Abstract
Mitochondrial pre-mRNAs undergo posttranscriptional RNA editing as directed by small guide RNAs (gRNAs) to produce functional mRNAs in kinetoplastid protozoa. The pre-mRNAs and gRNAs are encoded in the maxicircle and minicircle components, respectively, of the kinetoplastid mitochondrial DNA (kDNA), and editing is catalyzed by a multienzyme protein complex. Trypanosoma evansi AnTat3/3, which lacks maxicircles but retains a single class of minicircles, and a dyskinetoplastic mutant of Trypanosoma brucei EATRO164, which is devoid of kDNA, were both shown to retain genes and proteins for the editing complex. The proteins are present in complexes that immunoprecipitate and sediment indistinguishably from wild-type complexes. The complexes catalyze precleaved insertion and deletion editing as well as full-round deletion editing in vitro. Thus, mutants which lack the natural substrates for RNA editing and all or most gRNAs retain editing complexes that contain the four primary catalytic activities of editing and function in editing, at least in vitro. Therefore neither pre-mRNA nor gRNA is required to form functional RNA-editing complexes.
mRNA transcripts which are encoded in the maxicircle component of the unusual kinetoplastid protozoan mitochondrial DNA (kDNA) require maturation by the process of RNA editing, which involves multiple but specific uridylate (U) insertions and deletions (for recent reviews, see references 12, 23, and 42). Accurate editing is directed by short trans-acting guide RNAs (gRNAs) (3), which are encoded in the minicircle component of kDNA. Editing occurs by sequential activities of endoribonuclease, 3′-terminal uridylyl transferase (TUTase) for U insertion or 3′-exouridylase for U deletion, and, finally, RNA ligase.
RNA editing is catalyzed by a multiprotein complex, the editosome, that in T. brucei sediments at ∼20S (8, 30). The editosome was initially reported to contain from 7 to more than 20 major polypeptides depending on the purification protocol (24, 28, 31), and recent studies in this laboratory identified 21 proteins in the editosome (46). Two of the proteins, REL1 and REL2, have RNA ligase activities (18, 25, 32, 36). Interestingly, gene inactivation studies have shown that the REL1 ligase is essential for editing while REL2 ligase is not (10). The TbMP57 protein has TUTase activity (N. L. Ernst, personal communication), but whether it is essential for editing has not yet been tested. A 108-kDa TUTase is essential for editing (1) but has not yet been detected in the editosome and may function in gRNA U-tail addition (Ernst, personal communication). In addition, TbMP81 (10), TbMP63 (19), TbMP44 (B. Wang, personal communication) are present in the editosome and are essential for editing and for editosome integrity. Deletion of both alleles of the gene for the Hel61 helicase that is present in the editosome reduces but does not eliminate editing (26).
Trypanosomes that do not edit mitochondrial RNAs or have gross abnormalities in editing as a result of the absence of all or most kDNA sequences can grow as bloodstream forms (BFs) but not procyclic forms (PFs) (for reviews, see references 6, 16, and 35). Dyskinetoplastic (Dk) strains of Trypanosoma brucei EATRO164 that were produced by repetitive acriflavine treatment are devoid of all kinetoplastid DNA (44, 45) and lack edited mitochondrial mRNA (14, 15). The naturally occurring Dk strain, T. evansi AnTat 3/3, which is closely related to and perhaps a mutant of T. brucei, lacks maxicircle DNA and contains one minicircle sequence class, rather than hundreds, and also lacks edited mRNAs (4, 40). The lack of edited mRNAs reflects the absence of the maxicircle-encoded pre-mRNA and most, if not all, gRNAs. Neither strain survives as PFs, presumably due to their reliance for energy production on the oxidative phosphorylation system (5, 6, 7, 11, 47), many components of which are encoded in edited mRNAs (39, 43). The two strains grow well as BFs, and it was presumed that this was because the BFs produce most, if not all, of their energy by glycolysis (references 11 and 37 and references therein). However, blockage of editing is lethal to BF as well as PF stages (18, 19, 36). This suggests that RNA editing may be essential for the survival of both life cycle stages of T. brucei and therefore that DK mutants must have undergone significant compensatory changes for survival.
Functionally active editosomes have been isolated from T. brucei, and several of their component proteins and activities have been characterized. All editosome proteins identified to date are encoded in nuclear DNA and have a putative mitochondrial targeting signal sequence indicating cytoplasmic synthesis and import into the organelle. The reagents and assays used to characterize editosomes in wild-type T. brucei were used to assess the presence and general characteristics of editosomes in T. evansi AnTat 3/3 and a Dk mutant of T. brucei EATRO164. We show here that these Dk mutants contain editosome genes and editosomes with general physical and functional characteristics that are indistinguishable from those of wild-type trypanosomes. These studies indicate that mRNA and gRNA are not required for in vivo production of editosomes.
MATERIALS AND METHODS
Cell growth and enrichment of mitochondria.
T. brucei EATRO164wt is a cloned wild-type strain of T. brucei with functional RNA editing. EATRO164Dk clone IIa is a stable acriflavin-induced mutant of T. brucei EATRO164wt that lacks detectable maxicircle and minicircle DNA (44, 45). T. evansi AnTat 3/3 is a naturally occurring Dk mutant of T. brucei that lacks maxicircle DNA but has one of minicircle DNA class rather than hundreds (4, 40). All three strains are infectious to mice and rats and can be grown in axenic culture. EATRO164wt can be grown as procyclic insect stage forms, but the other two cannot. BFs of all three strains were grown in BALB/c mice or Wistar adult male rats and purified from blood with a DEAE ion-exchange column (22) or grown in vitro in supplemented HMI-9 and collected and washed by centrifugation as specified previously (17).
Mitochondrial enrichment was performed on cells isolated from rats by a method modified from those of Williams and Frank (48) and Bertrand and Hajduk (2). Briefly, cells were resuspended (2.4 × 109 cells/ml) in ice-cold DTE (1 mM Tris [pH 8.0], 1 mM EDTA) and disrupted by five strokes in a Dounce homogenizer, and 60% sucrose was added to a final concentration of 10% sucrose. The homogenate was then centrifuged at 15,800 × g for 10 min, resuspended in STM (20 mM Tris [pH 8.0], 0.25 M sucrose) with MgCl2 and CaCl2 at 3.0 and 0.3 mM final concentrations, respectively, and incubated on ice for 1 h after the addition of 30 U RNase-free DNase. After addition of an equal volume of STE (20 mM Tris [pH 8.0], 0.25 M sucrose, 2 mM EDTA), the homogenate was centrifuged at 15,800 × g for 10 min to pellet the mitochondria. The pellet was resuspended in STE and centrifuged at 1,500 × g for 5 min, and the resultant supernatant was centrifuged at 16,000 × g for 15 min; this was repeated twice to wash the mitochondria. The final preparation consisted mostly of mitochondria and flagella as observed by microscopy.
DNA and RNA manipulations.
Whole-cell DNA was prepared by standard methods from 120 ml of axenic BF cultures or from cells isolated from rats. Whole-cell RNA was isolated using Ultraspec RNA reagent (Biotecx Laboratories, Inc., Houston, Tex.) as specified by the manufacturer. The presence of maxicircle genes and TbMP52 (REL1) and TbMP48 (REL2) nuclear genes for editosome proteins was assayed by PCR under standard Taq polymerase conditions with the gene-specific primers shown in Table 1. Reverse transcription-PCR was performed using the same primers and conditions that were described previously (36). Briefly, contaminating DNA was removed from RNA isolated from axenic BF cultures by RNase-free DNase (Promega) treatment followed by phenol-chloroform extraction. Reverse transcription with 1 μg of RNA and the appropriate primer was performed at 42°C with Superscript II reverse transcriptase (Life Technologies). All bands detected were cloned into pGEM-T-EASY, and three clones isolated from each DNA band were sequenced.
TABLE 1.
Oligonucleotides used in diagnostic PCR amplifications for the maxicircle-encoded ATP synthase subunit 6 (A6), and NADH:ubiquinone oxidoreductase subunits (ND4 and ND7) and for the nucleus-encoded, editing-associated ligases (REL1 and REL2)
| Gene | Oligonucleotides | Fragment size (bp) |
|---|---|---|
| A6 | AAAAATAAGTATTTTGATATTATTAAAG | 381 |
| TATTATTAACTTATTTGATC | ||
| ND4 | TGTGTGACTACCAGAGAT | 256 |
| ATCCTATACCCGTGTGTA | ||
| ND7 | ATGACTACATGATAAGTA | 161 |
| CGGAAGACATTGTTCTACAC | ||
| REL1 | ACTGCAGATGCAACTCCAAAGG | 875 |
| CGTGACGAATGACC | ||
| REL2 | ATGTTGCGTCGCCTCGGTGTA | 1,252 |
| TCATTCGCTAAAGTCAGGAG |
Size exclusion liquid chromatography and glycerol gradient ultracentrifugation.
Whole-cell lysates from axenic cell cultures were fractionated by glycerol gradient sedimentation (28). Typically, 6 × 108 cells were lysed in 600 μl of buffer A (20 mM HEPES [pH 7.9], 10 mM MgCl2, 300 mM KCl, 1% Triton X-100) containing protease inhibitors (10 μg of leupeptin per ml, 5 μg of pepstatin per ml, 1 mM Pefabloc) and incubated for 30 min at 4°C. These cell lysates were cleared by centrifugation, and 500 μl of lysate was loaded on to a 11.2-ml, 10 to 30% linear glycerol gradient in 20 mM HEPES (pH 7.9)-10 mM MgCl2-100 mM KCl-1 mM dithiothreitol (DTT) in 15- by 95-mm Beckman polyallomer tubes. The gradients were centrifuged for 5 h at 4°C (38,000 rpm in an SW40 rotor [Beckman]), and 500-μl fractions were collected and assayed immediately or flash frozen at −70°C for storage. Mitochondrial lysates from trypanosomes isolated from infected rats were fractioned by size exclusion liquid chromatography using a Superose 6 HR (10/30) column (Pharmacia), as described previously (28). Briefly, a 250-μl sample was loaded onto the column at a 0.2-ml/min flow rate and eluted with 10 mM Tris (pH 7.0)-10 mM MgCl2-200 mM KCl-1 mM DTT. Fractions of 500 μl were collected, concentrated to 80 μl, and assayed as described below.
Immunoprecipitation.
Immunoprecipitations were performed with whole-cell lysates or glycerol gradient fractions using monoclonal antibody P1H3-D7, which is specific for the TbMP63 editosome component and which was coupled to goat anti-mouse immunoglobulin G (IgG) Dynabeads (M-450; Dynal) as described previously (29). Immunoprecipitations were performed in IP300 buffer (10 mM Tris [pH 7.6], 300 mM KCl, 0.1% Triton X-100) supplemented with 1% bovine serum albumin and then washed twice in the same buffer and twice in IP300 without bovine serum albumin.
Western analysis.
Protein samples were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10% polyacrylamide), transferred to polyvinylidene difluoride membranes, and examined by Western analysis using as primary antibodies monoclonal antibodies P4D8, P1H3, P3C1, and P3C12 which are specific for TbMP81, TbMP63, REL1, and TbMP42 editosome proteins, respectively (29). The primary antibodies were diluted 1:500, the secondary antibody, horseradish peroxidase-coupled goat anti-mouse IgG antibody, was used at a 1:1,000 dilution, and the blot was developed with enhanced chemiluminescence substrate (Pharmacia).
Enzyme assays.
RNA ligases were detected by autoadenylation using radiolabeled [α-32P]ATP as described previously (33). Protein samples were incubated for 15 min at 28°C with 2.5 μCi of [α-32P]ATP in 25 mm HEPES (pH 7.9)-10 mM magnesium acetate-50 mM KCl-0.5 mM DTT-10% dimethyl sulfoxide. The proteins were then resolved on sodium dodecyl sulfate-10% polyacrylamide gels, and the radiolabeled proteins were detected using a PhosphorImager.
The precleaved insertion in vitro editing assay was performed with gPCA6-2A gRNA with 3′CL13pp and 5′-end-labeled 5′CL18 substrates, and the precleaved deletion assay was performed with gA6[14]PC-del gRNA with U5-3′CL and 5′labeled U5-5′CL substrates, as described previously (20, 21). Full-round in vitro deletion editing was assayed using the mRNA substrate A6short/TAG.1 and the gRNA gA6[14]wt, which directs a 4U deletion reaction (38). The gRNA gA6[14]USDdccc, designed for improved in vitro deletion sensitivity, was also used (9). The gA6[14]wt and gA6[14]USDdccc sequences are GGAUAUACUAUAACUCCGAUAACGAAUCAGAUUUUGACAGUGAUAUGAUAAUUAUUUUUUUUUUUUUUUUU and GGAUAUACUAUAACUCCACCCUCACAACUUUCUU, respectively.
RESULTS
Dk mutants have genes for editosome proteins.
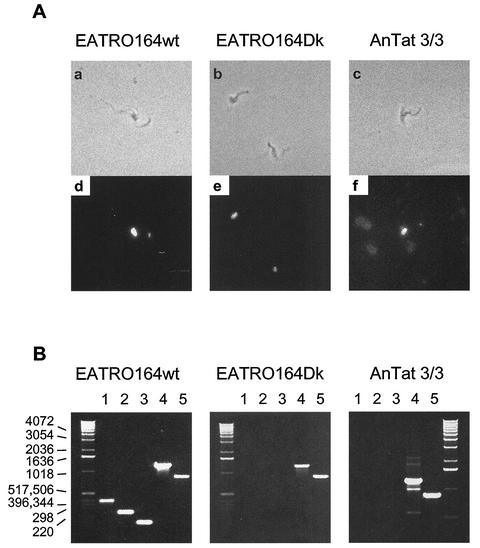
We confirmed that the Dk mutant strains used in this study lack maxicircle genes which encode the pre-mRNAs that are normally edited but found that they retain genes for editosome proteins. The kDNA can be observed directly by microscopy in 4′,6-diamidino-2-phenylindole (DAPI)-stained cells as a second smaller spot, compared to the stained nucleus, at one end of the trypanosome cell. The DAPI-stained kDNA is absent from T. brucei EATRO164Dk clone IIa but is evident both in its parental strain, T. brucei EATRO164wt, and in T. evansi AnTat 3/3, which contains a single class of minicircles (4, 40) (Fig. 1A). Reverse transcription-PCR of whole-cell RNA showed the presence of maxicircle transcripts in EATRO164wt but not in EATRO164Dk or T. evansi AnTat 3/3 (results not shown). PCR with whole-cell DNA using oligonucleotide primers specific for maxicircle genes confirmed the presence of maxicircle DNA in EATRO164wt and the absence of these genes in T. evansi AnTat 3/3 and EATRO164Dk (Fig. 1B). However, PCR using primers specific for the two editosome RNA ligases, REL1 and REL2, shows that these nucleus-encoded genes are present in the wild-type strain and both Dk strains.
FIG. 1.
Gene presence and absence in normal and mutant trypanosomes. (A) Phase-contrast (a to c) and DAPI-stained fluorescence (d to f) micrographs of T. brucei EATRO164 wild type (EATRO164wt), derived acriflavin-induced dyskinetoplastic T. brucei EATRO164 (EATRO164Dk), and T. evansi AnTat 3/3. (B) DNA products from PCR amplifications performed with whole-cell DNA, using oligonucleotide primers for maxicircle-encoded genes (lane 1, A6 [381 bp]; lane 2, ND4 [239 bp]; lane 3, ND7 [144 bp]) and nucleus-encoded RNA-editing-associated RNA ligase genes (lane 4, REL2 [1,252 bp]; lane 5, REL1 [859 bp]).
Dk mutants have editosomes.
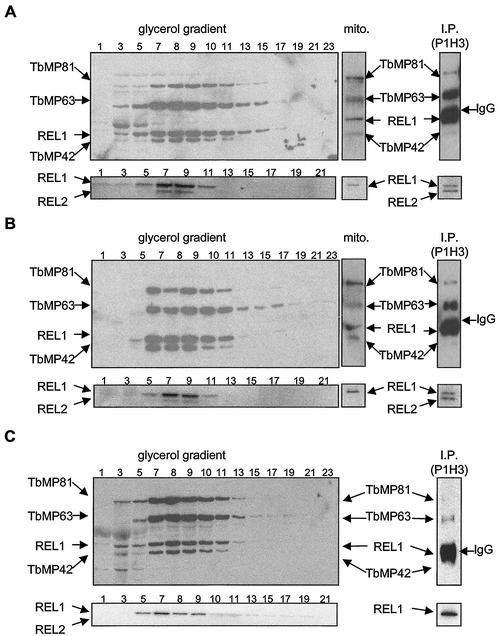
Western and autoadenylation analyses of glycerol gradient fractions of whole-cell lysates revealed that the wild-type and mutant cells all contain at least four of the five editosome proteins that can be detected using the available monoclonal antibodies or autoadenylation (Fig. 2). These proteins are localized within the mitochondrion, at least in BF EATRO164wt and EATRO164Dk, since mitochondria isolated from both contain similar amounts of TbMP81, TbMP63, TbMP42, and REL1 (Fig. 2). The lack of detection of REL2 probably reflects its reduced signal relative to REL1 and the difficulty in preparing mitochondria from BFs rather than its absence from the mitochondrion. Mitochondria were not prepared from T. evansi AnTat 3/3 because of the requirement for numerous cells. The relative abundances of the five proteins to each other are indistinguishable between the two EATRO strains, but the amount of TbMP42 may be somewhat reduced in AnTat 3/3. The REL2 protein was not detected by adenylation in T. evansi AnTat 3/3, although the gene is present. Thus, although no normal editing substrates (i.e., maxicircle-encoded pre-mRNAs) are present and no gRNAs are present in EATRO164Dk, all editosome proteins for which we have reagents for detection are present in the mitochondrion.
FIG. 2.
Editosomes in normal and Dk mutant trypanosomes. (A) T. brucei EATRO164wt; (B) T. brucei EATRO164Dk; (C) T. evansi AnTat 3/3. Editosome protein components were identified by Western analysis using monoclonal antibodies to proteins TbMP81, TbMP63, REL1 (TbMP52), and TbMP42 (top panels for each strain) and by autoadenylation of REL1 and REL2 (TbMP48) (bottom panels for each strain), as described in Materials and Methods. Fractions (0.5 ml) from the 10 to 30% glycerol gradients (glycerol gradient) of whole-cell lysates (bottom right), samples enriched for mitochondria (mito.), and immunoprecipitates using immobilized monoclonal antibody P1H3 to TbMP63 [I.P. (P1H3)] were examined. A marker with a sedimentation coefficient of ∼20S elutes over fractions 7 to 10 on the same glycerol gradient. IgG indicates the IgG heavy chain of P1H3, which reacts with the anti-mouse second antibody.
The five editosome proteins that we can detect by Western and autoadenylation assays cosediment in glycerol gradients in the mutants with a profile indistinguishable from that in EATRO164wt (Fig. 2). They peak at ∼20S, as does in vitro RNA editing. REL1 autoadenylation and RNA ligase activities in isolated mitochondria from wild-type and Dk mutant strains elute from 30-ml Superose S6 size exclusion gel filtration columns (5-ml void volume) over the volume range from 10 to 15 ml and 8 to 16 ml, respectively (results not shown). This corresponds closely to the REL1 elution profile and to in vitro editing (8 to 14 ml) seen in the purification of the ∼1.6-MDa editing complex from PFs (28). Western and autoadenylation analyses of immunoprecipitates prepared by using monoclonal antibody P1H3, which is specific for TbMP63, coprecipitated the other four editosome proteins that we assayed for, namely, TbMP81, REL1 (TbMP52), REL2 (TbMP48), and TbMP42, in EATRO164wt and EATRO164Dk. Coprecipitation of TbMP81, TbMP42, and REL1 but not REL2 was detected in T. evansi AnTat3/3, but the signal for each protein was generally lower, which may reflect the sensitivity of detection or actual absence of REL2. Thus, the editosome proteins are present in a complex that is very similar to that in wild-type T. brucei.
In vitro editing by editosomes from Dk mutants.
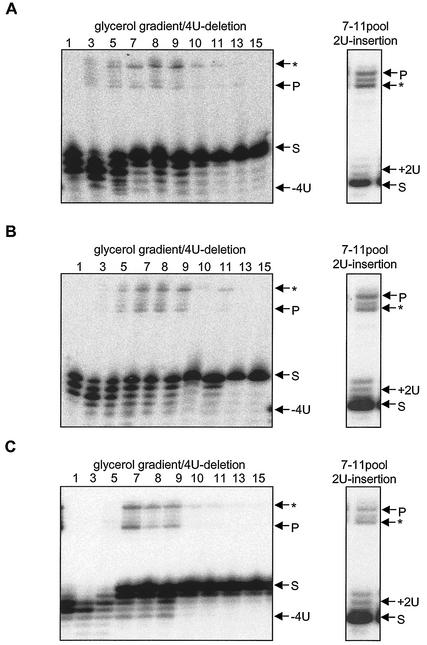
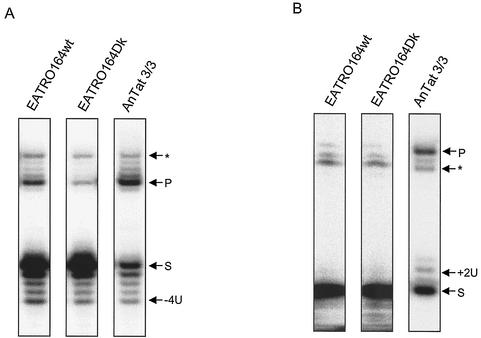
Glycerol gradient fractions from EATRO164wt, EATRO164Dk, and AnTat 3/3 were assayed for their ability to catalyze deletion and insertion editing in vitro. The ability of the editosomes to edit was examined using in vitro-precleaved editing assays (20, 21). In precleaved deletion assays, gRNAs are designed to direct the deletion of 4U in the substrate RNA, and in the precleaved insertion assays, gRNAs are designed to direct the insertion of 2U in the substrate RNA. The two assays test for the presence of exonuclease and TUTase activities, respectively, as well as RNA ligase activity. The glycerol gradient fractions from whole-cell lysates that contain the editosome proteins (Fig. 2) catalyzed specific 4U-precleaved deletion in all three strains (Fig. 3). This activity is dependent on the presence of the gRNA (data not shown) and indicates the presence of the U-specific 3′ exonuclease and RNA ligase activities that function in deletion editing. Similar activity profiles for gRNA-directed 2U insertion were observed for EATRO164wt and EATRO164Dk (data not shown). The nuclease activity in the top fractions of the T. evansi AnTat 3/3 total-cell lysate is greater than in the T. brucei lysates. It is not gRNA specific and is unlikely to be related to editing. Pooled glycerol gradient fractions 7 to 11 from whole-cell lysates of all three strains catalyzed gRNA-directed 2U insertion (Fig. 3), indicating the presence of the 3′ TUTase and RNA ligase activities that function in insertion editing. Immunoprecipitates from whole-cell lysates of all three strains made with monoclonal antibody P1H3, specific for TbMP63, also catalyzed gRNA-directed precleaved deletion and precleaved insertion editing in vitro (Fig. 4). The results with the normal and Dk T. brucei strains were quite similar, and the immunoprecipitates from T. evansi were quite active, especially in insertion editing. Thus, the editing gRNA-directed TUTase, exonuclease, and ligase activities are in an immunoprecipitable complex in the mutants as well as in the wild-type trypanosomes.
FIG. 3.
Precleaved deletion and insertion in vitro editing of glycerol gradient fractions. Fractions 1 to 15 (top to bottom) from whole-cell lysates of T. brucei EATRO164wt (A), T. brucei EATRO164Dk (B), and T. evansi AnTat 3/3 (C) were assayed for gRNA-directed deletion editing (left panels), and pooled fractions 7 to 11 were assayed for gRNA-directed insertion editing (right panels). The arrows indicate the radiolabeled 5′CL18 pre-mRNA substrate (S), the processed substrate from which 4U are removed (−4U) or to which 2U added (+2U), the edited product (P), and the background of the substrate ligated to the 3′ fragment without processing (∗).
FIG. 4.
Precleaved deletion (A) and insertion (B) in vitro editing of immunoprecipitates. Immunoprecipitates from whole-cell lysates of T. brucei EATRO164wt, T. brucei EATRO164Dk, and T. evansi AnTat 3/3 made with monoclonal antibody P1H3, which is specific for editosome protein TbMP63, were assayed for gRNA-directed precleaved deletion (A) and precleaved insertion (B) editing in vitro. The arrows indicate the radiolabeled 5′CL18 pre-mRNA substrate (S), the processed substrate from which 4U are removed (−4U) or to which 2U added (+2U), the edited product (P), and the background of the substrate ligated to the 3′ fragment without processing (∗).
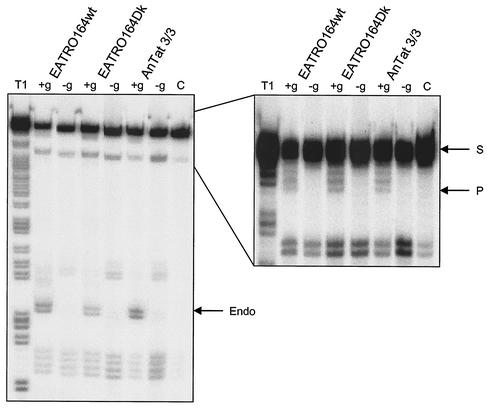
To assess gRNA-directed full-round editing, which includes gRNA-directed endonuclease activity, the 73-nucleotide mRNA A6short/TAG.1, along with gRNA gA6[14]wt (38) or gA6[14]USDdccc, which has increased complementarity and in vitro deletion editing activity (9), was used. These assays used 10 μl of glycerol gradient fraction 9 from whole-cell lysates, which corresponds to the ∼20S complex. Full-round editing was observed for both EATRO164wt and EATRO164Dk when the gA6[14]wt gRNA was used (data not shown), and all three strains exhibited gRNA-dependent full-round deletion editing with gA6[14]USDdccc (Fig. 5). The gRNA-specific cleavage product reveals the presence of the editing endonuclease activity. Thus, the editosomes present in the mutants contain the endonuclease, exonuclease, 3′TUTase, and RNA ligase activities required for RNA editing.
FIG. 5.
Full-round deletion editing by editosomes from Dk mutants. The32P-labeled A6short/TAG substrate was incubated with glycerol gradient fraction 9 from T. brucei EATRO164wt, T. brucei EATRO164Dk, and T. evansi AnTat 3/3 in the presence (+g) or absence (−g) of gA6[14]USDdccc gRNA. The substrate (S) and products of specific endonuclease cleavage (Endo) and editing (P) are indicated. The right panel is an enlarged section of the full gel in the left panel to show the substrate and product region. The endonuclease doublet products reflect the addition of one and two C's during the T4 RNA ligase radiolabeling of the substrate. A control (C) lane has no added fraction 9, and RNase T1-digested substrate is used as a marker (T1).
DISCUSSION
This study shows that mutant trypanosomes that lack the mitochondrial genes for the pre-mRNA that are normally edited and all or most genes for gRNAs that specify the editing contain editosomes that function in in vitro editing. The acriflavin-induced Dk mutant T. brucei EATRO164Dk contains no maxicircle or minicircle mitochondrial DNA, and naturally occurring T. evansi AnTat 3/3 has no maxicircle DNA and only a single class of minicircle DNA. However, both strains contain nuclear genes for editosome proteins, and the expression of five of these in EATRO164Dk and four in T. evansi AnTat 3/3 was directly demonstrated by the Western and/or autoadenylation assay. The proteins are present in the mitochondrion and in a complex that is indistinguishable from wild-type-isolated editosomes in sedimentation, gel permeation column elution, and immunoprecipitation characteristics. Fractions containing the complex catalyze both insertion and deletion editing in vitro. Mutants devoid of mitochondrially transcribed RNA-editing substrates and all or most gRNA have editosomes that function in vitro and thus do not require these RNAs for their assembly or activity.
The editosomes in the Dk mutants contain most, if not all, of the proteins known to be present in wild-type editosomes. Five components, TbMP81, TbMP63, TbMP42, REL1 (TbMP52), and REL2 (TbMP48), were directly demonstrated in T. brucei EATRO164Dk, and all but REL2, which may be present but not detected, were demonstrated in T. evansi AnTat 3/3. Down regulation of REL2 to undetectable levels in T. brucei 427 cells by RNA interference showed that this protein is not essential for cell survival or for RNA editing, suggesting that REL1 may be able to compensate for its loss in the editosome assembly and function (10). In addition, the ability to perform full-round editing indicates that the editosome in the mutants contains endonuclease, TUTase, and ExoUase activities that are likely to be catalyzed by other proteins. Furthermore, the sedimentation coefficient for both Dk mutants and the elution profile from Superose S6 of the editosomes from the EATRO164Dk mutants are indistinguishable from those of wild-type editosomes, suggesting a similar protein composition in the mutants. The wild-type editosomes are ∼20S, with a molecular mass of approximately 1.6 MDa, and contain about 21 proteins (46), suggesting that these proteins and their genes are present in the Dk mutants.
The editosomes in the Dk mutants are present in the mitochondrion as complexes that can catalyze editing, at least in vitro. This is evident from the presence of the proteins in mitochondrial fractions and sedimentation and immunoprecipitation data (Fig. 2). This indicates that targeting of the proteins to the mitochondrion through amphipathic N-terminal peptides (28, 29) and assembly into functional complexes occurs in the mutants. This does not exclude the possibility that some portions of the proteins remain and perhaps function outside of the mitochondrion, although no evidence for this exists to date.
The presence of functional editosomes in T. brucei strains which do not contain mitochondrial pre-mRNA or gRNA shows that these RNAs are not required for assembly of the functional complexes. There is no evidence for other RNA components of editosomes. Micrococcal nuclease treatment that digests most RNA associated with editosomes reduces but does not eliminate in vitro editing (34), and no RNA species other than mitochondrial pre-mRNA and gRNA have been identified in editosomes (24, 34). In addition, RNA-editing complexes purified from procyclic trypanosomes that catalyze in vitro editing did not contain detectable mRNA or gRNA (31). These experiments are not definitive, reflecting the difficulty in determining whether such RNA is present. Nevertheless, we cannot rule out the presence of mitochondrially imported tRNAs in the editosome. Specific physical and functional interactions have been found among editosome proteins. Although the investigation of these specific interactions is at an early stage, REL1 and TbMP63 have been shown to physically interact (29), as have REL2 and TbMP81, and these interactions enhance ligase activity (A. Schnaufer and S. S. Palazzo, unpublished data). Taken together, these data indicate the possibility that assembly of functional editosomes requires protein-protein interactions and that the editosomes may be able to be reconstituted in vitro. Similarly, it has recently been shown that the yeast spliceosomal small nuclear ribonucleoproteins associate prior to binding of their respective pre-mRNA substrates (41). This does not preclude the possibility that the Dk mutants are incapable of editing in vivo. Factors other than those contained in the editosome may be required for in vivo editing.
Dk mutants lack the maxicircle genes for components of the oxidative phosphorylation system including ubiquinone oxidoreductase subunits, cytochrome b, cytochrome oxidase subunits, and ATP synthase subunits (12, 13, 39, 43). They also lack the maxicircle genes for the mitochondrial rRNAs and the RPS12 mitochondrial ribosomal protein (13). This indicates that they are likely to be unable to produce a functional mitochondrial oxidative phosphorylation system or synthesize protein in the mitochondrion. The inability of the Dk mutants to grow as PFs (references 5, 6, 11, and 35 and references therein), for which energy generation relies on mitochondrial oxidative phosphorylation (reference 47 and references therein) is consistent with this. The ability of the Dk mutants to grow as BFs was presumed to reflect the reliance of BFs on glycolysis for energy production (5, 6, 11). It was also presumed that editing was not required in BFs since Dk mutants do not edit mitochondrial mRNAs. However, inactivation of the expression of each of several editosome genes is lethal in BF trypanosomes. This suggests that either editing is normally essential in BFs or the editosome proteins have functions other than in editing. The former possibility seems more likely since loss of expression of several editosome proteins tested is lethal in BFs. Inactivation of one of these genes in the Dk mutants should answer this issue. Several edited mRNAs accumulate preferentially in the BF stage over the PF stage. BFs may rely heavily on their protein products, such as those for the putative respiratory complex I for terminal electron transport or the A6 gene product for the ATP synthase complex, to maintain the mitochondrial membrane potential (35).
The ability of the Dk mutants to survive as BFs while inactivation of editosome gene expression can be lethal suggests that they have undergone either genetic or physiological compensation or both. A less likely possibility is that edited versions of the maxicircle genes were retrotransposed into the genome, perhaps in the nucleus, resulting in the ability to bypass editing rather than bypassing the loss of enzymes. The extensive selection used to generate T. brucei EATRO164Dk (44, 45) implies something other than a system of regulation of enzyme activity.
What events lead to viable Dk strains? The naturally occurring Dk species of trypanosomes and the high rate of occurrence of Dk forms in natural populations suggest that the mitochondrial DNA is not highly stable (references 16 and 27 and references therein). The partial or total loss of kDNA can be tolerated if the requirement for transmission, entailing the conversion to PFs in the insect vector, can be bypassed by other forms of transmission (mechanical or venereal) and the reliance on editing and its products is bypassed, as indicated above. It is possible that multiple events (e.g., mutations) are required to generate viable Dk strains, and conditions such as repeated transmission without an insect vector may be needed to allow these multiple events to occur. In any event, the origins of T. evansi AnTat 3/3 and T. brucei EATRO164Dk are different. These different origins may have resulted in different compensatory changes and perhaps different editosome compositions.
Not only do both the naturally occuring Dk T. evansi AnTat 3/3 and the chemically induced T. brucei EATRO164Dk express the major known components of the RNA-editing complex, but also these components assemble into a large multiprotein complex with all four enzyme activities characteristic of the mitochondrial RNA editing complex and with the ability to perform at least full-round in vitro deletion editing. Thus, gRNA and mRNA are not required for the assembly of a functional RNA-editing complex in vivo. The question whether some editing proteins have essential functions other than RNA editing still remains to be answered.
Acknowledgments
We thank Jean Feagin, Achim Schnaufer, and Aswini Panigrahi for their helpful advice and discussions. We are grateful to Nancy Ernst, Setareh Palazzo, and Achim Schnaufer for sharing unpublished results.
This work was supported by the NIH grant AI14102 to K.S.
REFERENCES
- 1.Aphasizhev, R., S. Sbicego, M. Peris, S. H. Jang, I. Aphasizheva, A. M. Simpson, A. Rivlin, and L. Simpson. 2002. Trypanosome mitochondrial 3′ terminal uridylyl transferase (TUTase): the key enzyme in U-insertion/deletion RNA editing. Cell 108:637-648. [DOI] [PubMed] [Google Scholar]
- 2.Bertrand, K. I., and S. L. Hajduk. 2000. Import of a constitutively expressed protein into mitochondria from procyclic and bloodstream forms of Trypanosoma brucei. Mol. Biochem. Parasitol. 106:249-260. [DOI] [PubMed] [Google Scholar]
- 3.Blum, B., and L. Simpson. 1990. Guide RNAs in kinetoplastid mitochondria have a nonencoded 3′ oligo(U) tail involved in recognition of the preedited region. Cell 62:391-397. [DOI] [PubMed] [Google Scholar]
- 4.Borst, P., F. Fase-Fowler, and W. C. Gibson. 1987. Kinetoplast DNA of Trypanosoma evansi. Mol. Biochem. Parasitol. 23:31-38. [DOI] [PubMed] [Google Scholar]
- 5.Borst, P., and J. H. J. Hoeijmakers. 1979. Extrachromosomal DNA. ICN-UCLA Symp. Mol. Cell. Biol. 15:533-548. [Google Scholar]
- 6.Borst, P., and J. H. J. Hoeijmakers. 1979. Kinetoplast DNA. Plasmid 2:20-40. [DOI] [PubMed] [Google Scholar]
- 7.Clayton, C. E., and P. Michels. 1996. Metabolic compartmentation in African trypanosomes. Parasitol. Today 12:465-471. [DOI] [PubMed] [Google Scholar]
- 8.Corell, R. A., L. K. Read, G. R. Riley, J. K. Nellissery, T. Allen, M. L. Kable, M. D. Wachal, S. Seiwert, P. J. Myler, K. D. Stuart, T. E. Allen, and S. D. Seiwert. 1996. Complexes from Trypanosoma brucei that exhibit deletion editing and other editing-associated properties. Mol. Cell. Biol. 16:1410-1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cruz-Reyes, J., A. Zhelonkina, L. Rusche, and B. Sollner-Webb. 2001. Trypanosome RNA editing: simple guide RNA features enhance U deletion 100-fold. Mol. Cell. Biol. 21:884-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drozdz, M., S. S. Palazzo, R. Salavati, J. O'Rear, C. Clayton, and K. Stuart. 2002. TbMP81 is required for RNA editing in Trypanosoma brucei. EMBO J. 21:1791-1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Englund, P. 1981. Kinetoplast DNA, p. 333-381. In M. Levandowsky and S. Hutner (ed.), Biochemistry and physiology of protozoa. Academic Press, Inc., New York, N.Y.
- 12.Estevez, A. M., and L. Simpson. 1999. Uridine insertion/deletion RNA editing in trypanosome mitochondria—a review. Gene 240:247-260. [DOI] [PubMed] [Google Scholar]
- 13.Feagin, J. E. 2000. Mitochondrial genome diversity in parasites. Int. J. Parasitol. 30:371-390. [DOI] [PubMed] [Google Scholar]
- 14.Feagin, J. E., J. M. Abraham, and K. Stuart. 1988. Extensive editing of the cytochrome c oxidase III transcript in Trypanosoma brucei. Cell 53:413-422. [DOI] [PubMed] [Google Scholar]
- 15.Feagin, J. E., D. P. Jasmer, and K. Stuart. 1987. Developmentally regulated addition of nucleotides within apocytochrome b transcripts in Trypanosoma brucei. Cell 49:337-345. [DOI] [PubMed] [Google Scholar]
- 16.Hajduk, S. 1978. Influence of DNA complexing compounds on the kinetoplast of Trypanosomatide. Prog. Mol. Subcell. Biol. 6:158-200.
- 17.Hirumi, H., and K. Hirumi. 1989. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 75:985-989. [PubMed] [Google Scholar]
- 18.Huang, C. E., J. Cruz-Reyes, A. G. Zhelonkina, S. O'Hearn, E. Wirtz, and B. Sollner-Webb. 2001. Roles for ligases in the RNA editing complex of Trypanosoma brucei: band IV is needed for U-deletion and RNA repair. EMBO J. 20:4694-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang, C. E., S. F. O'Hearn, and B. Sollner-Webb. 2002. Assembly and function of the RNA editing complex in Trypanosoma brucei requires band III protein. Mol. Cell. Biol. 22:3194-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Igo, R. P., Jr., S. S. Palazzo, M. L. K. Burgess, A. K. Panigrahi, and K. Stuart. 2000. Uridylate addition and RNA ligation contribute to the specificity of kinteoplastid insertion RNA editing. Mol. Cell. Biol. 20:8447-8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Igo, R. P., Jr., D. Weston, N. Ernst, A. K. Panigrahi, R. Salavati, and K. Stuart. 2002. Role of uridylate-specific exoribonuclease activity in kinetoplastid RNA editing. Eukaryot. Cell 1:112-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lanham, S. M. 1968. Separation of trypanosomes from the blood of infected rats and mice by anion-exchangers. Nature 218:1273-1274. [DOI] [PubMed] [Google Scholar]
- 23.Madison-Antenucci, S., J. Grams, and S. L. Hajduk. 2002. Editing machines: the complexities of trypanosome RNA editing. Cell 108:435-438. [DOI] [PubMed] [Google Scholar]
- 24.Madison-Antenucci, S., R. S. Sabatini, V. W. Pollard, and S. L. Hajduk. 1998. Kinetoplastid RNA-editing-associated protein 1 (REAP-1): a novel editing complex protein with repetitive domains. EMBO J. 17:6368-6376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McManus, M. T., M. Shimamura, J. Grams, and S. L. Hajduk. 2001. Identification of candidate mitochondrial RNA editing ligases from Trypanosoma brucei. RNA 7:167-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Missel, A., A. E. Souza, G. Norskau, and H. U. Göringer. 1997. Disruption of a gene encoding a novel mitochondrial DEAD-box protein in Trypanosoma brucei affects edited mRNAs. Mol. Cell. Biol. 17:4895-4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muhlpfordt, V. H. 1963. Uber die Bedeutung und Feinstruktur des Blepharoplasten bei parasitischen Flagellaten. Z. Tropenmed. Parasitol. 14:357-398. [PubMed] [Google Scholar]
- 28.Panigrahi, A. K., S. Gygi, N. Ernst, R. P. Igo, Jr., S. S. Palazzo, A. Schnaufer, D. Weston, N. Carmean, R. Salavati, R. Aebersold, and K. D. Stuart. 2001. Association of two novel proteins, TbMP52 and TbMP48, with the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol. 21:380-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panigrahi, A. K., A. Schnaufer, N. Carmean, R. P. Igo, Jr., S. Gygi, N. L. Ernst, S. S. Palazzo, D. Weston, R. Aebersold, R. Salavati, and K. D. Stuart. 2001. Four related proteins of the Trypanosoma brucei RNA editing complex. Mol. Cell. Biol. 21:6833-6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pollard, V. W., M. E. Harris, and S. L. Hajduk. 1992. Native mRNA editing complexes from Trypanosoma brucei mitochondria. EMBO J. 11:4429-4438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rusché, L. N., J. Cruz-Reyes, K. J. Piller, and B. Sollner-Webb. 1997. Purification of a functional enzymatic editing complex from Trypanosoma brucei mitochondria. EMBO J. 16:4069-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rusché, L. N., C. E. Huang, K. J. Piller, M. Hemann, E. Wirtz, and B. Sollner-Webb. 2001. The two RNA ligases of the Trypanosoma brucei RNA editing complex: cloning the essential band IV gene and identifying the band V gene. Mol. Cell. Biol. 21:979-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sabatini, R., and S. L. Hajduk. 1995. RNA ligase and its involvement in guide RNA/mRNA chimera formation. Evidence for a cleavage-ligation mechanism of Trypanosoma brucei mRNA editing. J. Biol. Chem. 270:7233-7240. [DOI] [PubMed] [Google Scholar]
- 34.Salavati, R., A. K. Panigrahi, B. A. Morach, S. S. Palazzo, R. P. Igo, and K. Stuart. 2002. Endoribonuclease activities of Trypanosoma brucei mitochondria. Mol. Biochem. Parasitol. 120:23-31. [DOI] [PubMed] [Google Scholar]
- 35.Schnaufer, A., G. J. Domingo, and K. D. Stuart. 2002. Natural and induced dyskinetoplastid trypanosomatids: how to live without mitochondrial DNA. Int. J. Parasitol. 32:1071-1084. [DOI] [PubMed] [Google Scholar]
- 36.Schnaufer, A., A. K. Panigrahi, B. Panicucci, R. P. Igo, Jr., R. Salavati, and K. D. Stuart. 2001. An RNA ligase essential for RNA editing and survival of the bloodstream form of Trypanosoma brucei. Science 291:2159-2162. [DOI] [PubMed] [Google Scholar]
- 37.Schneider, A. 2001. Unique aspects of mitochondrial biogenesis in trypanosomatids. Int. J. Parasitol. 31:1403-1415. [DOI] [PubMed] [Google Scholar]
- 38.Seiwert, S. D., S. Heidmann, K. D. Stuart, and K. Stuart. 1996. Direct visualization of uridylate deletion in vitro suggests a mechanism for kinetoplastid RNA editing. Cell 84:831-841. [DOI] [PubMed] [Google Scholar]
- 39.Simpson, L. 1987. The mitochondrial genome of kinetoplastid protozoa: genomic organization, transcription, replication, and evolution. Annu. Rev. Microbiol. 41:363-382. [DOI] [PubMed] [Google Scholar]
- 40.Songa, E. B., P. Paindavoine, E. Wittouck, N. Viseshakul, S. Muldermans, M. Steinert, and R. Hamers. 1990. Evidence for kinetoplast and nuclear DNA homogeneity in Trypanosoma evansi isolates. Mol. Biochem. Parasitol. 43:167-180. [DOI] [PubMed] [Google Scholar]
- 41.Stevens, S. W., D. E. Ryan, H. Y. Ge, R. E. Moore, M. K. Young, T. D. Lee, and J. Abelson. 2002. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell 9:31-44. [DOI] [PubMed] [Google Scholar]
- 42.Stuart, K., T. E. Allen, S. Heidmann, and S. D. Seiwert. 1997. RNA editing in kinetoplastid protozoa. Microbiol. Mol. Biol. Rev. 61:105-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stuart, K., and J. E. Feagin. 1992. Mitochondrial DNA of kinetoplastids. Int. Rev. Cytol. 141:65-88. [DOI] [PubMed] [Google Scholar]
- 44.Stuart, K., and S. R. Gelvin. 1980. Kinetoplast DNA of normal and mutant Trypanosoma brucei. Am. J. Trop. Med. Hyg. 29:1075-1081. [DOI] [PubMed] [Google Scholar]
- 45.Stuart, K. D. 1971. Evidence for the retention of kinetoplast DNA in an acriflavin-induced dyskinetoplastic strain of Trypanosoma brucei which replicates the altered central element of the kinetoplast. J. Cell. Biochem. 49:189-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stuart, K. D., A. K. Panigrahi, A. Schnaufer, M. Drozdz, C. Clayton, and R. Salavati. 2002. Composition of the editing complex of Trypanosoma brucei. Philos. Trans. R. Soc. Lond. Ser. B 357:71-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tielens, A. G., and J. J. Van Hellemond. 1998. The electron transport chain in anaerobically functioning eukaryotes. Biochim. Biophys. Acta 1365:71-78. [DOI] [PubMed] [Google Scholar]
- 48.Williams, N., and P. H. Frank. 1990. The mitochondrial ATP synthase of Trypanosoma brucei: isolation and characterization of the intact F1 moiety. Mol. Biochem. Parasitol. 43:125-132. [DOI] [PubMed] [Google Scholar]