Abstract
The Candida albicans vacuole has previously been observed to undergo rapid expansion during the emergence of a germ tube from a yeast cell, to occupy the majority of the parent yeast cell. Furthermore, the yeast-to-hypha switch has been implicated in the virulence of this organism. The class C vps (vacuolar protein sorting) mutants of Saccharomyces cerevisiae are defective in multiple protein delivery pathways to the vacuole and prevacuole compartment. In this study C. albicans homologues of the S. cerevisiae class C VPS genes have been identified. Deletion of a C. albicans VPS11 homologue resulted in a number of phenotypes that closely resemble those of the class C vps mutants of S. cerevisiae, including the absence of a vacuolar compartment. The C. albicans vps11Δ mutant also had much-reduced secreted lipase and aspartyl protease activities. Furthermore, vps11Δ strains were defective in yeast-hypha morphogenesis. Upon serum induction of filamentous growth, mutants showed delayed emergence of germ tubes, had a reduced apical extension rate compared to those of control strains, and were unable to form mature hyphae. These results suggest that Vps11p-mediated trafficking steps are necessary to support the rapid emergence and extension of the germ tube from the parent yeast cell.
The fungal vacuole is an acidic, membrane-bound compartment containing a variety of hydrolytic enzymes. The functions of the fungal vacuole have been well defined for model fungi such as Saccharomyces cerevisiae and Aspergillus nidulans. These include degradation of cellular proteins for the recycling of amino acids and storage of cellular metabolites such as amino acids, phosphate, and metal ions (25). A functional vacuole is necessary for cytoplasmic homeostasis, and several S. cerevisiae mutants lacking an intact vacuole are sensitive to external pH and osmotic pressure (4). Regulated uptake and release of stored nutrients by the vacuole also help to maintain stable nutrient concentrations within the cytoplasm (24, 31). While the vacuoles' functions are not essential for vegetative growth, they are of increased importance, and often essential, for survival during periods of stress such as starvation or growth at elevated temperatures (4, 45). In addition, the vacuole may play a central role during processes of differentiation; for example, S. cerevisiae mutants defective in vacuolar hydrolase activity are unable to undergo the process of sporulation (51). This suggests that the vacuole may function in adaptation to new environments.
To date little has been reported about the specific functions of the vacuole in pathogenic species. It was postulated that the vacuole is likely to play a key role during adaptation of a pathogenic fungus to different host environments. Furthermore, vacuolar functions may be required for processes of cellular differentiation. An intensively studied example of such a differentiation process is the yeast-hypha transition in the fungal pathogen Candida albicans, which has been strongly implicated in the pathogenicity of this organism (10). Previous work by Gow and Gooday (20) studied the morphology of the C. albicans vacuole through the yeast-hypha transition. It was observed that germ tube emergence is accompanied by a rapid increase in vacuolar volume (20). The rapid expansion of the vacuole coincides with migration of parental cytoplasmic material into the hyphal tip. As the germ tube extends, subapical compartments can also be identified; these are composed almost entirely of vacuole and contain little protoplasm, whereas the protoplasm migrates with the hyphal tip. After a delay period of apparent inactivity, the highly vacuolated compartments regenerate cytoplasm and the vacuoles recede. Formation of secondary germ tubes from mother cells or of branches from subapical compartments occurs only after cytoplasmic regeneration, when the cytoplasm again migrates into the newly synthesized hyphal tip, leaving behind a highly vacuolated parent cell. At present the mechanism by which vacuole expansion occurs during germ tube formation and its regulation remain obscure. However, one major advantage of this mechanism may be that it requires minimal amounts of de novo cytoplasmic biosynthesis while allowing C. albicans to germinate rapidly and explore its environment or forage for nutrients under limiting conditions. Such a developmental model is unusual in filamentous fungi, and this mechanism may be unique to C. albicans.
More than 40 S. cerevisiae vacuolar protein sorting (vps) mutants, which missort the vacuolar hydrolase carboxypeptidase Y (CPY) to the cell surface, have been identified (3, 35, 36, 37). The corresponding protein products define components required for the delivery of vacuolar hydrolases from the Golgi apparatus to the vacuole. The class C vps mutants, vps11 (pep5), vps16, vps18 (pep3), and vps33, exhibit the most severe phenotypic defects of all vps mutants (3, 35). The class C VPS genes of S. cerevisiae encode large hydrophilic proteins, which physically interact in a complex on the cytosolic surface of the vacuolar membrane (32). These proteins are required for the delivery of proteins to the vacuole from the Golgi apparatus, from the cell surface via endocytosis, from the cytoplasm via the cytoplasm-to-vacuole trafficking route, and through autophagy (32, 35, 49). The class C Vps complex mediates the docking of transport vesicles with the vacuolar membrane through the regulation of vacuolar t-SNARE (target soluble N-ethylmaleimide-sensitive factor attachment protein receptors) (Vam3p and Vam7p) and transport vesicle vehicular (v)-SNARE (Vti1p) interaction, to form the trans-SNARE complex (40), a critical step leading to membrane fusion. Furthermore, it has been shown that Vps11p interacts with Vps39p, the guanine nucleotide exchange factor for Ypt7p, a Rab GTPase that mediates the initial “tethering” of vesicle and target membranes (50). Thus, the Class C Vps complex has an essential function in coordinating the vesicle fusion machinery at the vacuolar surface.
In addition, Vps11p and Vps18p have been shown to function at earlier steps in the CPY and endocytotic delivery pathways (42). Woolford et al. (49) uncovered a genetic interaction between VPS11 and VPS8, a gene known to have functions in the recycling transport step from the prevacuole compartment (PVC) to the Golgi apparatus. This was later supported by analysis of vps11 and vps18 temperature-sensitive mutant strains (42). Synthetic interactions were also observed between vps11ts and vps18ts alleles and genes known to function in Golgi-to-PVC transport, including PEP12 and PEP7 (42), which encode an endosomal t-SNARE and SNARE regulator, respectively. The same study revealed that VPS18 function was required for endocytotic transport from the cell surface to the PVC. As such, deletion of the class C VPS genes results in a range of secondary phenotypes, including the missorting of multiple vacuolar hydrolases as inactive precursors, an inability to grow at 37°C, and sensitivity to osmotic stress (4, 5, 14, 30, 42). The class C mutants also show reduced activity of Kex2p (34), an endopeptidase localized to a trans-Golgi compartment, which proteolytically cleaves secreted peptides such as the α-factor precursor (17). Class C vps mutants lack a recognizable vacuole compartment and instead accumulate a range of different-sized vesicles, presumed to be transport intermediates destined for the vacuole (4, 32). In summary, both Vps11p and Vps18p function at multiple steps in vesicle-mediated transport to the vacuole and are essential for vacuolar biogenesis. Since this project was undertaken, a C. albicans strain with a deletion of a vps gene (C. albicans VPS34) was reported as defective in filamentation (8). This supports the hypothesis that vps trafficking pathways may be required for filamentation.
The aims of this study were, first, to generate a C. albicans mutant strain defective in vacuolar protein delivery pathways and vacuole biogenesis and, second, to assess how germ tube emergence is affected by these defects.
MATERIALS AND METHODS
Strains.
The C. albicans strains used in this study are listed in Table 1. Strain BWP17 was provided by Aaron Mitchell (Columbia University) (48). Strain YJB6284 (prototrophic strain derived from BWP17) was provided by Judy Berman (University of Minnesota) (6). C. albicans was transformed using the lithium acetate procedure (18). The PCR-based gene disruption method (48) was utilized to construct VPS11 heterozygous and null strains. Oligonucleotides (11D5 and 11D3 [Table 2 ]) for disruption were designed so that each primer contained 20 bp homologous to PCR disruption plasmids flanked with 70 bp of sequence to direct homologous integration into the VPS11 open reading frame (ORF). The primer set was used in PCR amplifications with plasmids pRS-ARGΔSpeI and pGEM-URA3. Two nanograms of plasmid DNA was amplified with forward and reverse primers, each at a 1 μM concentration, by using Life Technologies Taq polymerase and reagents according to the manufacturer's instructions or by using Ready-To-Go PCR Beads (Amersham Pharmacia Biotech). Amplification conditions were as follows: 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 1 min/kb, with a final step of 72°C for 10 min. The resulting vps11::ARG4 and vps11::URA3 PCR products consist of a 5′ targeting sequence (+17 to +87 nucleotides [nt] from the ATG start site of VPS11) and a 3′ targeting sequence (+3233 to +3302 nt), separated by either the ARG4 or the URA3 marker. The vps11::ARG4 product was transformed into BWP17 to generate four heterozygous strains, GPS1 to GPS4. Strains GPS1 to -4 were transformed with vps11::URA3 to generate three vps11::ARG4/vps11::URA3 double-knockout strains, GPD1 to GPD3. Arg+ and Arg+ Ura+ transformants were screened for the correct integration event by Southern hybridization using the VPS11-specific probe excised from plasmid pGP115. VPS11 revertant strains were constructed by reintroducing a copy of VPS11 into the his1 locus of the null strain GPD1. Plasmids pGEM-HIS1 and pGH11 were linearized at the unique NruI site in the HIS1 flanking sequences to target integration into the his1 locus of the vps11Δ double-mutant strain GPD1. GPD1 was transformed with linearized pGH11 (GPR1 to GPR3) or linearized pGEM-HIS1 (GPH1 to GPH3). Integration of these plasmids at his1 was confirmed by PCR using a primer flanking the his1 locus (HIS3AMP [Table 2]) combined with either a primer within the VPS11 insert sequences (11HISR [Table 2]) or a primer within the pGEM-HIS1 sequences (GEMHISR [Table 2]). Multiple isolates for each constructed strain were subject to phenotypic analysis. As expected, no phenotypic differences were observed for any given genotype.
TABLE 1.
C. albicans strains used in this study
| Strain | Parent | Relevant genotype | Source or reference |
|---|---|---|---|
| BWP17 | SC5314 | ura3Δ/ura3Δ arg4Δ/arg4Δ his1Δ/his1Δ VPS11/VPS11 | 48 |
| GPS1-4 | BWP17 | ura3Δ/ura3Δ arg4Δ/arg4Δ his1Δ/his1Δ VPS11/vps11Δ::ARG4 | This study |
| GPD1-3 | GPS1-2 | ura3Δ/ura3Δ arg4Δ/arg4Δ his1/his1Δ vps11Δ::ARG4/vps11Δ::URA3 | This study |
| GPR1-3 | GPD1 | ura3Δ/ura3Δ arg4Δ/arg4Δ his1Δ/his1Δ::HIS1::VPS11 vps11Δ::ARG4/vps11Δ::URA3 | This study |
| GPH1-3 | GPD1 | ura3Δ/ura3Δ arg4Δ/arg4Δ his1Δ/his1Δ::HIS1 vps11Δ::ARG4/vps11Δ::URA3 | This study |
| YJB6284 | BWP17 | ura3Δ/ura3Δ arg4Δ/arg4Δ::ARG4URA3 his1Δ/his1Δ::HIS1 VPS11/VPS11 | 6 |
| S/01 | Clinical isolate | URA3/URA3 ARG4/ARG4 HIS1/HIS1 VPS11/VPS11 | R. Matthews, University of Manchester, Manchester, United Kingdom |
TABLE 2.
Primers used in this study
| Primer | Sequencea |
|---|---|
| CAEND1 | CTAACCATTCCTCCTTGTCGCC |
| CAEND2 | CTTCCACCATGAACCAACCCC |
| CAPEP31 | GATCAAGCCTTAGAACTTGCCC |
| CAPEP32 | CCTTGTGGAACGCATGATGGC |
| 11D5 | TTCTCCTTCGAGGACAATATCATCTCCTCTATTATCATTATCATCATGG AGACAGTTCCAATTGTTTGATTGTGGAATTGTGAGCGGATA |
| 11D3 | CTATTGATCTTCCATTACACCTTTACCAATATAATCAGAAACAAATTTA AATTTATCATTACTAGAATGCTTTCCCAGTCACGACGTT |
| 11PB5 | CCTATCATTTTCGGTGTTAGG |
| 11PB3 | GGTGTAATGAATGTAATTGGC |
| 11R5 | TCAGGATCCAGGCAATACATGTGATTCG |
| 11R3 | TCAGGATCCATTGGGAGTCCCCCCACTCC |
| GEMHISR | CTCCCGGCCGCCATGGCC |
| 11HISR | ACTGGGTGCTAGTTCTTTCC |
| HIS3AMP | GTTGGTGTGGCCCAGAGAC |
The introduced BamHI site is underlined.
Media and growth conditions.
Strains were routinely grown on yeast extract-peptone-dextrose (YPD) supplemented with uridine (50 μg/ml) (YPD + uri) at 30°C. Minimal medium (yeast nitrogen base [YNB]) was supplemented with the appropriate auxotrophic requirements as described for S. cerevisiae (9) except for uridine (50 μg/ml). The yeast-to-hypha transition was induced in either liquid or solid (inclusion of 2% agar) medium. The transition medium was either M199 medium containing Earle's salts and glutamine (Invitrogen), buffered with 150 mM HEPES and adjusted to pH 7.5, or 10% fetal calf serum (FCS) in YPD. Filamentation in liquid media was induced by inoculating 2 × 106 to 5 × 106 cells/ml and incubating at 37°C with shaking (200 rpm). For solid surfaces, 5 × 105 cells in 5 μl of water were spotted onto plates and incubated for 2 to 5 days at 37°C.
The growth of vps11Δ double-knockout, heterozygous, and revertant strains was compared with that of the parental strain, BWP17, and the prototrophic strain, YJB6284, under a range of growth conditions described below. Strains were grown overnight in YPD + uri at 30°C, pelleted, and resuspended in sterile distilled water at 1 × 107, 2 × 106, 4 × 105, or 8 × 104 cells/ml. Each cell suspension was then transferred to a 96-well plate and applied to a solid test agar by using a sterile multipronged applicator. To test temperature sensitivity, cells were plated onto YPD + uri agar and incubated at 30, 37, or 42°C. To test osmotic sensitivity, cells were plated onto YPD + uri agar supplemented with either 1 M NaCl or 2.5 M glycerol and were incubated at 30°C.
Plasmids.
The plasmids used for templates in PCR amplifications, pGEM-HIS1, pRS-ARGΔSpeI, and pGEM-URA3, were kindly provided by Aaron Mitchell (Columbia University) (48). The VPS11 ORF, including 338 nt of upstream promoter and 201 nt of downstream 3′ sequence, was PCR amplified from C. albicans strain BWP17 genomic DNA with primers 11R5 and 11R3 (Table 2), and the 3.8-kb product was ligated into pGEM-TEasy (Promega) to produce plasmid pGP11. Plasmid pGP11 was digested with BamHI, and the VPS11 insert was ligated into the BamHI site of pGEM-HIS1 to form pGH11. A plasmid containing the 5′ upstream region of VPS11 was constructed by PCR amplification from C. albicans BWP17 genomic DNA with primers 11PB5 (nt −413 to −393 from the ATG start codon) and 11PB3 (nt −10 to −30) (Table 2). The PCR product was ligated into pGEM-TEasy to yield plasmid pGP115.
DNA manipulations and sequence analysis.
Plasmid isolation and enzymatic reactions were performed either by standard methods (38) or according to the manufacturer's instructions. Bacterial transformations were performed using chemically competent Escherichia coli DH5α (Invitrogen). Genomic DNA from C. albicans was isolated using glass beads (9). Southern blot analysis was performed as previously described (43). Probes were radiolabeled on purified plasmid inserts with [α-32P]dCTP by using Ready-To-Go DNA Labeling Beads (Amersham Pharmacia Biotech). DNA sequences of selected clones were determined by the Protein and Nucleic Acid Chemistry Laboratory at the University of Leicester (Leicester, United Kingdom). Sequence data were analyzed by using the University of Wisconsin Genetics Computer Group (GCG) sequence analysis software programs (13). Nucleotide database searches were performed using BLAST (1). PCR was performed under routine conditions unless otherwise described. The sequences of oligonucleotides used in PCR amplifications are listed in Table 2. Putative protein sequences were analyzed in BLAST searches and with other programs including GCG (13) and PSORT2 (http://bioweb.pasteur.fr/seqanal/interfaces/psort2.html).
Isolation of C. albicans VPS11 and VPS18.
Incomplete VPS11- and VPS18-like ORF sequences were identified on the C. albicans sequence database (http://alces.med.umn.edu/candida.html) through BLAST searches using the S. cerevisiae VPS11 and VPS18 ORF sequences. Oligonucleotides were designed for the specific amplification of these C. albicans sequences (CAEND1 and CAEND2 for VPS11; CAPEP31 and CAPEP32 for VPS18 [Table 2]) from C. albicans strain S/01 genomic DNA, by PCR. This produced a 393-nt VPS11 fragment and a 607-nt VPS18 fragment, which were used as probes for the isolation of complete gene sequences. VPS11 and VPS18 were isolated from a C. albicans strain S/01 genomic DNA library cloned into the S. cerevisiae plasmid vector YEp213 (kindly provided by P. Meacock, University of Leicester). This library was maintained in E. coli (strain DH5α), and 8,000 individual clones were screened by colony hybridization with C. albicans VPS11- and VPS18-specific probes by using routine procedures (38). Plasmid DNA was isolated from each “positive” clone and used as template DNA in PCR amplifications using primer set CAEND1-CAEND2 or CAPEP31-CAPEP32 to confirm the presence of C. albicans VPS11- or VPS18-specific sequences, respectively. The inserts of selected clones were then sequenced.
Northern blot analysis.
Cells grown at 30°C overnight in YPD + uri were subcultured at 5 × 106 cells/ml into 50 ml of fresh YPD + uri and incubated at 30°C with shaking for 5 h. RNA was then isolated from each culture by using glass beads and phenol (39). Ten to 30 μg of total RNA was loaded per lane and electrophoresed in formaldehyde gels containing morpholinepropanesulfonic acid (MOPS) (38). Gels were blotted and hybridized as described for Southern blot analysis (43). A 2-kb DNA fragment internal to the VPS11 ORF was excised with SpeI from pGP11, radiolabeled with [α-32P]dCTP, and used as a specific VPS11 probe. The C. albicans actin gene was used as an RNA integrity and loading control probe. In this case a 2-kb SalI internal fragment was isolated from pPAC5 (kindly provided by W.A. Fonzi, Georgetown University).
Enzyme assays.
Secreted aspartyl proteinase (SAP) activity was induced in liquid culture with 0.2% bovine serum albumin (BSA) as a nitrogen source and was assayed on agar containing BSA as described by Crandall and Edwards (12). Secreted lipase activity was detected on egg yolk-containing agar as described by Fu et al. (16). In both assays an opaque ring around the colony is indicative of secreted enzymatic activity. A second test for secreted lipase activity was growth on YNB agar containing 2.5% Tween 20 as a carbon source. CPY activity was quantified by a method modified from the work of Jones (22). The CPY-specific substrate N-benzoyl-l-tyrosine p-nitroanilide (NTPNA) was diluted 1:5 from a 2.5-mg/ml stock solution in dimethyl formamide with 0.1 M Tris-HCl (pH 7.5). A 190-μl volume of assay buffer was added to the wells of a microtiter plate, and 5 × 107 cells suspended in 10 μl of sterile distilled H2O were added to each well. Assay plates were incubated at 37°C before absorbance at 405 nm was measured at 5 and 20 h. Controls with cells but no NTPNA were also measured, and these values were subtracted from those for reactions with NTPNA. Each experiment was performed in triplicate, and statistical analysis was performed using the t test. Data are given as mean A405 ± standard deviation.
Vacuolar morphology.
The vacuolar marker dye 5- (and 6-) carboxy-2′,7′-dichlorofluorescein diacetate (carboxy-DCFDA; Molecular Probes, Inc.) was used to visualize vacuolar morphology according to the manufacturer's instructions. Cells grown at 30°C overnight in liquid YPD medium were harvested and resuspended at 106/ml in 50 mM sodium citrate buffer, pH 5, containing 2% glucose. Carboxy-DCFDA was then added to a final concentration of 10 μM from a 10 mM stock (made in dimethyl formamide) and incubated at 30°C for 15 min. Stained cells were then applied to polylysine-coated slides and examined using a confocal fluorescence microscope. As a control for organelle integrity, nuclei were also stained with the specific dye 4′,6′-diamidino-2-phenylindole (DAPI), as described by Burke et al. (9).
RESULTS
Identification of C. albicans class C VPS genes.
Initial attempts to isolate C. albicans class C VPS homologues through the functional complementation of the four S. cerevisiae class C vps mutants were unsuccessful. BLAST searches of the C. albicans sequence database (http://alces.med.umn.edu/gbsearch/ybc.html) using the S. cerevisiae class C VPS ORFs as a query sequence identified sequences with homology to VPS11, VPS18, and VPS33. These were used as query sequences in further BLAST searches to identify overlapping sequences, and contigs were constructed from the available data for each of the ORFs identified. However, at this time the database-derived contigs spanned only short regions of each of these ORFs. From the available sequence data, oligonucleotides were designed for specific amplification of each of the VPS11-, VPS18-, and VPS33-like ORF fragments by PCR. Then the products of these amplifications were used to probe a C. albicans genomic DNA library. This led to the identification of C. albicans genomic clones encoding VPS11 and VPS18 homologues. Sequencing of these clones revealed the entire C. albicans VPS11 and VPS18 ORFs (accession numbers AJ492193 and AJ289080, respectively).
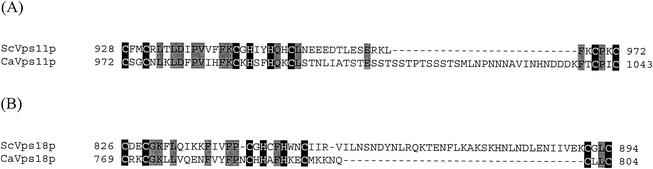
The C. albicans VPS11 ORF encodes an 1,100-amino-acid protein that shares 35% identity and 48% similarity with Vps11p of S. cerevisiae. The VPS18-like ORF encodes a protein of 810 amino acids, sharing 31% identity and 45% similarity with its S. cerevisiae homologue. A putative cysteine-rich RING (really interesting new gene) domain has been identified in the C-terminal regions of both S. cerevisiae Vps11p and Vps18p (7, 14, 30, 34), and it is likely that these mediate protein-protein interactions to form large complexes. Site-directed mutagenesis has demonstrated that at least the first cysteine residue of the proposed Vps18p motif is required for normal function (34). Notably, many of the cysteine and histidine residues of the previously proposed RING domain were conserved in the C. albicans homologues (Fig. 1). The putative C. albicans VPS11 gene product is predicted to possess a CX2CX12CXHX2HX2CX43CX2C sequence near its C terminus. Similarly, the putative C. albicans protein Vps18p has a CX2CX14CXHX2HX2CX5CX2C sequence near its C terminus. Both of these sequences conform to the H2 type RING domain consensus CX2CX9-39HX1-3HX2-3CX2C4-48CX2C (7). Neither protein is predicted to have transmembrane domains or N-terminal signal peptides, suggesting that they do not enter the secretory pathway, and likely reside in the cytoplasm. A possible vacuolar targeting motif was identified at amino acid 418 of Vps11p (KLPN), but given the lack of a signal peptide for endoplasmic reticulum translocation, this is unlikely to be of importance. Lupas's algorithm (26) detected heptad repeats likely to form α-helices within residues 530 to 558 of Vps11p and 722 to 750 of Vps18p, which through interaction with other similar motifs may participate in the formation of coiled-coil structures (41). S. cerevisiae Vps11p is predicted to form a similar helical region (residues 850 to 914), while S. cerevisiae Vps18p is not. The functional significance of the S. cerevisiae Vps11p heptad repeat has not yet been reported, but it may mediate protein-protein interactions. Similar helical domains present on v- and t-SNAREs are known to mediate the tight interaction of these proteins through the formation of coiled-coils in the trans-SNARE complex (46). Through interaction with similar structures on v- and t-SNAREs, the heptad repeats present on Vps11p and Vps18p may regulate the formation of trans-SNARE complexes and thereby regulate vesicle-target membrane fusion.
FIG. 1.
C. albicans Vps11p and Vps18p have conserved C-terminal RING domains. C-terminal sequences from S. cerevisiae and C. albicans Vps11p proteins (A) and from S. cerevisiae and C. albicans Vps18p proteins (B) were aligned using MultAlin (11). A series of cysteine and histidine residues (highlighted on a solid background) which conform to the H2 type RING consensus sequence are conserved in both sets of homologues. Other conserved residues are highlighted on a shaded background. Amino acid residue numbers are given at each end of the sequences.
More recently, the sequences of ORFs with homology to all four class C VPS genes have been made available through the C. albicans genome sequencing project (http://www-sequence.stanford.edu/group/candida).
Construction of C. albicans vps11Δ mutant.
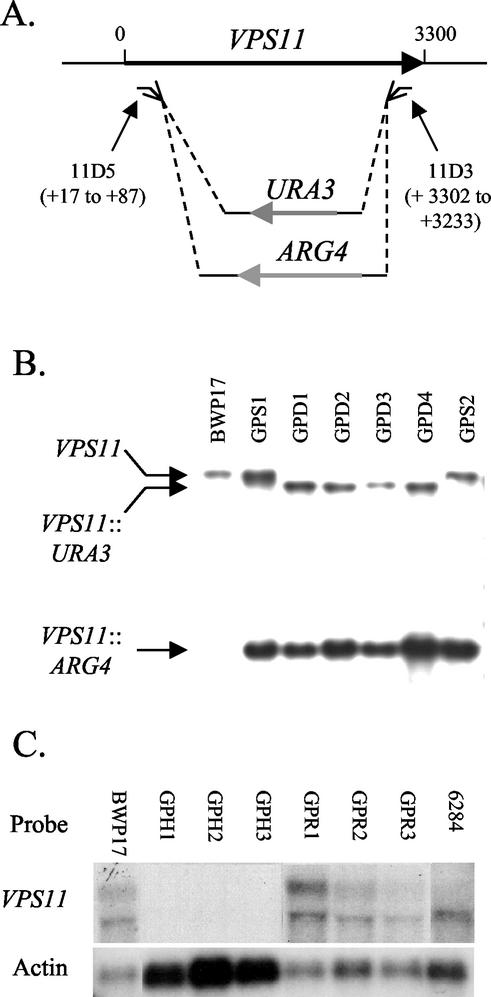
The PCR-based gene disruption method described by Mitchell and colleagues (48) was utilized for construction of a vps11Δ null strain as described in Materials and Methods. Correct integration of either marker at the VPS11 chromosomal locus should result in the deletion of 3,144 bp of the 3,300-bp ORF (Fig. 2A). Initially the ARG4 cassette was transformed into the BWP17 recipient strain (arg4Δ/arg4Δ his1Δ/his1Δ ura3Δ/ura3Δ), and Arg+ transformants were screened by Southern blot hybridization for the desired integration event by using a VPS11-specific probe (Fig. 2B). VPS11/vps11Δ::ARG4 “single mutants” (GPS1 to GPS4) were then transformed with the vps11::URA3 cassette, and Arg+ Ura+ transformants were screened for a second integration event, resulting in vps11Δ::ARG4/vps11Δ::URA3 “double mutants” (GPD1 to GPD3) (Fig. 2B).
FIG. 2.
Deletion of C. albicans VPS11. (A) Line drawing of C. albicans VPS11 locus and location of region deleted in vps11Δ::ARG4 and vps11Δ::URA3. (B) Southern blot analysis of constructed heterozygotes VPS11/vps11Δ (GPS1 and -2) and vps11Δ/vps11Δ strains (GPD1 to -4). The parental strain, BWP17 (VPS11/VPS11), is shown in the leftmost lane. (C) RNA isolated from cells grown to log phase (1 × 107 to 2 × 107 cells/ml) in YPD medium was subjected to Northern blot analysis using a VPS11-specific probe. Two low-abundance RNA species of approximately 3.0 and 3.6 kb were detected in each of the parent strain (BWP17), the prototrophic control strain (YJB6284), and three revertant strains (GPR1 to -3). No VPS11 transcript was detected in the double-mutant strains GPH1 to -3. The blot was stripped and reprobed for the actin transcript as a loading and RNA integrity control.
In order to confirm that the loss of VPS11 function was responsible for any observed phenotypes, a “wild-type” copy of VPS11 was reintroduced into the his1 locus of the double mutant GPD1 on plasmid pGEM-HIS1 to form revertant strains GPR1 to GPR3. Plasmid control strains were also constructed by transforming GPD1 with linearized pGEM-HIS1, generating strains GPH1 to GPH3.
RNA prepared from strains BWP17, GPS1, GPD1 and -2, GPR1 to -3, GPH1 to -3, and YJB6284 (a prototroph derived from BWP17) was subjected to Northern blot analysis with a VPS11-specific probe (Fig. 2C). This resulted in the detection of two low-abundance transcripts of approximately 3.6 and 3.0 kb in the parental strain BWP17 (Fig. 2C, leftmost lane) and the prototrophic control strain YJB6284 (Fig. 2C, rightmost lane). The 3.0-kb band comigrated with the larger rRNA species and may result from “trapping” of the VPS11 transcript in the rRNA. It is unlikely that the second transcript is the product of a second VPS11 homologue, since neither transcript was detected in our vps11Δ strains. The same species were detected in the heterozygous strain GPS1 (data not shown) and in each of three revertant strains (GPR1, GPR2, and GPR3) (Fig. 2C). As expected, this transcript was not detected in the null strains GPD1 and -2 (data not shown) and GPH1 to -3 (Fig. 2C), even when a threefold increase in RNA was probed (30 μg instead of 10 μg).
Deletion of C. albicans VPS11 causes defects in vacuole function and biogenesis.
C. albicans strains with deletions in both VPS11 alleles grew significantly more slowly than parental strains (the generation time was approximately twice that of the parental strain) under all conditions tested. Furthermore, the mutant was more heterogeneous in cell size, with approximately one-third of cells appearing oversized when grown in YPD broth at 30°C. In contrast, when grown in the less-rich YNB medium, mutant cells were smaller than Vps11+ cells.
Initially the C. albicans vps11Δ mutant strains were examined for phenotypes indicative of defective vacuole function. The double mutants exhibited an inability to grow on YPD agar supplemented with 2.5 M glycerol or 1 M NaCl2 (Fig. 3), indicating sensitivity to osmotic stress. The VPS11 null mutant was also sensitive to growth on YNB medium supplemented with 400 μM CuSO4 (data not shown), consistent with defects in metal ion homeostasis. The parental and heterozygous strains grew well at either 30 or 37°C and grew less abundantly at 42°C. However, while the double mutant grew well at either 30 or 37°C, it was completely unable to grow at 42°C (Fig. 3). Each of these phenotypes closely resembles those of the S. cerevisiae class C vps mutants (4, 5, 28, 30), with the exception that the S. cerevisiae mutants are sensitive to growth at 37°C rather than 42°C. Furthermore, each phenotype was fully complemented by the reintroduction of a wild-type copy of VPS11 (GPR1 to -3) but not by that of the pGEM-HIS1 vector alone (GPH1 to -3) (Fig. 3). These observed phenotypes are consistent with defects in multiple vacuolar functions.
FIG. 3.
The C. albicans vps11Δ mutant is sensitive to osmotic and temperature stress. Serial dilutions of strains BWP17 (VPS11/VPS11), GPS1 (VPS11/vps11Δ), GPD1 (vps11Δ/vps11Δ), GPR1 (vps11Δ/vps11Δ/VPS11), GPR2 (vps11Δ/vps11Δ/VPS11), GPH1 (vps11Δ/vps11Δ), GPH2 (vps11Δ/vps11Δ), and YJB6284 (VPS11/VPS11) were spotted onto YPD + uri agar and incubated as described in Materials and Methods. Cell suspensions were also spotted onto YPD + uri agar supplemented with either 2.5 M glycerol or 1 M NaCl2 and incubated at 30°C.
The S. cerevisiae class C vps mutants are defective in the delivery, processing, and activity of several vacuolar hydrolases including CPY (21, 35). CPY activity was assayed to provide an indication of vacuolar hydrolase function in the C. albicans vps11Δ mutant. The vps11Δ mutant, GPH1, showed a threefold reduction in CPY activity (0.189 ± 0.057 [mean A405 ± standard deviation]) compared to the prototrophic control strain YJB6284 (0.618 ± 0.105; P < 0.01). The revertant, GPR1, showed restored CPY activity and actually had higher levels of CPY activity (1.126 ± 0.163) than YJB6284.
The vacuole morphology of the prototrophic strains YJB6284, GPR1, and GPH1 was determined by using carboxy-DCFDA (Molecular Probes, Inc), a fluorescent dye which specifically accumulates within the vacuole (33). The control strain YJB6284 had typical wild-type vacuole morphology, with one to three well-defined, brightly stained subcellular compartments (Fig. 4A). The revertant strain GPR1 exhibited a staining pattern indistinguishable from that of the control strain, indicating a normal vacuole morphology (Fig. 4C). However, the double mutant GPH1 did not localize the fluorophore to any distinct subcellular compartment (Fig. 4B), instead exhibiting staining of a lower intensity dispersed throughout the cell. This diffuse staining pattern confirms the absence of an intact vacuole in the C. albicans vps11Δ mutant. A normal nucleus morphology was observed for each strain by DAPI staining (data not shown), indicating that nuclear structure is unaffected in the vps11Δ mutant.
FIG. 4.
C. albicans VPS11 is involved in vacuole biogenesis. Strains YJB6284 (VPS11/VPS11), GPH1 (vps11Δ/vps11Δ), and GPR1 (vps11Δ/vps11Δ/VPS11) were stained with the vacuole-specific dye carboxy-DCFDA to analyze vacuole morphology. (A) YJB6284 cells have one to three distinctly stained subcellular vacuole compartments. (B) The double mutant GPH1 exhibits diffuse staining throughout the cytoplasm, indicating the absence of a vacuole compartment. (C) The revertant strain GPR1 has a staining pattern similar to that of YJB6284, indicating that vacuole biogenesis is restored.
The C. albicans vps11 mutant has reduced secreted proteinase and lipase activities.
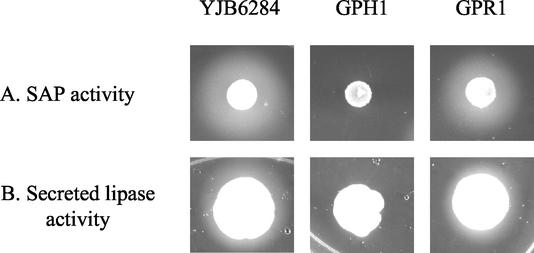
S. cerevisiae vps11 and vps18 mutants are defective in Kex2p-mediated maturation of the mating pheromone α-factor (34). In C. albicans the Kex2p endopeptidase is required for the maturation and full activity of SAP (27). A C. albicans kex2 mutant strain has been described as having reduced SAP activity and furthermore is defective in yeast-hypha morphogenesis (27). We therefore examined SAP activity in our C. albicans vps11Δ mutant as an indicator of Kex2p activity by observing BSA hydrolysis at the peripheries of colonies grown on solid BSA-plus-yeast extract agar. Colonies of the prototrophic control (YJB6284) and revertant (GPR1) strains had large opaque zones at their peripheries, indicating SAP-mediated BSA hydrolysis (Fig. 5A). The double mutant (GPH1) had no visible opaque zone, indicating a loss of SAP activity (Fig. 5A). The reduction in SAP activity was much more severe than that for the C. albicans kex2 mutant (27); thus, loss of Kex2p activity alone cannot account for the SAP defects in GPH1. This suggests that there are either different or additional defects in the processing or secretion of SAP in GPH1. C. albicans is also known to secrete a number of lipase enzymes (16). As a further marker of secretion, secreted lipase activity was assessed on egg yolk medium. The absence of a white precipitation zone around strain GPH1 (Fig. 5B) is indicative of little or no secreted lipase activity in the vps11Δ mutant. The revertant GPR1 had opaque zones of a size similar to that of the YJB6284 zones (Fig. 5B). Reduced secreted lipase activity was confirmed by the severely retarded growth of the vps11Δ mutant on YNB medium containing Tween 20 as a carbon source (data not shown).
FIG. 5.
The C. albicans vps11Δ mutant has reduced secreted lipase and protease activities. Shown are results for strains YJB6284 (VPS11/VPS11), GPH1 (vps11Δ/vps11Δ), and GPR1 (vps11Δ/vps11Δ/VPS11). (A) SAP expression was induced in liquid culture using BSA as a nitrogen source before SAP activity was assessed on BSA-plus-yeast extract agar. The white precipitation zone indicates SAP-mediated BSA hydrolysis. (B) Cell suspensions were spotted onto egg yolk medium in order to detect lipase activity, indicated by a white precipitation zone at the colony periphery.
The C. albicans VPS11 null mutant is defective in the yeast-hypha switch.
The vps11Δ strain was examined for defects in morphogenesis under both solid agar and liquid inducing conditions. Mutant strains GPD1 and -2 and GPH1 to -3 failed to filament on either M199 (Fig. 6A) or 10% FCS (data not shown) solid medium at 37°C, conditions which resulted in extensive filamentation of the parental strain (BWP17), control strain (YJB6284), and revertant strains (GPR1 to -3) (Fig. 6A).
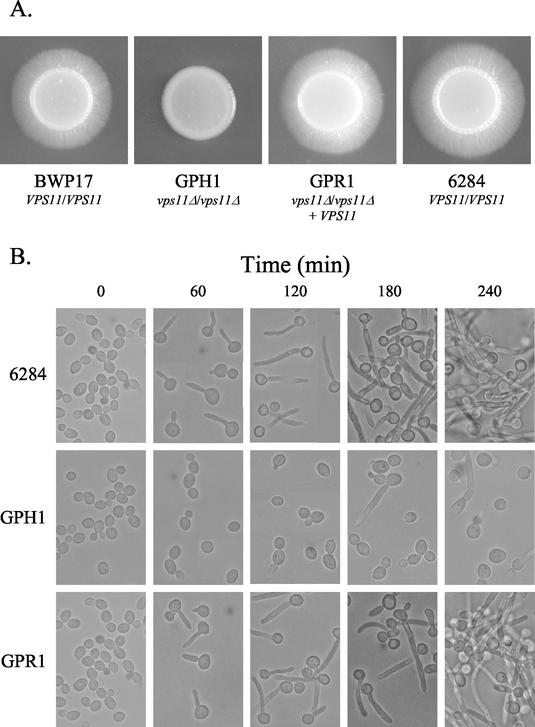
FIG. 6.
C. albicans VPS11 is required for normal filamentation. (A) Cell suspensions of BWP17 (VPS11/VPS11), GPH1 (vps11Δ/vps11Δ), GPR1 (vps11Δ/vps11Δ/VPS11), and YJB6284 (prototrophic control strain) were spotted onto M199 agar medium and incubated at 37°C to induce filamentation. After 4 days, BWP17, YJB6284, and GPR1 all had well defined filamentous borders. No filamentous border was observed for the double mutant GPH1. (B) Prototrophic strains YJB6284, GPH1, and GPR1 grown in YPD medium were subcultured to YPD medium with 10% (vol/vol) fetal bovine serum and incubated at 37°C to induce filamentation.
Yeast-hypha morphogenesis was next examined in liquid culture. Each strain was grown to saturation in YPD medium at 30°C, then subcultured into fresh YPD supplemented with 10% (vol/vol) FCS, and incubated at 37°C to induce filamentation. Under these conditions the majority of both the control (YJB6284) and revertant (GPR1) cells rapidly formed germ tubes, clearly visible after 60 min (Fig. 6B). By 120 min, the majority of germ tubes had elongated to form parallel walled hyphae with some branches. Some pseudohyphae were also observed (Fig. 6B). In contrast, few cells of the double-mutant strain GPH1 were observed to form true germ tubes after 60 or even 120 min of serum induction (Fig. 6B). A proportion of GPH1 cells were observed in elongated forms, but many of these elongated forms had visible constrictions along the cell wall (Fig. 6B), characteristic of pseudohyphal growth, or else formed aberrant short projections which tapered toward the end. Of the true germ tubes that were present, none were observed of more than two or three cell compartments in length, even after 18 h. By 180 min, YJB6284 and GPR1 hyphae had begun to aggregate and form clumps that were visible to the naked eye at 240 min. In contrast, no clumping was observed in the GPH1 culture, even after 18 h of growth in serum culture.
Thus, upon serum induction of filamentous growth, only a small proportion (<10%) of cells of the C. albicans vps11Δ mutant strain were able to produce true germ tubes. In addition, there was a significant delay in the emergence of thesegerm tubes, which had a reduced apical extension rate compared to those of parental and revertant strains. Finally, the mutant cells were unable to sustain filamentous growth to form mature hyphae.
DISCUSSION
In this study, we have demonstrated that Vps11p is required for vacuole biogenesis, SAP and secreted lipase activities, and normal yeast-hypha morphogenesis in C. albicans. As expected, the C. albicans vps11Δ mutant lacked normal vacuole morphology and exhibited a diffuse staining pattern with the vacuole-specific dye carboxy-DCFDA. This contrasts with the S. cerevisiae class C vps mutants, which produce a punctate staining pattern with similar dyes (30, 44). This could be due to a difference either in the actual morphology of these mutants or in the ability of carboxy-DCFDA to localize to small vesicles in C. albicans.
Several possibilities exist to explain the retarded filamentation in vps11Δ mutants. Firstly, the vacuole has previously been observed to undergo rapid expansion upon serum-induced morphogenesis (20). The S. cerevisiae class C VPS gene products form a complex required for the docking and fusion of multiple transport intermediates with the vacuolar membrane (32). During filamentation in C. albicans, the rapid increase in vacuolar volume must result from a net gain of membranous material at the vacuole membrane. The incorporation of new membrane material at the vacuole membrane during expansion may well depend on the putative fusion machinery encoded by the class C VPS homologues identified in this study. Therefore, in addition to maintaining the structural integrity of the vacuole in the yeast form, the C. albicans class C homologues may be required to facilitate vacuolar expansion upon hyphal induction. In agreement with this hypothesis, during initial studies of C. albicans VPS18, it was found that transcript expression increased severalfold following induction of hyphal growth in serum culture (28). It is possible that this expansion fulfills a physical requirement, perhaps to aid the rapid extension of the germ tube. The vacuolar expansion observed upon germ tube emergence may reduce or at least delay the requirement for cytoplasmic biosynthesis while supporting the rapid emergence of the germ tube. An ability to undergo this transition quickly, even under nutrient-poor conditions, may be advantageous to C. albicans, permitting a quick response to conditions that favor filamentous growth. It has also been suggested that the rapid transition from yeast to hyphal growth triggered upon phagocytosis can result in lysis of the host phagocyte (2) and may represent a mechanism of escape from the host's defense.
Secondly, S. cerevisiae Vps11p and Vps18p are known to function at multiple transport steps in vacuolar protein delivery (42), and loss of these proteins' activities results in defects in multiple PVC and vacuolar functions. It is likely that the C. albicans homologues described here operate at equivalent steps in this organism. Thus, a further possibility is that a specific function of the vacuole and/or PVC, rather than their physical expansion, is required for filamentation. The C. albicans vps11Δ strain described in this study was defective in a range of vacuolar functions that resulted in temperature and osmotic sensitivity. S. cerevisiae class C vps mutants are also known to be defective in the processing of multiple vacuolar hydrolases and to missort these hydrolases as inactive precursors (35). Vacuolar hydrolase activity is essential for the process of sporulation in S. cerevisiae (51), an example of cellular differentiation. It therefore remains possible that it is the reduced hydrolase activity of C. albicans vps11Δ strains rather than a lack of vacuolar expansion which accounts for the yeast-hypha differentiation defects. This explanation of the defects observed in the Candida vps11Δ mutant is less favored by us, mainly because the yeast-hypha transition is a very rapid process in C. albicans, whereas the degradation mediated by the vacuolar hydrolases during sporulation and autophagy in S. cerevisiae occurs much more slowly, over many hours (15).
Thirdly, S. cerevisiae vps11 and vps18 mutants are also defective in Kex2p-dependent maturation of α-factor (34). Kex2p is an endopeptidase localized to a trans-Golgi compartment (17, 23). Ordinarily it transits to the PVC via the CPY transport pathway before returning to the Golgi apparatus in a retrograde transport step (29). Due to the transport defects in the vps11 and vps18 mutants, Kex2p is unlikely to be returned to the Golgi apparatus, resulting in depletion of Kex2p activity in the trans-Golgi compartment (47). The loss of SAP activity in our C. albicans vps11Δ mutant could be due in part to similar defects in Kex2p function. This is of particular interest in light of the fact that a C. albicans kex2Δ mutant has been reported as defective in hyphal formation (27). Thus, the filamentation defects of our vps11Δ mutant could result from a depletion of Kex2p activity in the trans-Golgi compartment. More generally, the function of the Golgi “donor” may be perturbed or compromised in our vps11Δ mutant due to the absence of a PVC or vacuole “recipient” compartment.
Additional studies are required to elucidate which of the possibilities discussed above accounts for the morphogenesis defects observed in our vps11Δ mutant. At present it is not clear which of the Vps11p-mediated trafficking steps are required for filamentation. The construction of C. albicans mutants defective in individual transport steps to the vacuole will begin to identify which transport steps, and by inference which compartment functions, are required for filamentation.
Somewhat unexpectedly, the C. albicans vps11Δ mutant exhibited very little or no SAP activity in the BSA plate assay. This defect was more severe than that of a kex2Δ mutant (27), so it is unlikely to be accounted for by Kex2p function alone. This could suggest a defect in either SAP biosynthesis or secretion. A similar loss of secreted lipase activity was also demonstrated on egg yolk medium. The almost total loss of these activities could be caused by defects in the secretory pathway from Golgi apparatus to cell surface. The secretory pathway of S. cerevisiae vps11 mutants is apparently unaffected, as judged by invertase and α-factor secretion as well as CPY mislocalization to the cell surface (34, 35, 37). This may indicate that C. albicans Vps11p functions as part of a general fusion complex required for multiple delivery pathways from the Golgi apparatus. Alternatively, the SAP and lipase defects may be a secondary consequence of the absence of a vacuolar compartment. This may result in the mislocalization of proteolytic enzymes to the cell surface, as is the case for S. cerevisiae vps11 mutants (35), potentially causing degradation of secreted proteins. However, further investigation is required to determine the cause of the SAP and lipase defects.
The importance of vacuolar inheritance during hyphal development has been the subject of a separate investigation (C. Barelle, R. Mathias, C. Gaillardin, N. Gow, and A. Brown, Abstr. Yeast Gen. Mol. Biol. Meet., abstr. 103, 2000). A C. albicans vac8Δ mutant was constructed and shown to be defective in vacuole inheritance. Surprisingly, this has little effect on germ tube formation or extension rate but results in an increase in branching frequency. Although these mutants do not inherit vacuolar material from the parent cell, after a short delay they are able to generate a new vacuole compartment. These results suggest that vacuole protein-sorting pathways, rather than inheritance, are important during the yeast-hypha switch.
The observations made by Gow and Gooday (19) raise several important questions. First, from where is the extra membrane derived which is incorporated into the vacuole during germination? Is there a “reservoir” of membrane vesicles that fuse to the vacuole upon hyphal induction, or is membrane material derived from other membrane-bound organelles such as the PVC, Golgi apparatus, or endoplasmic reticulum? How is this process of vacuolar expansion regulated? This may prove more difficult to investigate and may be resolved only with further elucidation of the signal transduction pathways that regulate morphogenesis. While the present study was in progress, it was reported in the literature that a C. albicans VPS34 homologue had been disrupted (8). The Candida vps34Δ mutant is phenotypically very similar to the vps11Δ strain described here. The vps34Δ strain is hypersensitive to temperature and osmotic stress, it shows delayed emergence of germ tubes, and only a small proportion of cells form true germ tubes in response to serum induction (8). However, there are several differences between the vps34Δ mutant and the vps11Δ mutant analyzed in this study. First, it was not reported if the vps34Δ strain has a reduced germ tube extension rate. Also, no defects in CPY, SAP, or secreted lipase activity were reported. Moreover, while the vps11-deficient strain lacks a central vacuole, the vps34-null strain has a grossly enlarged vacuole, occupying approximately 80% of cell volume (8). The C. albicans vps34Δ mutant is predicted to have defects in anterograde and retrograde traffic between the Golgi apparatus and the vacuole. Thus, upon induction of hyphal growth, it is predicted that the vacuole would neither expand nor contract. This may account for the observed defects in filamentous growth. The similarities between the C. albicans vps34Δ and vps11Δ mutants could suggest that it is not just the presence of a vacuole but the trafficking pathways to the vacuole that are important for morphogenesis. It is likely that these pathways mediate vacuole expansion, which may be required for germination (19).
The results of this study demonstrate that one or more of the trafficking steps mediated by Vps11p are required for normal filamentation in the pathogen C. albicans. However, further investigation is required to identify which of the transportation steps affected in our vps11Δ mutant accounts for the defect in filamentous growth.
Acknowledgments
The work at Leicester University was supported through a BBSRC postgraduate studentship. Work at Georgetown University and the LSUHSC School of Dentistry was supported by NIH grant NIAIDAI46142, awarded to J.S.
We thank Peter Meacock (Leicester University) for providing the C. albicans genomic DNA library used in these studies. We also thank A. P. Mitchell, W. A. Fonzi, and J. Berman for providing plasmids and C. albicans strains.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Arai, T., Y. Mikami, and K. Yokoyama. 1977. Phagocytosis of Candida albicans by rabbit alveolar macrophages and guinea pig neutrophils. Sabouraudia 15:171-177. [PubMed] [Google Scholar]
- 3.Bankaitis, V. A., L. M. Johnson, and S. D. Emr. 1986. Isolation of yeast mutants defective in protein targeting to the vacuole. Proc. Natl. Acad. Sci. USA 83:9075-9079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banta, L. M., J. S. Robinson, D. J. Klionsky, and S. D. Emr. 1988. Organelle assembly in yeast: characterization of yeast mutants defective in vacuolar biogenesis and protein sorting. J. Cell Biol. 107:1369-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banta, L. M., T. A. Vida, P. K. Herman, and S. D. Emr. 1990. Characterization of yeast Vps33p, a protein required for vacuolar protein sorting and vacuole biogenesis. Mol. Cell. Biol. 10:4638-4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bensen, E. S., S. G. Filler, and J. Berman. 2002. A Forkhead transcription factor is important for true hyphal as well as yeast morphogenesis in Candida albicans. Eukaryot. Cell 1:787-798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borden, K. L. 2000. RING domains: master builders of molecular scaffolds? J. Mol. Biol. 295:1103-1112. [DOI] [PubMed] [Google Scholar]
- 8.Bruckmann, A., W. Kunkel, A. Hartl, R. Wetzker, and R. Eck. 2000. A phosphatidylinositol 3-kinase of Candida albicans influences adhesion, filamentous growth and virulence. Microbiology 146:2755-2764. [DOI] [PubMed] [Google Scholar]
- 9.Burke, D., D. Dawson, and T. Stearns. 2000. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 10.Calderone, R. A., and W. A. Fonzi. 2001. Virulence factors of Candida albicans. Trends Microbiol. 9:327-335. [DOI] [PubMed] [Google Scholar]
- 11.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crandall, M., and J. E. Edwards, Jr. 1987. Segregation of proteinase-negative mutants from heterozygous Candida albicans. J. Gen. Microbiol. 133:2817-2824. [DOI] [PubMed] [Google Scholar]
- 13.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dulic, V., and H. Riezman. 1989. Characterization of the END1 gene required for vacuole biogenesis and gluconeogenic growth of budding yeast. EMBO J. 8:1349-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Egner, R., M. Thumm, M. Straub, A. Simeon, H. J. Schuller, and D. H. Wolf. 1993. Tracing intracellular proteolytic pathways. Proteolysis of fatty acid synthase and other cytoplasmic proteins in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 268:27269-27276. [PubMed] [Google Scholar]
- 16.Fu, Y., A. S. Ibrahim, W. Fonzi, X. Zhou, C. F. Ramos, and M. A. Ghannoum. 1997. Cloning and characterization of a gene (LIP1) which encodes a lipase from the pathogenic yeast Candida albicans. Microbiology 143:331-340. [DOI] [PubMed] [Google Scholar]
- 17.Fuller, R. S., A. J. Brake, and J. Thorner. 1989. Intracellular targeting and structural conservation of a prohormone-processing endoprotease. Science 246:482-486. [DOI] [PubMed] [Google Scholar]
- 18.Gietz, D., A. St Jean, R. A. Woods, and R. H. Schiestl. 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20:1425.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gow, N. A., and G. W. Gooday. 1984. A model for the germ tube formation and mycelial growth form of Candida albicans. Sabouraudia 22:137-144. [DOI] [PubMed] [Google Scholar]
- 20.Gow, N. A., and G. W. Gooday. 1982. Vacuolation, branch production and linear growth of germ tubes in Candida albicans. J. Gen. Microbiol. 128:2195-2198. [DOI] [PubMed] [Google Scholar]
- 21.Jones, E. W. 1977. Proteinase mutants of Saccharomyces cerevisiae. Genetics 85:23-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, E. W. 2002. Vacuolar proteases and proteolytic artifacts in Saccharomyces cerevisiae. Methods Enzymol. 351:127-150. [DOI] [PubMed] [Google Scholar]
- 23.Julius, D., A. Brake, L. Blair, R. Kunisawa, and J. Thorner. 1984. Isolation of the putative structural gene for the lysine-arginine-cleaving endopeptidase required for processing of yeast prepro-alpha-factor. Cell 37:1075-1089. [DOI] [PubMed] [Google Scholar]
- 24.Kitamoto, K., K. Yoshizawa, Y. Ohsumi, and Y. Anraku. 1988. Dynamic aspects of vacuolar and cytosolic amino acid pools of Saccharomyces cerevisiae. J. Bacteriol. 170:2683-2686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klionsky, D. J., P. K. Herman, and S. D. Emr. 1990. The fungal vacuole: composition, function, and biogenesis. Microbiol. Rev. 54:266-292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lupas, A., M. Van Dyke, and J. Stock. 1991. Predicting coiled coils from protein sequences. Science 252:1162-1164. [DOI] [PubMed] [Google Scholar]
- 27.Newport, G., and N. Agabian. 1997. KEX2 influences Candida albicans proteinase secretion and hyphal formation. J. Biol. Chem. 272:28954-28961. [DOI] [PubMed] [Google Scholar]
- 28.Palmer, G. E. 2001. Functional analysis of the vacuole in Candida albicans. Ph.D. thesis. University of Leicester, Leicester, United Kingdom.
- 29.Piper, R. C., A. A. Cooper, H. Yang, and T. H. Stevens. 1995. VPS27 controls vacuolar and endocytic traffic through a prevacuolar compartment in Saccharomyces cerevisiae. J. Cell Biol. 131:603-617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Preston, R. A., M. F. Manolson, K. Becherer, E. Weidenhammer, D. Kirkpatrick, R. Wright, and E. W. Jones. 1991. Isolation and characterization of PEP3, a gene required for vacuolar biogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:5801-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raguzzi, F., E. Lesuisse, and R. R. Crichton. 1988. Iron storage in Saccharomyces cerevisiae. FEBS Lett. 231:253-258. [DOI] [PubMed] [Google Scholar]
- 32.Rieder, S. E., and S. D. Emr. 1997. A novel RING finger protein complex essential for a late step in protein transport to the yeast vacuole. Mol. Biol. Cell 8:2307-2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberts, C. J., C. K. Raymond, C. T. Yamashiro, and T. H. Stevens. 1991. Methods for studying the yeast vacuole. Methods Enzymol. 194:644-661. [DOI] [PubMed] [Google Scholar]
- 34.Robinson, J. S., T. R. Graham, and S. D. Emr. 1991. A putative zinc finger protein, Saccharomyces cerevisiae Vps18p, affects late Golgi functions required for vacuolar protein sorting and efficient alpha-factor prohormone maturation. Mol. Cell. Biol. 11:5813-5824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Robinson, J. S., D. J. Klionsky, L. M. Banta, and S. D. Emr. 1988. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 8:4936-4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rothman, J. H., I. Howald, and T. H. Stevens. 1989. Characterization of genes required for protein sorting and vacuolar function in the yeast Saccharomyces cerevisiae. EMBO J. 8:2057-2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rothman, J. H., and T. H. Stevens. 1986. Protein sorting in yeast: mutants defective in vacuole biogenesis mislocalize vacuolar proteins into the late secretory pathway. Cell 47:1041-1051. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook, J., and D. W. Russell (ed.). 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Academic Press, Cold Spring Harbor, N.Y.
- 39.Saporito-Irwin, S. M., C. E. Birse, P. S. Sypherd, and W. A. Fonzi. 1995. PHR1, a pH-regulated gene of Candida albicans, is required for morphogenesis. Mol. Cell. Biol. 15:601-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sato, T. K., P. Rehling, M. R. Peterson, and S. D. Emr. 2000. Class C Vps protein complex regulates vacuolar SNARE pairing and is required for vesicle docking/fusion. Mol. Cell 6:661-671. [DOI] [PubMed] [Google Scholar]
- 41.Skehel, J. J., and D. C. Wiley. 1998. Coiled coils in both intracellular vesicle and viral membrane fusion. Cell 95:871-874. [DOI] [PubMed] [Google Scholar]
- 42.Srivastava, A., C. A. Woolford, and E. W. Jones. 2000. Pep3p/Pep5p complex: a putative docking factor at multiple steps of vesicular transport to the vacuole of Saccharomyces cerevisiae. Genetics 156:105-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sturtevant, J., R. L. Cihlar, and R. A. Calderone. 1998. Disruption studies of a Candida albicans gene, ELF1, a member of the ATP binding cassette family. Microbiology 144:2311-2321. [DOI] [PubMed] [Google Scholar]
- 44.Szczypka, M. S., Z. Zhu, P. Silar, and D. J. Thiele. 1997. Saccharomyces cerevisiae mutants altered in vacuole function are defective in copper detoxification and iron-responsive gene transcription. Yeast 13:1423-1435. [DOI] [PubMed] [Google Scholar]
- 45.Takeshige, K., M. Baba, S. Tsuboi, T. Noda, and Y. Ohsumi. 1992. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119:301-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weis, W. I., and R. H. Scheller. 1998. Membrane fusion. SNARE the rod, coil the complex. Nature 395:328-329. [DOI] [PubMed] [Google Scholar]
- 47.Wilsbach, K., and G. S. Payne. 1993. Vps1p, a member of the dynamin GTPase family, is necessary for Golgi membrane protein retention in Saccharomyces cerevisiae. EMBO J. 12:3049-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woolford, C. A., G. S. Bounoutas, S. E. Frew, and E. W. Jones. 1998. Genetic interaction with vps8-200 allows partial suppression of the vestigial vacuole phenotype caused by a pep5 mutation in Saccharomyces cerevisiae. Genetics 148:71-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wurmser, A. E., T. K. Sato, and S. D. Emr. 2000. New component of the vacuolar class C-Vps complex couples nucleotide exchange on the Ypt7 GTPase to SNARE-dependent docking and fusion. J. Cell Biol. 151:551-562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zubenko, G. S., and E. W. Jones. 1981. Protein degradation, meiosis and sporulation in proteinase-deficient mutants of Saccharomyces cerevisiae. Genetics 97:45-64. [DOI] [PMC free article] [PubMed] [Google Scholar]