Abstract
The dinoflagellates have very large genomes encoded in permanently condensed and histoneless chromosomes. Sequence alignment identified significant similarity between the dinoflagellate chromosomal histone-like proteins of Crypthecodinium cohnii (HCCs) and the bacterial DNA-binding and the eukaryotic histone H1 proteins. Phylogenetic analysis also supports the origin of the HCCs from histone-like proteins of bacteria.
The evolutionary development of histones and nucleosomes led to more efficient packaging of DNA into structured chromosomes, which was probably a key step in the evolution of eukaryotes from prokaryotes (13). The dinoflagellates represent an exception to this histone-based increase of genome size among eukaryotes (2). The dinoflagellate genome ranges from 2 to 3 pg (similar to the haploid human genome) to 200 pg (18), but ultrastructural and biochemical studies of the dinoflagellate chromatin identified an absence of nucleosome structures (3, 9) and histone proteins (17). Even more surprisingly, despite the lack of histones the dinoflagellate chromosomes are permanently condensed throughout interphase and have been proposed to exist in a liquid crystal state (4). Proteins associated with DNA have been isolated and characterized as predominantly low-molecular-mass proteins (14 to 17 kDa) rich in basic amino acids (17). Six chromosomal proteins were identified that represented only 10% of the DNA mass (25), a ratio even smaller than that of the prokaryotes. Immunoscreening of a cDNA expression library constructed from mRNA isolated from Crypthecodinium cohnii identified two clones (HCC1 and HCC2) with high similarity to the previously purified proteins. These clones confirmed the small size and basic nature of the proteins and allowed localization of the encoded proteins at the periphery of the permanently condensed chromosomes (19). Database searches at the time found no convincing homology to the histones and other chromosomal proteins. The ciliates and the apicomplexans, which together with the dinoflagellates form the alveolates, possess chromosomal proteins similar to histones of other eukaryotes (16), which generates an intriguing question as to the origin of the dinoflagellate histone-like proteins of C. cohnii (HCCs). A thorough investigation of the relationship between dinoflagellate HCCs and other prokaryotic and eukaryotic chromosomal binding proteins is necessary to facilitate a more complete understanding of the evolution of histones.
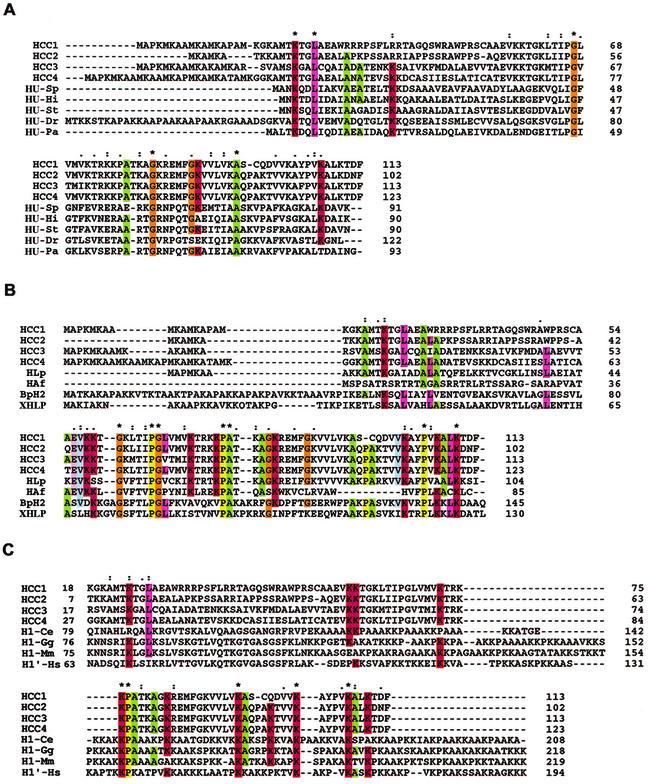
Accordingly, we designed two PCR primers against the 5′ and 3′ ends of the previously identified clones HCC1 and HCC2: pHCC1 sense, 5′-TATGGATCCATGGCCCCCAAGATG-3′, and antisense, 5′-TATAAGCTTGAAGTCTGTCTTGAGGGC-3′. Reverse transcription-PCR with mRNA isolated from C. cohnii amplified a number of DNA fragments of the expected size. Subsequent sequencing of these fragments allowed identification of two clones, HCC3 and HCC4, with homology to HCC1-HCC2 (Fig. 1A). The alignment of all four HCC variants enabled the proteins to be separated into two regions (Fig. 1A). The first 40 to 45 amino acids of each protein are highly variable apart from a region of 10 amino acids that is 100% conserved among HCC1, -2, and -4. Conversely, the final 70 to 75 amino acids of all four variants are highly homologous, with 64% absolute identity and 77% similarity when conservative substitutions are considered. HLP, an HCC-like gene product isolated by PCR from the dinoflagellate Lingulodinium polyedrum (6), has a high degree of homology to the HCCs (Fig. 1B).
FIG. 1.
Alignment of HCCs with HUs, histone-like proteins, and histone H1. (A) Multiple alignments of HCCs and selective bacterial HUs: HCC1 (A56581), HCC2 (B56581), HCC3 (AY128510), and HCC4 (AY128511); HU-Pa (Pseudomonas aeruginosa, JC4061); HU-Dr (Deinococcus radiodurans, Q9RZ89); HU-St (Salmonella enterica serovar Typhimurium, S01732); HU-Hi (Haemophilus influenzae, P43722); and HU-Sp (Streptococcus pneumoniae, AAK75224). (B) Multiple alignments of dinoflagellate and bacterial histone-like proteins: HLP (L. polyedrum, AF482694), HAF (A. fundyense, AAD20317), BpH2 (B. pertussis, AAB40156), and XHLP (X. fastidiosa 9a5c, AAF84745). (C) Multiple alignments of HCCs and selective eukaryotic histone H1 proteins: H1-Ce (Caenorhabditis elegans, NP_510410), H1-Gg (Gallus gallus, P09987), H1′-Hs (Homo sapiens, XP_009973), and H1-Mm (Mus musculus, AAA37814). Asterisks indicate positions which have a single, fully conserved residue; colons indicate that one of the “strong” groups in parentheses is fully conserved (STA/NEQK/NHQK/NDEQ/QHRK/MILV/MILF/HY/FYW); periods indicate that one of the “weaker” groups in parentheses is fully conserved (CSA/ATV/SAG/STNK/STPA/SGND/SNDEQK/NDEQHK/NEQHRK/FVLIM/HFY).
Our database searches with BLASTX revealed similarity to a number of bacterial DNA-binding proteins, especially the bacterial HUs (Fig. 1A). HUs are the most abundant small molecular multifunctional proteins in the Escherichia coli nucleoid (10). With the help of the software program ClustalX, we aligned the putative amino acid sequences of the HCCs with five randomly selected HUs (Fig. 1A). HUs are polypeptides of about 100 amino acids, while the HCCs are about 110 amino acids in length. However, since our PCR strategy was based on the amino and carboxyl termini of HCC1 and HCC2, we cannot completely rule out the possibility of extra sequences at the C terminus. Similarly, since the amino-terminal region of the protein contains a number of methionine residues, further library construction and screening as well as mRNA extension experiments will be required to determine whether the isolated genes encode full-length proteins. Given this, compared with the HUs, the HCCs have an extra amino-terminal sequence of about 20 to 30 amino acids in which the peptide sequence MAPK appears frequently. The highly variable nature of the amino termini suggests that these proteins are not redundant in their functions. A recent study (11) identified regions of similarity between the basic regions of the sea urchin H1b protein and the HCC2 protein. We have also been able to identify similar regions of homology between histone H1 proteins and HCCs (Fig. 1C). There is an approximately 10% identity between the two groups of proteins, predominantly in C-terminal positions occupied by basic residues. Some of these residues are also conserved between HCCs and HUs (Fig. 1A). The repeated occurrence of these residues has been postulated previously to result in a proline-kinked AK α-helix organization (7). Prototypical examples of this structure in histones of animals confer the ability to bind to DNA and to facilitate chromatin folding and condensation (22). One of the basic residues, K-107 of HCC1, is conserved in all eukaryotic H1 proteins and bacterial HUs analyzed.
The extent of similarity (23%) between HCCs and HUs is not restricted to the basic residues at the C terminus, as is found in the histones. Three glycine residues are identical in all HUs and HCCs but not in the H1 histones. One of these glycine residues (Gly-66 in HUs) lies within the DNA-binding β-ribbon arms of the dimeric HUs (23) and is located at the arm's flexible tip, which has been postulated to encircle the DNA (26).
Interestingly, histone H1-like sequences from Bordetella pertussis (BpH2) and Xylella fastidiosa (XHLP) were identified during the search for HCC homologues. Sequence comparison reveals that BpH2-XHLP has more than 35% sequence homology with all HCCs. Similar to sequence identities among the HCCs themselves, the C-terminal 65 amino acids show the highest identity with BpH2-XHLP (over 50%), including the three glycines. Based on sequence similarities, BpH2 and XHLP are more closely related to the HCCs, and together they form a group that is intermediate between the histone H1 proteins and the bacterial HUs. HCCs have higher homology to BpH2-XHLP than to HAF, the other histone-like protein from the dinoflagellate Alexandrium fundyense (Fig. 1C). The genomes of B. pertussis and the intercellular bacterium Chlamydia trachomatis contain other histone H1-like gene products, BpH1 and Hc1, respectively (8, 21), which are more closely affiliated with the histone H1 proteins and do not contain the glycine residues (sequences not shown). It is possible that the Hc1-BpH1 group was derived from an ancestral form between the ancestral BpH2-HCC and the eukaryotic histone H1.
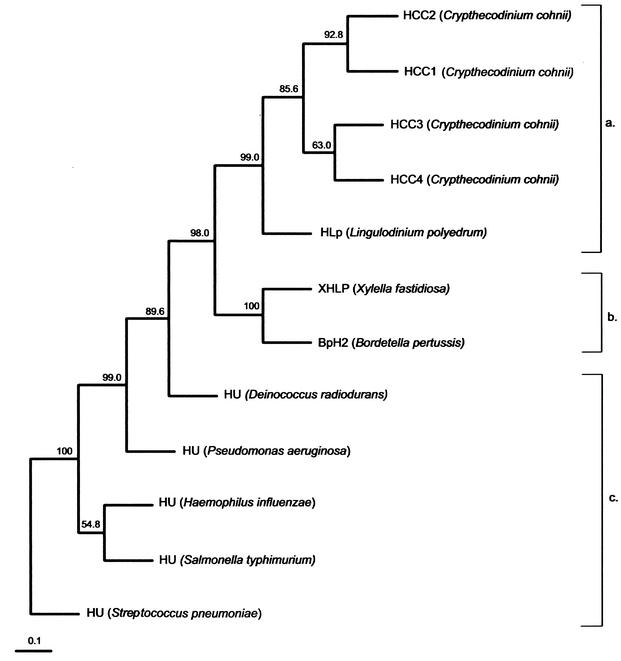
Using the HCC sequences and randomly chosen published sequences of HUs and HLPs, we constructed an unrooted UPGMA (unweighted pair group method with averages) tree by use of the PHYLIP program (Fig. 2). As expected, the three families form distinct clades. The HLPs form an intermediate clade between the HCCs and the bacterial HUs, confirming the intermediate nature of their primary sequences. The bootstrap values for the major nodes are highly significant. Assuming the evolution of histone H1 from the HUs, the dinoflagellate HCCs probably evolved from ancestral bacterial BpH2-like genes and represent an intermediate form before the loss of some bacterium-specific residues (e.g., the conserved glycines) and the acquisition of the wing domain. The ancestral alveolates did develop eukaryotic histones, as observed for the ciliates and the apicomplexans. Dinoflagellates may have subsequently lost these proteins, and a set of primitive bacterial BpH2-XHLP-like proteins may have been reintroduced via gene transfer from a prokaryotic source. This was supported by the isolation of the nucleus-encoded dinoflagellate RuBisCo (ribulose-1,5-biphosphate carboxylase-oxygenase) gene in Gonyaulax polyedra, which is more homologous to those of the proteobacteria than to other eukaryotes (14). Based on the condensed states and birefringence, three different chromosomal types can be observed in dinoflagellates (5), suggesting that different groups of dinoflagellates may have different basic chromosomal proteins. Interestingly, the homology between the HCCs and HAF, the putative DNA-binding protein from the dinoflagellate A. fundyense (24), is actually lower than the homology between the HCCs and BpH2-XHLP. One possible explanation may be the occurrence of a high evolutionary rate after the initial establishment of the BpH2-like gene in the alveolates.
FIG. 2.
A phylogenetic tree (UPGMA) inferred from the multiple alignments of amino acid sequences of histone-like proteins from dinoflagellate HCCs (a), bacterial histone-like protein family (b), and bacterial DNA-binding HU proteins (c) was constructed with PHYLIP version 3.5. The numbers at the nodes represent the bootstrap percentages by creation of 500 replicated data sets with SEQBOOT. The accession number for each protein sequence is as follows: HCC1 (C. cohnii), A56581; HCC2 (C. cohnii), B56581; BpH2 (B. pertussis), AAB40156; XHLP (X. fastidiosa), AAF84745; HU (Salmonella enterica serovar Typhimurium), S01732; HU (Streptococcus pneumoniae), AAK75224; HU (Haemophilus influenzae), P43722; HU (Pseudomonas aeruginosa), JC4061; HU (Deinococcus radiodurans), Q9RZ89.
The conservation of histones and the nucleosomes in the majority of eukaryotes implies that the eukaryotic histone-DNA complex probably existed in their last common ancestor. Both primary sequence data and three-dimensional structural comparisons support the origin of the eukaryotic core histones from the archaeal histones (15, 20). This contrasts with the evolution of the histone H1 as predicted from the sequence homologies with the eubacterial HU family. Both the conserved “histone fold” motif (three α-helices) of the core histones and the N-terminal α-helices of the eubacterial HUs, while having no primary sequence homology, function to enable dimer formation (1, 26). Interestingly, the lack of the histone fold motif in “histone-like proteins” in eubacteria, mitochondria, and dinoflagellates is correlated with the aggregation sensitivity of their DNA during conventional cytological fixation for electron microscopy (12). The HCCs and BpH2, BpH1, and other bacterial “histone-like” sequences probably represent a continuum in the evolution of the eukaryotic histone H1 from the prokaryotic HU families (including integration host factor and HN-S). Further phylogenetic analysis with these additional histone-like sequences will be required to give a more accurate account of the evolution of chromosomal proteins and chromosome structures.
Acknowledgments
The present study was partly supported by a CERG grant (HKUST6096/02 M) from the Research Grant Council of Hong Kong to J.T.Y.W.
REFERENCES
- 1.Arents, G., and E. N. Moudrianakis. 1995. The histone fold: a ubiquitous architectural motif utilized in DNA compaction and protein dimerization. Proc. Natl. Acad. Sci. USA 92:11170-11174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bendich, A. J., and K. Drlica. 2000. Prokaryotic and eukaryotic chromosomes: what's the difference? Bioessays 22:481-486. [DOI] [PubMed] [Google Scholar]
- 3.Bodansky, S., L. B. Mintz, and D. S. Holmes. 1979. The mesokaryote Gyrodinium cohnii lacks nucleosomes. Biochem. Biophys. Res. Commun. 88:1329-1336. [DOI] [PubMed] [Google Scholar]
- 4.Bouligand, Y., and V. Norris. 2001. Chromosome separation and segregation in dinoflagellates and bacteria may depend on liquid crystalline states. Biochimie 83:187-192. [DOI] [PubMed] [Google Scholar]
- 5.Cachon, J., H. Sato, M. Cachon, and Y. Sato. 1989. Analysis by polarizing microscopy of chromosomal structure among dinoflagellates and its phylogenetic involvement. Biol. Cell 65:51-60. [Google Scholar]
- 6.Chudnovsky, Y., J. F. Li, P. J. Rizzo, J. W. Hastings, and T. F. Fagan. 2002. Cloning, expression, and characterization of a histone-like protein from the marine dinoflagellate Lingulodinium polyedrum (Dinophyceae). J. Phycol. 38:543-550. [Google Scholar]
- 7.Churchill, M. E. A., and A. A. Travers. 1991. Protein motifs that recognize structural features of DNA. Trends Biochem. Sci. 16:92-97. [DOI] [PubMed] [Google Scholar]
- 8.Hackstadt, T., T. J. Baehr, and Y. Ying. 1991. Chlamydia trachomatis developmentally regulated protein is homologous to eukaryotic histone H1. Proc. Natl. Acad. Sci. USA 88:3937-3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herzog, M., and M. O. Soyer. 1981. Distinctive features of dinoflagellate chromatin. Absence of nucleosomes in a primitive species Prorocentrum micans E. Eur. J. Cell Biol. 232:295-302. [PubMed] [Google Scholar]
- 10.Hodges-Garcia, Y., P. J. Hagerman, and D. E. Pettijohn. 1989. DNA ring closure mediated by protein HU. J. Biol. Chem. 264:14621-14623. [PubMed] [Google Scholar]
- 11.Kasinsky, H. E., J. D. Lewis, J. B. Dacks, and J. Ausio. 2001. Origin of H1 linker histones. FASEB J. 15:34-42. [DOI] [PubMed] [Google Scholar]
- 12.Li, J.-Y., B. Arnold-Schulz-Gahmen, and E. Kellenberger. 1998. Histones and histone-like DNA-binding proteins: correlation between structural differences, properties and functions. Microbiology 145:1-2. [DOI] [PubMed] [Google Scholar]
- 13.Minsky, A., R. Ghirlando, and Z. Reich. 1997. Nucleosomes: a solution to a crowded intracellular environment. J. Theor. Biol. 188:379-385. [DOI] [PubMed] [Google Scholar]
- 14.Morse, D., P. Salois, P. Markovic, and J. W. Hastings. 1995. A nuclear-encoded form II RuBisCo in dinoflagellates. Science 268:1622-1624. [DOI] [PubMed] [Google Scholar]
- 15.Pereira, S. L., and J. N. Reeve. 1999. Archaeal nucleosome positioning sequence from Methanothermus fervidus. J. Mol. Biol. 289:675-681. [DOI] [PubMed] [Google Scholar]
- 16.Rizzo, P. J. 1976. Basic chromosomal proteins in lower eukaryotes: relevance to the evolution and function of histones. J. Mol. Evol. 8:79-94. [DOI] [PubMed] [Google Scholar]
- 17.Rizzo, P. J. 1981. Comparative aspects of basic chromatin proteins in dinoflagellates. Biosystems 14:433-443. [DOI] [PubMed] [Google Scholar]
- 18.Rizzo, P. J. 1991. The enigma of the dinoflagellate chromosomes. J. Protozool. 38:246-252. [Google Scholar]
- 19.Sala-Rovira, M., M. L. Geraud, D. Caput, F. Jaques, M. O. Soyer-Gobillard, G. Vernet, and M. Herzog. 1991. Molecular cloning and immunolocalization of two variants of the major basic nuclear protein (HCC) from the histone-less eukaryote Crypthecodinium cohnii (Pyrrhophyta). Chromosoma 100:510-518. [DOI] [PubMed] [Google Scholar]
- 20.Sandman, K., and J. N. Reeve. 2000. Structure and functional relationships of archaeal and eukaryal histones and nucleosomes. Arch. Microbiol. 173:165-169. [DOI] [PubMed] [Google Scholar]
- 21.Scarlato, V., B. Arico, S. Goyard, S. Ricci, R. Manetti, A. Prugnola, R. Manetti, P. Polverino-De-Laureto, A. Ullmann, and R. Rappuoli. 1995. A novel chromatin-forming histone H1 homologue is encoded by a dispensable and growth-regulated gene in Bordetella pertussis. Mol. Microbiol. 15:871-881. [DOI] [PubMed] [Google Scholar]
- 22.Subirana, J. A. 1990. Analysis of the charge distribution in the carboxyl-terminal region of histone H1 as related to its interactions with DNA. Biopolymers 29:1351-1357. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka, I., K. Appelt, J. Dijk, S. W. White, and K. S. Wilson. 1984. 3-Å resolution structure of a protein with histone-like properties in prokaryotes. Nature 310:376-381. [DOI] [PubMed] [Google Scholar]
- 24.Taroncher-Oldenburg, G., and D. M. Anderson. 2000. Identification and characterization of three differentially expressed genes, encoding S-adenosylhomocysteine hydrolase, methionine aminopeptidase, and a histone-like protein, in the toxic dinoflagellate Alexandrium fundyense. Appl. Environ. Microbiol. 66:2105-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vernet, G., M. Sala-Rovira, M. Maeder, F. Jaques, and M. Herzog. 1990. Basic nuclear proteins of the histone-less eukaryote Crypthecodinium cohnii (Pyrrhophyta): two-dimensional electrophoresis and DNA-binding properties. Biochim. Biophys. Acta 1048:281-289. [DOI] [PubMed] [Google Scholar]
- 26.Vism, H., R. Boelens, M. Mariani, R. Stroop, C. E. Vorgias, K. S. Wilson, and R. Kaptein. 1994. 1H, 13C, and 15N resonance assignments and secondary structure analysis of the HU protein from Bacillus stearothermophilus using two- and three-dimensional double- and triple-resonance heteronuclear magnetic resonance spectroscopy. Biochemistry 33:14858-14870. [DOI] [PubMed] [Google Scholar]