giardia cathepsin Bs (29) and a similar sequence is found in most of the E. histolytica enzymes (EhCP1-14 and EhCP18-19 [Fig. 1]).
Another class of receptors that may support the cellular targeting of proteins are integrins. In higher eukaryotes, these transmembrane heterodimers bind various matrix proteins, all of which have a tripeptide consensus sequence, Arg-Gly-Asp (RGD), in common. This motif serves to attach proteins to the surface without transmembrane-spanning domains or GPI anchors (28, 41). Interestingly, three of the E. histolytica enzymes (EhCP5, EhCP18, and EhCP112) contain an RGD motif, which lie within the catalytic domain in two of the proteins (Fig. 1).
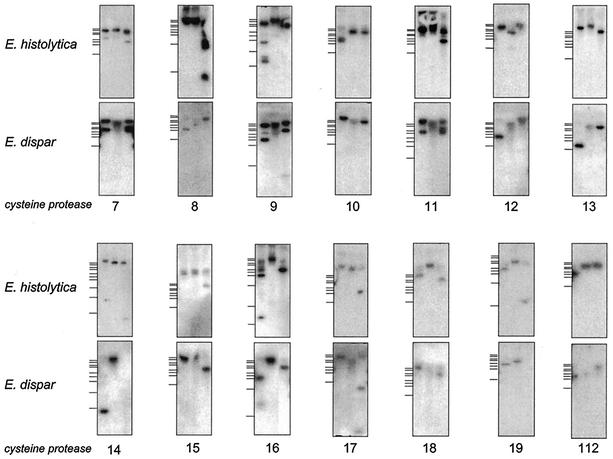
Previous studies have shown that on average, protein-coding regions in E. histolytica and the closely related but nonpathogenic species E. dispar have 95% sequence identity (46). This has allowed the identification and cloning of corresponding cp genes from E. dispar (edcp2, edcp3, edcp4, and edcp6) by using ehcp sequences as cross-hybridizing probes (8). Interestingly, this approach revealed that functional orthologs corresponding to ehcp1 and ehcp5 are absent in E. dispar (8, 46). Accordingly, comparative Southern blot analyses were performed using ehcp7 and the newly identified ehcp genes as probes. The results indicate that orthologous sequences to all of these genes are present in E. dispar (Fig. 6). Thus, with regard to the pathogenic property of E. histolytica, EhCP1 and EhCP5 appear to be of particular importance.
FIG. 6.
Southern blot analysis of E. histolytica and E. dispar genomic DNA. Total genomic DNAs of either the E. histolytica isolate HM-1:IMSS or the E. dispar isolates SAW 760 were digested using HincII, HindIII, and NdeI, separated on agarose gels, and blotted onto nylon membranes. The blots were sequentially hybridized with the coding regions of all 20 E. histolytica cysteine protease genes. Hybridization and washing were performed as described in Materials and Methods. Size markers are indicated on the left. They represent 10, 8, 6, 5, 4, 3.5, 3, 2, 1, and 0.5 kb.
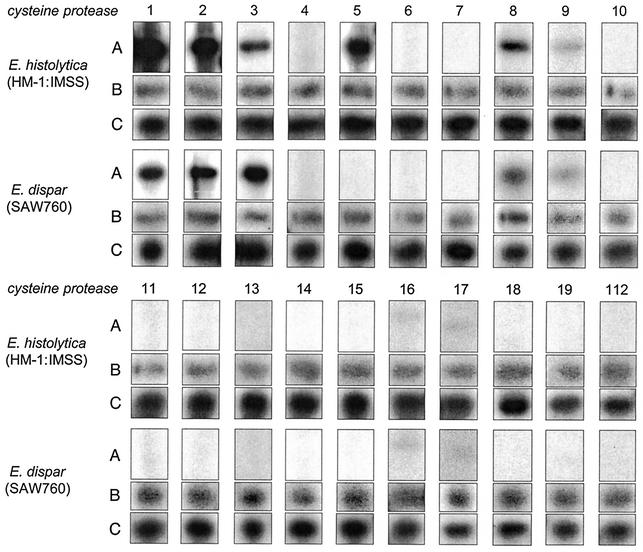
Based on protein and RNA analyses, previous studies have suggested that EhCP1, EhCP2, and EhCP5 are the most abundantly expressed cysteine proteases in cultured E. histolytica trophozoites, whereas the highest expression in E. dispar was found for EdCP3 (8). To extend these analyses, comparative Northern blots were prepared using RNA isolated from cultured E. histolytica and E. dispar trophozoites. Subsequently, these blots were hybridized with the various ehcp-coding sequences. As sampling controls, all of the blots were subsequently hybridized with probes derived from a highly expressed and from a poorly expressed E. histolytica gene (Fig. 7). The results confirmed previous observations that ehcp1, ehcp2, and ehcp5 are indeed strongly expressed in E. histolytica. In addition, a medium level of expression was found for ehcp3, ehcp8, and ehcp9 since the hybridization signals became positive within 24 h of exposure whereas ehcp16 and ehcp17 revealed only very low levels of expression since the hybridization signals became visible only after at least 5 days of exposure. Interestingly, 12 of the 20 ehcp genes revealed no hybridization signal at all, even after extended exposure of the blots for more than 10 days, indicating that the majority of ehcp genes are not expressed in cultured E. histolytica trophozoites. Likewise, a minority of only 6 cp genes were found to be expressed in cultured E. dispar trophozoites. As seen in E. histolytica, hybridization signals were obtained with ehcp1, ehcp2, ehcp3, ehcp8, ehcp9, ehcp16, and ehcp17. (Note that the signal with ehcp1 is due to cross-hybridization to edcp2, since the two genes have 86% sequence identity.) With the exception of ehcp3, the intensities of the signals in E. dispar were generally lower than in E. histolytica (Fig. 7). Thus, these Northern blot results are fully consistent with previous protein data, which indicated that the amount of cysteine protease is considerablely larger in E. histolytica than in E. dispar (13, 27).
FIG. 7.
Expression of the various cysteine protease genes in the E. histolytica isolate HM-1: IMSS and the E. dispar isolate SAW 760. (A) Total cellular RNA of each isolate were separated on formaldehyde-agarose gels, blotted onto nylon membranes, and hybridized with the coding region of the various E. histolytica cysteine protease genes as indicated. (B and C) As controls, the blots were stripped and sequentially hybridized with ehcp9 (B) and an E. histolytica actin gene probe (C).
Gene expression in E. histolytica is performed in a monocistronic fashion (9), and both 5′ upstream and 3′ downstream regions contribute to the regulation of transcription (25). Thus, sequences within the flanking regions might contribute to the differences in expression of the various ehcp genes. Interestingly, analysis of the flanking regions revealed that the six ehcp genes found to be substantially expressed in cultured ameba trophozoites (ehcp1, ehcp2, ehcp3, ehcp5, ehcp8, and ehcp9) contain an extended TATA box-like motif of the sequence TATTTAAACT, about 35 bp upstream of the translation initiation ATG. In contrast, this motif is absent in all of the remaining genes, which revealed no hybridization signal on Northern blots or which required extended exposure for at least 5 days.
Questions remain about the role of the various cysteine proteases in Entamoeba. Obviously, only a small subset of the enzymes is required to support growth during in vitro cultivation. However, because all 20 ehcp genes contain full-length open reading frames, it is likely that all of the genes and the corresponding enzymes are functional. Genes that are no longer required in a given organism are thought to become inactive and rapidly degenerate, resulting in the destruction of the open reading frame, as has been shown for the ehcp5 homolog in E. dispar (46). Most of the current knowledge about the function of cysteine proteases in Entamoeba has been accumulated based on the analysis of cultured trophozoites. Thus, any possible role of enzymes that are not expressed during in vitro cultivation remains to be determined. However, it is tempting to speculate that these enzymes are important for the parasite during infection of the human host or for completion of the life cycle. They may be required to support the growth and/or survival of ameba during colonization of the human intestine and may enable the digestion of material which is usually absent from the culture medium, e.g., red blood cells and bacteria. Some of the molecules, in particular those predicted to be located on the membrane, may play a role during parasite invasion and destruction of host tissue. Another function could be the involvement in the process of parasite encystation and/or excystation, as has been recently shown for a cathepsin C-like enzyme in Giardia lamblia (42).
In summary, the results presented here indicate that E. histolytica contains considerably more cysteine proteases than previously suggested. The multitude of enzymes, their differences in primary structure, and the fact that they are conserved across different ameba species most probably indicate that many of these enzymes have distinct functions within the organism. Elucidation of the precise role of each of the various enzymes appears to be a major challenge but may help us understand some of the unique properties of this intestinal protozoan parasite.
Acknowledgments
This work was supported by the Deutschen Forschungsgemeinschaft (grant BR 1744/1-5).
REFERENCES
- 1.Ankri, S., T. Stolarsky, R. Bracha, F. Padilla-Vaca, and D. Mirelman. 1999. Antisense inhibition of expression of cysteine proteinases affects Entamoeba histolytica-induced formation of liver abscess in hamsters. Infect Immun. 67:421-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ankri, S., T. Stolarsky, and D. Mirelman. 1998. Antisense inhibition of expression of cysteine proteinases does not affect Entamoeba histolytica cytopathic or haemolytic activity but inhibits phagocytosis. Mol. Microbiol. 28:777-785. [DOI] [PubMed] [Google Scholar]
- 3.Avila, E. E., and J. Calderon. 1993. Entamoeba histolytica trophozoites: a surface-associated cysteine protease. Exp. Parasitol. 76:232-241. [DOI] [PubMed] [Google Scholar]
- 4.Barrett, A. J. 1998. Cysteine peptidase. In A. J. Barrett, N. D. Rawlings, and J. F. Woessner (ed.), Handbook of proteolytic enzymes. Academic Press, Inc., San Diego, Calif.
- 5.Bassi, D. E., H. Mahloogi, and A. J. Klein-Szanto. 2000. The proprotein convertases furin and PACE4 play a significant role in tumor progression. Mol. Carcinog. 28:63-69. [PubMed] [Google Scholar]
- 6.Beinfeld, M. C. 1998. Prohormone and proneuropeptide processing. Recent progress and future challenges. Endocrine 8:1-5. [DOI] [PubMed] [Google Scholar]
- 7.Berti, P. J., and A. C. Storer. 1995. Alignment/phylogeny of the papain superfamily of cysteine proteases. J. Mol. Biol. 246:273-283. [DOI] [PubMed] [Google Scholar]
- 8.Bruchhaus, I., T. Jacobs, M. Leippe, and E. Tannich. 1996. Entamoeba histolytica and Entamoeba dispar: differences in numbers and expression of cysteine proteinase genes. Mol. Microbiol. 22:255-263. [DOI] [PubMed] [Google Scholar]
- 9.Bruchhaus, I., M. Leippe, C. Lioutas, and E. Tannich. 1993. Unusual gene organization in the protozoan parasite Entamoeba histolytica. DNA Cell Biol. 12:925-933. [DOI] [PubMed] [Google Scholar]
- 10.Dwek, R. A. 1998. Biological importance of glycosylation. Dev. Biol. Stand. 96:43-47. [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1989. PHYLIP—Phylogeny Inference Package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 12.Garcia-Rivera, G., M. A. Rodriguez, R. Ocadiz, M. C. Martinez-Lopez, R. Arroyo, A. Gonzalez-Robles, and E. Orozco. 1999. Entamoeba histolytica: a novel cysteine protease and an adhesin form the 112 kDa surface protein Mol. Microbiol. 33:556-568. [DOI] [PubMed] [Google Scholar]
- 13.Hellberg, A., R. Nickel, H. Lotter, E. Tannich, and I. Bruchhaus. 2001. Overexpression of cysteine proteinase 2 in Entamoeba histolytica or Entamoeba dispar increases amoeba-induced monolayer destruction in vitro but does not augment amoebic liver abscess formation in gerbils Cell. Microbiol. 3:13-20. [DOI] [PubMed] [Google Scholar]
- 14.Huete-Perez, J. A., J. C. Engel, L. S. Brinen, J. C. Mottram, and J. H. McKerrow. 1999. Protease trafficking in two primitive eukaryotes is mediated by a prodomain protein motif. J. Biol. Chem. 274:16249-16256. [DOI] [PubMed] [Google Scholar]
- 15.Jacobs, T., I. Bruchhaus, T. Dandekar, E. Tannich, and M. Leippe. 1998. Isolation and molecular characterization of a surface-bound proteinase of Entamoeba histolytica. Mol. Microbiol. 27:269-276. [DOI] [PubMed] [Google Scholar]
- 16.Karrer, K. M., S. L. Peiffer, and M. E. DiTomas. 1993. Two distinct gene subfamilies within the family of cysteine protease genes. Proc. Natl. Acad. Sci. USA 90:3063-3067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kornfeld, S., and I. Mellman. 1989. The biogenesis of lysosomes. Annu. Rev. Cell Biol. 5:483-525. [DOI] [PubMed] [Google Scholar]
- 18.Li, E., W. G. Yang, T. Zhang, and S. L. Stanley, Jr. 1995. Interaction of laminin with Entamoeba histolytica cysteine proteinases and its effect on amebic pathogenesis. Infect. Immun. 63:4150-4153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lushbaugh, W. B., A. F. Hofbauer, and F. E. Pittman. 1985. Entamoeba histolytica: purification of cathepsin B. Exp. Parasitol. 59:328-336. [DOI] [PubMed] [Google Scholar]
- 20.Mai, J., D. M. Waisman, and B. F. Sloane. 2000. Cell surface complex of cathepsin B/annexin II tetramer in malignant progression. Biochim. Biophys. Acta 1477:215-230. [DOI] [PubMed] [Google Scholar]
- 21.McIntyre, G. F., G. D. Godbold, and A. H. Erickson. 1994. The pH-dependent membrane association of procathepsin L is mediated by a 9- residue sequence within the propeptide. J. Biol. Chem. 269:567-572. [PubMed] [Google Scholar]
- 22.McKerrow, J. H. 1989. Parasite proteases. Exp. Parasitol. 68:111-115. [DOI] [PubMed] [Google Scholar]
- 23.Nakayama, K. 1997. Furin: a mammalian subtilisin/Kex2p-like endoprotease involved in processing of a wide variety of precusor proteins. Biochem. J. 327:625-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Navarro-Garcia, F., L. Chavez-Duenas, V. Tsutsumi, F. Posadas del Rio, and R. Lopez-Revilla. 1995. Entamoeba histolytica: increase of enterotoxicity and of 53- and 75-kDa cysteine proteinases in a clone of higher virulence. Exp. Parasitol. 80:361-372. [DOI] [PubMed] [Google Scholar]
- 25.Nickel, R., and E. Tannich. 1994. Transfection and transient expression of chloramphenicol acetyltransferase gene in the protozoan parasite Entamoeba histolytica. Proc. Natl. Acad. Sci. USA 91:7095-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reed, S., J. Bouvier, A. S. Pollack, J. C. Engel, M. Brown, K. Hirata, X. Que, A. Eakin, P. Hagblom, F. Gillin, and J. H. Mckerrow. 1993. Cloning of a virulence factor of Entamoeba histolytica. Pathogenic strains possess a unique cysteine proteinase gene. J. Clin. Investig. 91:1532-1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reed, S. L., W. E. Keene, and J. H. McKerrow. 1989. Thiol proteinase expression and pathogenicity of Entamoeba histolytica. J. Clin. Microbiol. 27:2772-2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruoslahti, E. 1996. RGD and other recognition sequences for integrins. Annu. Rev. Cell Dev. Biol. 12:697-715. [DOI] [PubMed] [Google Scholar]
- 29.Sajid, M., and J. H. McKerrow. 2002. Cysteine proteases of parasitic organisms. Mol. Biochem. Parasitol. 120:1-21. [DOI] [PubMed] [Google Scholar]
- 30.Scholze, H., and W. Schulte. 1988. On the specificity of a cysteine proteinase from Entamoeba histolytica. Biomed. Biochim. Acta 47:115-123. [PubMed] [Google Scholar]
- 31.Scholze, H., and E. Tannich. 1994. Cysteine endopeptidases of Entamoeba histolytica. Methods Enzymol. 244:512-523. [DOI] [PubMed] [Google Scholar]
- 32.Schulte, W., and H. Scholze. 1989. Action of the major protease from Entamoeba histolytica on proteins of the extracellular matrix. J. Protozool. 36:538-543. [DOI] [PubMed] [Google Scholar]
- 33.Sloane, B. F., J. Rozhin, J. S. Hatfield, J. D. Crissman, and K. V. Honn. 1987. Plasma membrane-associated cysteine proteinases in human and animal tumors. Exp. Cell Biol. 55:209-224. [DOI] [PubMed] [Google Scholar]
- 34.Sloane, B. F., J. Rozhin, K. Johnson, H. Taylor, J. D. Crissman, and K. V. Honn. 1986. Cathepsin B: association with plasma membrane in metastatic tumors. Proc. Natl. Acad. Sci. USA 83:2483-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spice, W. M., and J. P. Ackers. 1993. Influence of bacteria on electrophoretic proteinase patterns of Entamoeba histolytica isolates. Int. J. Parasitol. 23:671-674. [DOI] [PubMed] [Google Scholar]
- 36.Spinella, G. M. 1999. Purification and biochemical characterization of a novel cysteine protease of Entamoeba histolytica. Eur. J. Biochem. 266:170-180. [DOI] [PubMed] [Google Scholar]
- 37.Stanley, S. L., Jr., T. Zhang, D. Rubin, and E. Li. 1995. Role of the Entamoeba histolytica cysteine proteinase in amebic liver abscess formation in severe combined immunodeficient mice. Infect. Immun. 63:1587-1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tannich, E., R. Nickel, H. Buss, and R. D. Horstmann. 1992. Mapping and partial sequencing of the genes coding for two different cysteine proteinases in pathogenic Entamoeba histolytica. Mol. Biochem. Parasitol. 54:109-111. [DOI] [PubMed] [Google Scholar]
- 39.Tannich, E., H. Scholze, R. Nickel, and R. D. Horstmann. 1991. Homologous cysteine proteinases of pathogenic and nonpathogenic Entamoeba histolytica. Differences in structure and expression. J. Biol. Chem. 266:4798-4803. [PubMed] [Google Scholar]
- 40.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Torshin, I. 2002. Structural criteria of biologically active RGD-sites for analysis of protein cellular function D a bioinformatics study. Med. Sci. Monit. 8:BR301-BR312. [PubMed] [Google Scholar]
- 42.Touz, M. C., M. J. Nores, I. Slavin, C. Carmona, J. T. Conrad, M. R. Mowatt, T. E. Nash, C. E. Coronel, and H. D. Lujan. 2002. The activity of a developmentally regulated cysteine proteinase is required for cyst wall formation in the primitive eukaryote Giardia lamblia. J. Biol. Chem. 277:8474-8481. [DOI] [PubMed] [Google Scholar]
- 43.Turk, B., D. Turk, and V. Turk. 2000. Lysosomal cysteine proteases: more than scavengers. Biochim. Biophys. Acta 1477:98-111. [DOI] [PubMed] [Google Scholar]
- 44.Van de Ven, W. J., J. W. Creemers, and A. J. Roebroek. 1991. Furin: the prototype mammalian subtilisin-like proprotein-processing enzyme. Endoproteolytic cleavage at paired basic residues of proproteins of the eukaryotic secretory pathway. Enzyme 45:257-270. [DOI] [PubMed] [Google Scholar]
- 45.Willhoeft, U., E. Campos-Gongora, S. Touzni, I. Bruchhaus, and E. Tannich. 2001. Introns of Entamoeba histolytica and Entamoeba dispar. Protist 152:149-156. [DOI] [PubMed] [Google Scholar]
- 46.Willhoeft, U., L. Hamann, and E. Tannich. 1999. A DNA sequence corresponding to the gene encoding cysteine proteinase 5 in Entamoeba histolytica is present and positionally conserved but highly degenerated in Entamoeba dispar. Infect. Immun. 67:5925-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang, Z., L. Wang, K. B. Seydel, E. Li, S. Ankri, D. Mirelman, and S. L. Stanley, Jr. 2000. Entamoeba histolytica cysteine proteinases with interleukin-1 beta converting enzyme (ICE) activity cause intestinal inflammation and tissue damage in amoebiasis. Mol. Microbiol. 37:542-548. [DOI] [PubMed] [Google Scholar]
- 48.Zhu, Y., and G. E. Conner. 1994. Intermolecular association of lysosomal protein precursors during biosynthesis. J. Biol. Chem. 269:3846-3851. [PubMed] [Google Scholar]