Abstract
Cytokinesis in the fission yeast Schizosaccharomyces pombe is regulated by a signaling pathway termed the septation initiation network (SIN). The SIN is essential for initiation of actomyosin ring constriction and septum formation. In a screen to search for mutations that can rescue the sid2-250 SIN mutant, we obtained scw1-18. Both the scw1-18 mutant and the scw1 deletion mutant (scw1Δ mutant), have defects in cell separation. Both the scw1-18 and scw1Δ mutations rescue the growth defects of not just the sid2-250 mutant but also the other temperature-sensitive SIN mutants. Other cytokinesis mutants, such as those defective for actomyosin ring formation, are not rescued by scw1Δ. scw1Δ does not seem to rescue the SIN by restoring SIN signaling defects. However, scw1Δ may function downstream of the SIN to promote septum formation, since scw1Δ can rescue the septum formation defects of the cps1-191β-1,3-glucan synthase mutant, which is required for synthesis of the primary septum.
A major function of the cell cycle and mitosis is to achieve accurate allocation of the two sets of duplicated sister chromatids to each daughter cell. At the end of each cell cycle, physical separation of the two daughter cells, a process known as cytokinesis, occurs and marks the completion of the whole cell cycle. It is key for the cell to execute all of these events in the correct order, at the right time, at the right place, and with high fidelity.
The fission yeast Schizosaccharomyces pombe provides an excellent eukaryotic model organism for the study of cytokinesis. Recent work with S. pombe has shed light on how septum formation and cytokinesis are regulated both spatially and temporally. The timing of cytokinesis in fission yeast is regulated by a signaling pathway termed the septation initiation network (SIN). The SIN is a spindle pole body (SPB)-localized signaling network that transmits a signal to the medial cortex at the end of anaphase to initiate actomyosin ring constriction and septum formation (33). An analogous pathway in Saccharomyces cerevisiae, termed the mitotic exit network, is required for mitotic exit and cytokinesis (4, 33). The SIN consists of a number of structural and signaling components (4, 33). Sid4p and Cdc11p form a complex at the SPB that is required for localization of all other SIN components (9, 24, 49). The Spg1p GTPase (43) functions upstream of the three protein kinases Cdc7p (13), Sid1p (19), and Sid2p (47), and both Sid1p and Sid2p have associated factors called Cdc14p (14) and Mob1p (21, 41), respectively. Spg1p is negatively regulated by a two-component GTPase-activating protein complex consisting of Cdc16p and Byr4p (17). Inactivation of the SIN results in failed cytokinesis and the formation of elongated and multinucleated cells that cannot form a division septum, while cells form several septa when the SIN is hyperactivated by inactivation of Cdc16p (34) or Byr4p (46).
The SIN becomes active in mitosis, and SIN components are recruited to the SPB or cell division site sequentially. The Cdc16p-Byr4p GTPase-activating protein complex localizes to the SPB in interphase (7, 27). As the mitotic spindle forms at metaphase, Cdc16p-Byr4p leaves the SPBs, and Spg1p at both SPBs switches to the active GTP-bound form (7, 27, 45). Cdc7p is then recruited to the SPB(s) by the GTP-bound form of Spg1p (45). During anaphase B, Spg1p is inactivated at one of the two SPBs by Cdc16p-Byr4p (7, 27), which causes loss of Cdc7p from that SPB. Sid1p-Cdc14p localizes to the Cdc7p-containing SPB and is required for activation of Sid2p-Mob1p, which then translocates to the actomyosin ring to trigger ring constriction and septation (19, 47). Targets of Sid2p-Mob1p at the cell division site required for cytokinesis are not known. One candidate target of the SIN, based on mutant phenotypes and genetic interactions, is the β-glucan synthase enzyme Cps1p, which is required for primary septum formation (26, 29).
Septum formation and cell separation require a number of distinct steps, including assembly and constriction of an actomyosin ring as in animal cells, septum formation, and septum disassembly to generate two equal-size daughter cells. The medially placed actomyosin ring structure is assembled in early mitosis (1) and then constricts at the end of anaphase. The division septum is assembled in a centripetal manner concomitant with actomyosin ring constriction. The main component of the S. pombe division septum is 1,3-β-glucan, which is synthesized by the β-glucan synthase Cps1p, which localizes to the actomyosin contractile ring concomitant with septum synthesis (10, 28). The secondary septum is then synthesized and the primary septum is degraded, allowing cell separation. At present, very little is known at a molecular level about how cell separation is achieved. However, the isolation and characterization of one transcription factor, Sep1p, whose mutations interfere with cell separation raised the possibility that expression of certain genes late in the cell cycle is required for efficient cell separation (40).
Presently nothing is known about how the SIN in S. pombe transmits the signal to initiate cytokinesis. Because Sid2p and Mob1p localize to the cell division site (21, 41, 47), they presumably transmit the signal to divide to the division machinery. Therefore, we screened for mutations that can suppress the growth defects in sid2 mutants. Here we describe the characterization of one of these suppressors, scw1-18, which on its own causes defects in cell separation. scw1-18 rescues all known SIN mutants but does not do so by restoring signaling through the SIN. Thus, the wild-type scw1+ gene may function downstream of or parallel with the SIN in regulating septum formation and stability in the final steps of cytokinesis.
MATERIALS AND METHODS
S. pombe growth conditions and genetic manipulations.
The fission yeast strains used in this study are listed in Table 1. Genetic crosses and general yeast techniques were performed as previously described (36). S. pombe strains were grown in rich medium (yeast extract [YE]) or Edinburgh minimal medium (EMM) with appropriate supplements (36). EMM with 5 μg of thiamine per ml was used to repress expression from the nmt1+ promoter. YE containing 100 mg of G418 (Calbiochem) per liter was used for selecting kanR-expressing cells. Microtubule formation was inhibited by the addition of various concentrations of methyl-2-benzimidazolecarbamate (MBC) in solid or liquid media. Synchronous populations of cells were generated by centrifugal elutriation with a Beckmann JE 5.0 rotor.
TABLE 1.
S. pombe strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| DM1560 | scw1-18 sid2-250 ura4-D18 h+ | Lab stock |
| DM105 | leu1-32 ura4-D18 ade6-210 h− | Lab stock |
| DM108 | leu1-32 ura4-D18 ade6-216 h− | Lab stock |
| DM1559 | scw1-18 ura4-D18 leu1-32 ade6-210 h+ | This study |
| DM1300 | scw1-18/scw1+ ade6-210/ade6-216 ura4-D18/ura4-D18 leu1-32/leu1-32 h+/h− | This study |
| DM1301 | scw1-18/scw1Δ::ura4+ ade6-210/ade6-216 ura4-D18/ura4-D18 leu1-32/leu1-32 h+/h− | This study |
| DM1274 | scw1Δ::ura4+ leu1-32 ura4-D18 ade6-210 h− | This study |
| DM1349 | scw1Δ::ura4+ leu1-32 ura4-D18 ade6 h+ | This study |
| DM1392 | scw1-3HA::kanR leu1-32 ura4-D18 ade6-210 h− | This study |
| DM1394 | scw1-13Myc::kanR leu1-32 ura4-D18 ade6-210 h− | This study |
| DM115 | sid4-A1 leu1-32 ura4-D18 ade6 h+ | Lab stock |
| DM1322 | scw1Δ::ura4+ sid4-A1 ade6 ura4-D18 leu1-32 h− | This study |
| DM274 | cdc11-123 ura4-D18 h+ | Lab stock |
| DM1326 | scw1Δ::ura4+ cdc11-123 ade6-210 ura4-D18 leu1-32 h− | This study |
| DM430 | spg1-106 ade6-210 ura4-D18 leu1-32 h+ | Lab stock |
| DM1412 | scw1Δ::ura4+ spg1-106 ade6-210 ura4-D18 leu1-32 h− | This study |
| DM1239 | cdc7-24 h+ | K. Gould |
| DM1364 | scw1Δ::ura4+ cdc7-24 leu1-32 h− | This study |
| DM458 | sid1-125 ade6-210 ura4-D18 leu1-32 h+ | Lab stock |
| DM1318 | scw1Δ::ura4+ sid1-125 ade6-210 ura4-D18 leu1-32 h− | This study |
| DM75 | sid1-239 ade6 ura4-D18 leu1-32 h+ | Lab stock |
| DM1366 | scw1Δ::ura4+ sid1-239 ura4-D18 leu1-32 ade6 h− | This study |
| DM436 | cdc14-118 ura4-D18 leu1-32 ade6-210 h+ | Lab stock |
| DM1328 | scw1Δ::ura4+ cdc14-118 ade6-210 ura4-D18 leu1-32 h+ | This study |
| DM429 | sid2-250 ade6 ura4-D18 leu1-32 h+ | Lab stock |
| DM1320 | scw1Δ::ura4+ sid2-250 ade6 ura4-D18 leu1-32 h− | This study |
| DM670 | mob1-1 ura4-D18 leu1-32 ade6 his3-D1 + pBGMob1-ts h− | Lab stock |
| DM1368 | scw1Δ::ura4+ mob1-1 ura4-D18 leu1-32 ade6 his3-D1 h− | This study |
| DM322 | cdc12-112 ura4-D18 leu1-32 ade6-210 h+ | Lab stock |
| DM1370 | scw1Δ::ura4+ cdc12-112 ura4-D18 leu1-32 ade6-210 h− | This study |
| DM2 | cdc15-140 ura4-D18 h+ | Lab stock |
| DM1372 | scw1Δ::ura4+ cdc15-140 ura4-D18 ade6-210 h+ | This study |
| DM916 | nda3-KM311 leu1-32 ura4-D18 ade6-21X h− | Lab stock |
| DM1268 | scw1-18 nda3-KM311 ade6-210 leu1-32 ura4-D18 h+ | This study |
| DM1459 | cdc11-123 GFP-mob1 ade6 ura4-D18 leu1-32 h+ | This study |
| DM1461 | scw1Δ::ura4+ cdc11-123 GFP-mob1 ade6 ura4-D18 leu1-32 h− | This study |
| DM1465 | cdc11-123 cdc7-GFP::ura4+ ade6-210 ura4-D18 leu1-32 h+ | This study |
| DM1467 | scw1Δ::ura4+ cdc11-123 cdc7-GFP::ura4+ ade6-210 ura4-D18 leu1-32 h+ | This study |
| DM497 | sid2-13Myc::kan ura4-D18 leu1-32 ade6-210 h− | Lab Stock |
| DM1440 | scw1Δ::ura4+ sid2-13Myc::kanR ade6 ura4-D18 leu1-32 h+ | This study |
| DM1443 | cdc11-123 sid2-13Myc::kanR ade6 ura4-D18 leu1-32 h+ | This study |
| DM1439 | scw1Δ::ura4+ cdc11-123 sid2-13Myc::kanR ade6 ura4-D18 leu1-32 h+ | This study |
| DM1447 | cdc7-24 sid2-13Myc::kanR leu1-32 ade6 h+ | This study |
| DM1445 | scw1Δ::ura4+ cdc7-24 sid2-13Myc::kanR leu1-32 h+ | This study |
| DM1214 | cps1-191 leu1-32 lys1-131 ura4-D18 ade6-21X h− | Lab stock |
| DM1535 | scw1Δ::ura4+ cps1-191 leu1-32 ura4-D18 ade6-21X h− | This study |
| DM1569 | cps1-UV1 leu1-32 ura4-D18 ade6-210 h− | Balasubramanian lab |
| DM1622 | scw1Δ::ura4+ cps1-UV1 leu1-32 ura4-D18 ade6-21X h+ | This study |
| DM1570 | cps1-UV2 leu1-32 ura4-D18 ade6-216 h− | Balasubramanian lab |
| DM1624 | scw1Δ::ura4+ cps1-UV2 leu1-32 ura4-D18 ade6-21X h− | This study |
| DM878 | sep1-1 leu1-32 ura4-D18 h− | Sipiczki lab |
| DM1587 | sep1-1 cps1-191 leu1-32 ura4-D18 h+ | This study |
To delete scw1+, the whole scw1+ open reading frame (ORF) was replaced by the ura4+ gene via a PCR-based procedure (2), using the oligonucleotides 5′-GGT TAC TTT ATC AAC CAC TTT GTC ATT CTT TTT TCT CTT CTT TTC AAT TAC CAT TAT ATA TAA TTT GCA AAC GCC AGG GTT TTC CCA GTC ACG AC-3′ and 5′-GGA CCT AAA GTC CTT GCA AGG TAT TGA TGA ATA ATG ATA AAA TGA AGA CGA GAA AAT GCT AGA TGA GCT ATT TGC CAG CGG ATA ACA ATT TCA CAC AGG A-3′.
Strains expressing Scw1p carboxy-terminally tagged with green fluorescent protein (GFP) and 13Myc were generated by PCR-based gene targeting (2) with the oligonucleotides 5′-GAC TCT TTG CTT AAT CAT ACT GGT GGA CAT AAC GAA GTC CAC GCC AGT CCC AGT TGG GGT AAT AAT CTA ATG TAT GGC AAA CGG ATC CCC GGG TTA ATT AA-3′ and 5′-GCT TAA CAG ATG GTT AAA GTT GCA TGC AGT CAA AGT GGA ATA GAT CGC AAC TTT TGA TTA ACA AAG AAT CAA TAT GCA AAA CGA ATT CGA GCT CGT TTA AAC-3′. Correct chromosomal integration in the resultant kanR transformants was confirmed by PCR analysis.
Diploid strains were constructed by crossing haploid strains carrying the ade6-complementing mutation ade6-210 or ade6-216 and selecting for ade+ white colonies.
Isolation of scw1-18 and cloning of scw1+.
The scw1 mutation was isolated in a screen for sid2-250 suppressors. Approximately 2 × 108 cells of a sid2-250 ura4-D18 leu1-32 ade6 h+ strain (DM429) were mutagenized for 15 min with nitrosoguanidine as described previously (36) and plated at 36°C. This screen yielded several hundred colonies, of which 125 were initially picked for further characterization. Many of these were discarded after further testing due to poor rescue of the sid2-250 mutation. From the remaining strains, 43 with representative phenotypes were picked and crossed to the wild type to determine if they represented single mutations and whether they had phenotypes on their own. Twenty-one of these mutants displayed a multiseptate phenotype (Fig. 1) and were kept for further study. The other mutants either had no phenotype on their own or had multiple mutations that contributed to the sid2-250 suppression. Complementation analysis of the remaining 21 mutants revealed that 19 fell into a single complementation group, which we later named scw1. One of these mutants, carrying scw1-18, was picked for further study. The other two mutants each fell into separate complementation groups and are not described here.
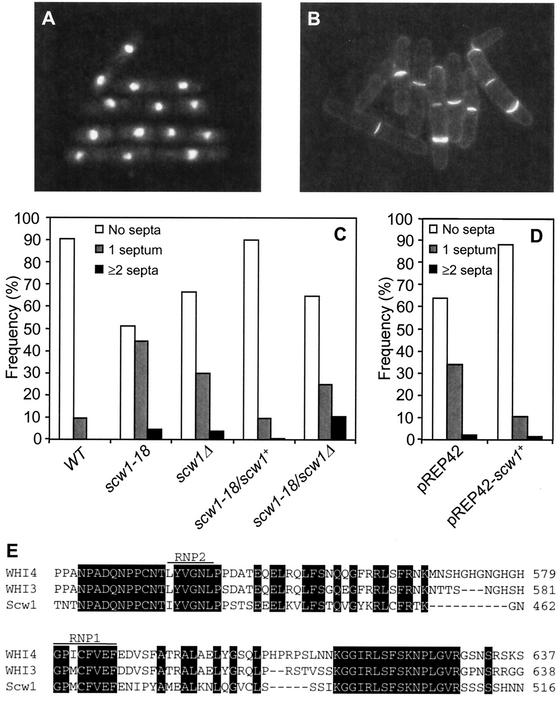
FIG. 1.
Characterization of scw1-18 and scw1Δ mutant strains. (A and B) scw1-18 (DM1559) cells from a log-phase culture at 30°C were fixed and stained with DAPI to visualize nuclei (A) or stained with Calcofluor to visualize cell wall and septa (B). (C) Wild-type (DM105), scw1-18 (DM1559), scw1Δ (DM1274), scw1-18/scw1+ (DM1300), and scw1-18/scw1Δ (DM1301) strains were grown in YE to mid-log phase at 30°C and then scored for the septation index. WT, wild type. (D) scw1-18 mutant cells (DM1559) containing either the pREP42 or pREP42-scw1+ plasmid were grown in EMM without uracil and thiamine for 24 h at 30°C, at which time the septation index was scored. (E) Alignment of the RNP domain of Scw1p with those of two related budding yeast proteins, Whi3 and Whi4. Conserved residues are marked with black boxes. The octamer RNP1 and hexamer RNP2 are labeled as previously defined (39).
To clone the scw1+ gene, we first mapped its approximate chromosomal location in a swi5 mutant background, which reduces recombination frequencies and allows for a crude map position to be determined (31). This analysis demonstrated a weak linkage to the ura4+ locus. Further mapping in a wild-type (non-swi5) background was carried out by crossing scw1-18 to strains bearing mutations in the region of ura4+. This analysis showed that scw1-18 was tightly linked (1.1 map units; 44 parental ditype and 1 tetratype) to the cut1-205 mutation. We then obtained cosmids in the region of the cut1+ locus from the Sanger Center. The his7+ gene was inserted into these cosmids as previously described (37), and they were transformed into scw1-18 his7-306 cells and tested for rescue of the multiseptate phenotype of scw1-18. Two of these cosmids, c5E4 and c16C4, were able to rescue the scw1-18 phenotype. Candidate genes from the region of overlap between these two cosmids were cloned into the pREP42 vector and tested for rescue of scw1-18. This analysis showed that the SPCC16C4.07 ORF was capable of rescuing scw1-18.
To clone scw1+ into the pREP42 vector (5), the coding region for the scw1+ gene was amplified by PCR from the wild-type S. pombe genome (using oligonucleotides 5′-CAT GCA TAT GTT TGT GGG ATC ACC G-3′ and 5′-CAT GGG ATC CCT ATT TGC CAT ACA TTA G-3′), and the product was digested with NdeI and BamHI and then subcloned into the pREP42 vector containing the thiamine-repressible nmt1 promoter (32).
Microscopy.
Immunofluorescence microscopy was done as described previously (3). For tubulin staining, primary monoclonal antitubulin antibody TAT1 (52) was followed by secondary anti-mouse Texas red or Alexa 594-immunoglobulin G (Molecular Probes). GFP fusion proteins were observed in cells after fixation with 3.7% formaldehyde. DNA was visualized with 4′,6-diamidino-2-phenylindole (DAPI) (Sigma) at 2 μg/ml. Photomicrographs were obtained with a Nikon Eclipse E600 fluorescence microscope coupled to a cooled charge-coupled device camera (ORCA-ER; Hamamatsu), and image processing and analysis were carried out with IPLab Spectrum software (Signal Analytics Corporation, Vienna, Va.).
RESULTS
Isolation and characterization of scw1-18.
In order to identify potential targets and/or regulators of Sid2p, we screened for mutations that could suppress the temperature-sensitive growth defect of sid2-250 mutant cells (see Materials and Methods). The majority of the suppressors identified had defects in cell separation, resulting in a high percentage of cells with single or multiple septa and multiple nuclei. Complementation analysis of 21 of these suppressors revealed that all but 2 fell into a single complementation group. In the course of these experiments, the same gene was isolated and called scw1 (23), and hence we have maintained this nomenclature. The other two mutations defined their own complementation groups and have not yet been further characterized. One scw1 allele, scw1-18, was chosen for further analysis. The major phenotypes of scw1-18 included multiple septa and a relatively high septation index, with 40 to 50% of log-phase cells having one or more septa (Fig. 1). We never observed more than two nuclei in each cell compartment in scw1-18 mutant cells, suggesting that the placement of the actomyosin ring and septum formation occur normally but the cells have a defect in cell separation. These cells did not show obvious heat or cold sensitivity (data not shown).
Identification of the scw1+ gene.
The scw1+ gene was cloned through a combination of genetic and physical mapping (see Materials and Methods). Expression of scw1+ in the scw1-18 strain rescued the cell separation defect in these cells (Fig. 1D). scw1+ is predicted to encode a protein of 561 amino acids with a molecular mass of 60 kDa. A database search revealed that Scw1p shows homology to RNA binding proteins, especially to two budding yeast proteins, WHI3 and WHI4, with the highest identity in the RNA binding domain (39) (Fig. 1E). Interestingly, WHI3 also seems to be involved in cell cycle regulation by causing localized translation of the CLN3 cyclin RNA (18). Deletion of the whole scw1+ ORF showed that the gene is not essential (see Materials and Methods). Closer examination revealed that the scw1Δ null mutant showed a cell separation defect similar to that of the scw1-18 mutant (Fig. 1A and data not shown), and it also could rescue the sid2-250 mutant at 30 and 36°C (see Fig. 3). Therefore, scw1-18 and scw1Δ behave similarly, suggesting that scw1-18 is a loss-of-function mutation. To confirm that scw1-18 represents a mutation in the gene scw1+, we tested whether scw1-18 and scw1Δ were complementing mutations. We constructed an scw1-18/scw1Δ diploid strain to test whether these cells showed a multiseptate phenotype. These diploid cells showed an increased percentage with single and multiple septa compared to the control diploid scw1-18/scw1+ cells or wild-type cells (Fig. 1C), consistent with scw1-18 being a mutant allele of scw1+.
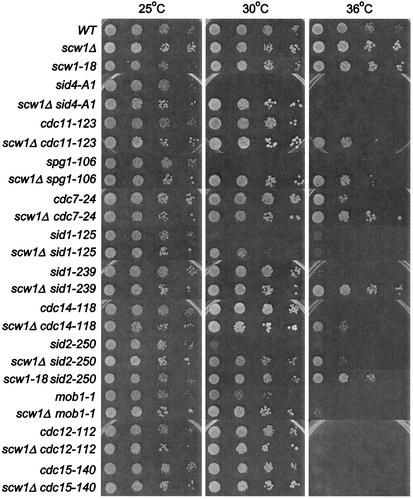
FIG. 3.
scw1Δ can rescue SIN mutants but not the actomyosin ring mutants. The indicated single and double mutant strains were tested by serial dilution patch test for growth. (Note that the single and double mutant strains used are listed in the same order in Table 1.) Dilutions shown were 10-fold, starting with 104 cells. Strains were pregrown in liquid YE at 25°C and then spotted onto YE plates at the indicated temperatures and incubated for 3 to 5 days before photography. WT, wild type.
Scw1p localizes to the cytoplasm.
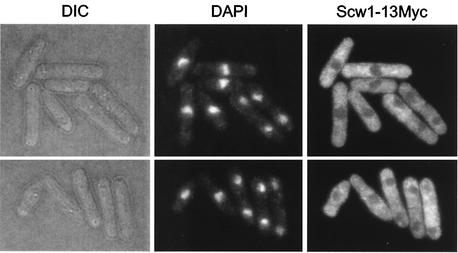
To determine the cellular localization of Scw1p, we tagged Scw1p by fusing the genomic scw1+ ORF to either GFP or 13Myc. Both Scw1p fusions were functional, since the strains expressing them were wild type in morphology, and the tagged alleles were unable to rescue the sid2-250 mutation (data not shown). Direct visualization of GFP fusion proteins and indirect immunofluorescence of Myc-fused proteins demonstrated that the proteins localized diffusely to the cytoplasm and were excluded from the nucleus at all stages of the cell cycle (Fig. 2 and data not shown). Scw1p was not observed at the SPB or cell division site.
FIG. 2.
Intracellular localization of epitope-tagged Scw1p. Wild-type cells expressing scw1-13Myc (DM1394) were grown in YE to log phase and then fixed and subjected to indirect immunofluorescence with anti-Myc antibody. DIC, differential interference contrast.
scw1Δ can rescue all SIN mutants but not actomyosin ring mutants.
We next tested whether the scw1Δ mutation specifically rescued the sid2-250 mutant or was capable of rescuing other SIN mutants. We constructed double mutants between scw1-18 or scw1Δ and all the other available temperature-sensitive SIN mutants, including the sid4-A1, cdc11-123, spg1-106, cdc7-24, sid1-125, sid1-239, cdc14-118, sid2-250, and mob1-1 mutants. Interestingly, serial dilution drop tests on plates at different temperatures showed that both scw1-18 and scw1Δ rescued the growth defects of all of these SIN mutants. The degree of rescue varied depending on the allele, with very strong mutant alleles, such as sid4-A1 and sid1-125, being rescued at 30 but not 36°C (Fig. 3 and data not shown), whereas other mutants were rescued at both 30 and 36°C. This analysis suggested that the scw1Δ mutation was not able to bypass the SIN to promote cytokinesis but required some degree of residual SIN signaling to promote rescue. Microscopic examination of double mutant cells (see below) showed a strong correlation between the ability of scw1Δ to rescue the temperature-sensitive growth defects of the SIN and its ability to rescue the SIN septation defects. For example, we found that the scw1Δ mutation rescues the septation defect of sid4-A1 and sid1-125 mutant cells at 30 but not 36°C, consistent with its ability to rescue these mutants growth defects at 30 but not 36°C, (Fig. 3 and data not shown). A similar correlation was observed for other SIN mutants, suggesting that scw1Δ rescues the growth defect of SIN mutants by restoring septum formation in these mutants.
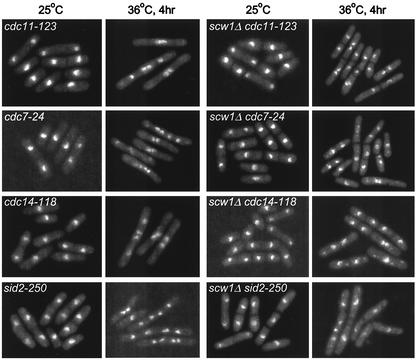
Microscopic analysis of these double mutant cells in liquid cultures at 36°C revealed that these cells could form septa, although sometimes not in a very efficient manner, leaving some cell compartments without nuclei while others had multiple nuclei (Fig. 4). In contrast, double mutants between scw1Δ and temperature-sensitive actin ring formation mutants, such as the cdc3-124, cdc12-112, and cdc15-140 mutants, failed to show any rescue of growth defects at 36°C (Fig. 3 and data not shown) and did not suppress the septum formation defects leading to multiple nuclei (data not shown). In addition, scw1-18 was also unable to suppress the growth defects of other temperature-sensitive mutants such as the alp4-1891, alp6-719, and alp16::ura4+ mutants (data not shown) (16, 51), further demonstrating that the suppression of the SIN is specific.
FIG. 4.
Microscopic analysis of double mutants between scw1Δ and SIN mutations. Cells of the indicated strains were grown in YE at 25°C to log phase and then shifted to 36°C for 4 h before being fixed and stained with DAPI.
scw1-18 can stabilize microtubules.
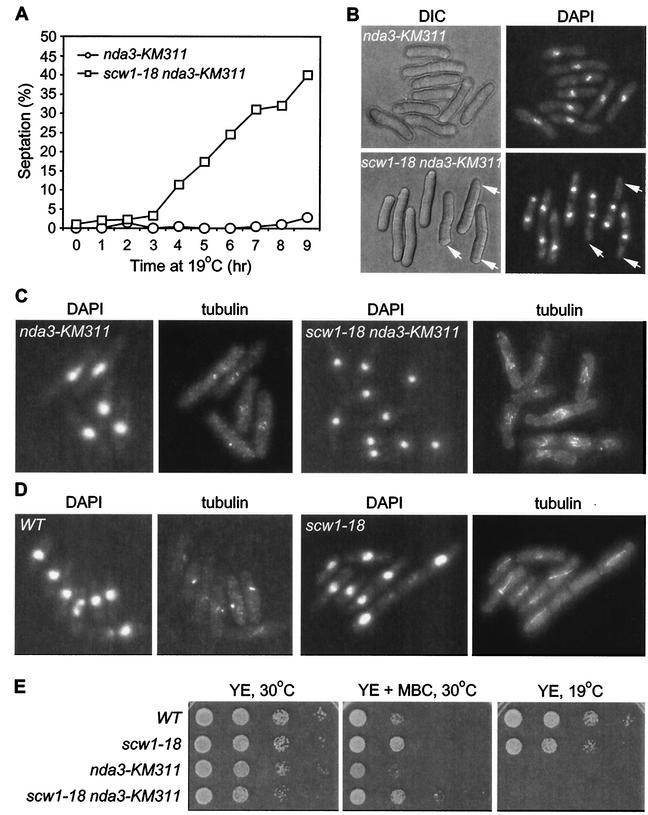
The fact that scw1Δ suppresses the SIN implies that wild-type Scw1p antagonizes the SIN. Mutants with mutations in other antagonists of the SIN, such as dma1, zfs1, and cdc16, have defects in the spindle checkpoint (6, 12, 38), and inappropriate activation of the SIN can cause spindle checkpoint defects (20). Therefore, we tested whether the scw1-18 mutation, like other SIN suppressors, also compromises the spindle checkpoint-mediated arrest caused by inactivation of the cold-sensitive nda3-KM311 β-tubulin mutant (50). The nda3-KM311 mutant normally arrests in early mitosis at the restrictive temperature due to the failure to form a mitotic spindle and does not exit mitosis and septate although the medial actomyosin ring has been formed (8, 35). We generated synchronous cultures, in early G2 phase, of both nda3-KM311 and scw1-18 nda3-KM311 mutants by elutriation and then shifted them to the restrictive temperature of 19°C. Septation was scored at 1-h intervals for both cultures, as a convenient way to monitor exit from mitosis. As expected, the nda3-KM311 control cells arrested without a septum and with a single nucleus (Fig. 5A and B). In sharp contrast, the scw1-18 nda3-KM311 mutant started to accumulate cells with one or more septa after 3 h, and by 9 h 40% of the cells had septated (Fig. 5A and B). Interestingly, the scw1-18 nda3-KM311 mutant cells after 9 h at 19°C could sometimes accomplish chromosome segregation but failed to fully separate their chromosomes, often resulting in anucleate cell compartments (Fig. 5B). This implies that the scw1-18 mutation may rescue the nda3-KM311 cell cycle block by partially restoring microtubule function in these cells. To test this, we examined the microtubules of asynchronous nda3-KM311 and scw1-18 nda3-KM311 cells that had been shifted to the restrictive temperature for 6 h. As expected, nda3-KM311 cells had no microtubules and showed staining only at the SPB. In contrast, scw1-18 nda3-KM311 cells displayed many short microtubules around or across the nucleus (Fig. 5C), suggesting that the scw1-18 mutation is not spindle checkpoint defective but partially restores microtubules in the nda3-KM311 mutation. The increased stability of microtubules in scw1-18 nda3-KM311 mutant cells is not sufficient to restore viability of nda3-KM311 cells at 19°C (Fig. 5E). The increased stability of microtubules in the scw1-18 nda3-KM311 mutant is not specific to nda3-KM311 mutant cells, because when we compared wild-type and scw1-18 cells treated with the microtubule-depolymerizing drug MBC, we found that the scw1-18 cells displayed more microtubules than wild-type cells (Fig. 5D). However, in terms of cell viability, the scw1-18 cells were only slightly resistant to MBC compared to wild-type cells (Fig. 5E).
FIG. 5.
The scw1-18 mutation can stabilize microtubules. (A) nda3-KM311 and scw1-18 nda3-KM311 cells were synchronized at early G2 by centrifugal elutriation from log-phase cultures grown at 30°C. Synchronized cells were then shifted to 19°C, and septation was scored for both cultures at 1-h intervals. (B) DAPI-stained cells at the 9-h time point. scw1-18 nda3-KM311 mutant cells often only partially segregated their DNA, leading to anucleate cell compartments (arrows). DIC, differential interference contrast. (C) DAPI and tubulin staining with TAT1 antibody of asynchronous cells of the indicated genotypes 6 h after a shift to 19°C. (D) DAPI and tubulin staining with TAT1 antibody of cells treated with 25 mg of MBC per ml for 2 h at 30°C. WT, wild type. (E) Serial dilution patch test for growth of the indicated single and double mutant strains. Dilutions shown were 10-fold, starting with 104 cells. Strains were pregrown in liquid YE at 25°C and then spotted onto YE plates or on YE with 10 mg of MBC per ml and incubated at the indicated temperatures for 3 to 5 days before photography.
scw1Δ does not rescue SIN mutants by restoring SIN protein localization and activity.
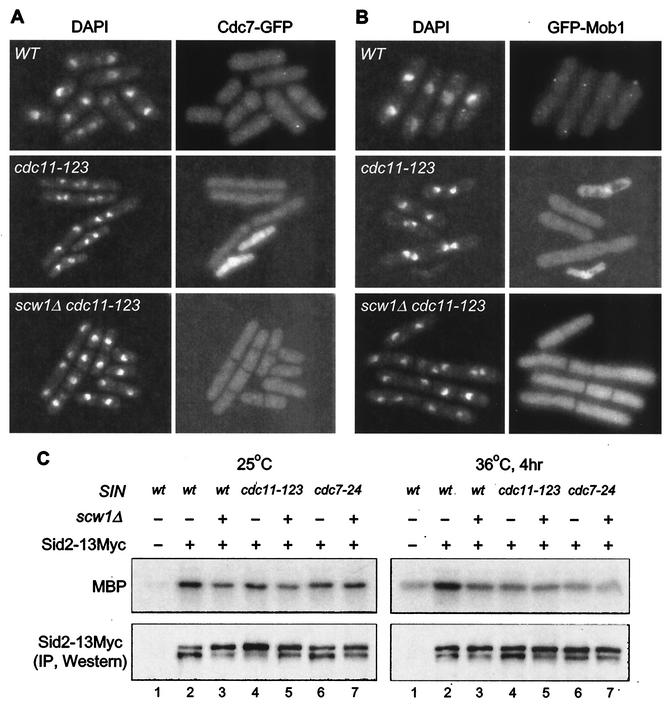
Interestingly, scw1Δ rescues cdc11-123 mutants that are defective for localization of downstream SIN components and activation of Sid2p kinase activity (19, 21, 24, 41, 47, 49). We tested whether either of these defects were restored by the scw1Δ mutation. We first examined whether the absence of scw1+ restored localization of SIN components in cdc11-123 mutants. Because the scw1Δ mutation was able to rescue cdc11-123 well only in liquid medium at 33.5°C, localization experiments were carried out at this temperature. Both Cdc7p-GFP and GFP-Mob1p were readily observed at SPBs in late anaphase or telophase wild-type cells at both 25 and 33.5°C (Fig. 6A and B and data not shown). The intensity of Cdc7p-GFP and GFP-Mob1p signals at the SPB in both cdc11-123 single and cdc11-123 scw1Δ double mutants at 25°C was reduced compared to that in the wild type (data not shown). In cdc11-123 cells incubated at 33.5°C, faint Cdc7-GFP and GFP-Mob1p signals at the SPB could only occasionally be observed (Fig. 6A and B). Similar results were observed for the cdc11-123 scw1Δ double mutant strain, and quantification of the SPB signals showed that the scw1Δ mutation was unable to restore the Cdc7-GFP and GFP-Mob1p SPB localization defect of cdc11-123 cells (Fig. 6A and B and data not shown). At higher temperatures, scw1Δ did not rescue cdc11-123 cells and no SPB localization of Cdc7p and Mob1p was observed, consistent with scw1Δ-mediated suppression of the SIN requiring a low level of SIN function. Similar results were observed when Sid1p and Sid2p were examined in cdc11-123 single or scw1Δ cdc11-123 double mutant cells (data not shown). Thus, the scw1Δ mutation does not rescue the cdc11-123 mutation by promoting localization of SIN components to the SPB.
FIG. 6.
scw1Δ does not rescues SIN mutants by restoring SIN localization and activity. (A and B) The indicated strains were grown in YE at 25°C to log phase and then shifted to 33.5°C for 4 h before being fixed and stained with DAPI and photographed for GFP fluorescence. WT, wild type. (C) The scw1Δ mutation does not promote Sid2p kinase activity. Various strains were harvested either in log phase at 25°C or after being shifted for 4 h to 36°C. The presence (+) or absence (−) of either Sid2-13Myc or the scw1Δ mutation is indicated, as is the presence of SIN mutations. Immune complexes were prepared from lysates with anti-Myc antibody, and then a kinase assay was performed with MBP as an artificial substrate, as previously described (47). Each sample was split in two, and phosphorylation of MBP (upper panels) or Sid2p-13Myc levels (lower panel) were detected by phosphorimager and Western analysis, respectively. IP, immunoprecipitation.
Since the experiments described above suggested that the scw1Δ mutation does not rescue SIN mutants by promoting localization of SIN components, we next wanted to test whether it could be functioning by increasing signaling through the pathway. Since Sid2p kinase activity depends on all other SIN proteins (47), we analyzed Sid2p kinase activity in scw1Δ cdc11-123 and scw1Δ cdc7-24 mutants. 13Myc epitope-tagged Sid2p was first immunoprecipitated with an anti-Myc antibody, and then in vitro Sid2 kinase assays were performed with myelin basic protein (MBP) as an artificial substrate. Sid2p-13Myc immune complexes were prepared from lysates of cells incubated at the permissive (25°C) and restrictive (36°C) temperatures for the cdc11-123 and cdc7-24 mutant strains. As previously observed, Sid2p kinase activity is reduced in cdc11-123 and cdc7-24 mutant strains compared to wild-type cells (Fig. 6C, lanes 4, 6, and 2, respectively). The presence of the scw1Δ mutation did not restore Sid2p kinase activity to cdc11-123 and cdc7-24 mutants (Fig. 6C, lanes 5 and 7, respectively), and in fact, the scw1Δ single mutant had somewhat reduced Sid2p kinase activity (Fig. 6C, lane 3). Similar results were obtained at a reduced restrictive temperature, where better rescue by the scw1Δ mutation is observed (data not shown). Taken together, these results suggest that the scw1Δ mutation does not rescue SIN mutants by restoring localization of SIN proteins or increasing signaling through the SIN.
The scw1 mutation restores septum formation in the cps1-191 β-glucan synthase mutant.
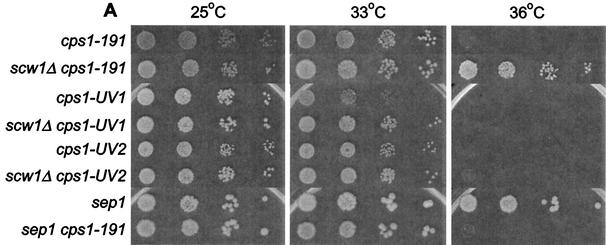
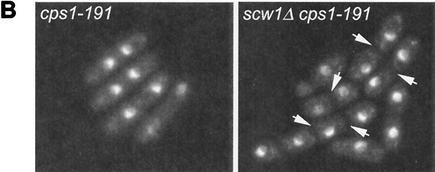
The analysis described above indicated that the scw1 mutation does not rescue the SIN mutants by restoring signaling through the SIN but may be promoting septum formation by acting downstream of the SIN. Previous genetic studies have suggested that the β-glucan synthase enzyme Cps1p may function downstream of the SIN to promote septum formation (29). Like SIN mutants, temperature-sensitive cps1 mutants fail to form septa at the restrictive temperature and arrest as binucleate cells. To test whether Scw1p could be affecting septum formation more directly, we tested whether the scw1Δ mutation could rescue the cps1 mutant strains. Interestingly, the scw1Δ mutation could rescue the temperature-sensitive growth defect of cps1-191 cells (Fig. 7A). This effect was allele specific, since scw1Δ was unable to rescue cps1-UV2 and could only weakly rescue cps1-UV1 at the reduced restrictive temperature of 33°C (Fig. 7A). Examination of single and double mutant cells after incubation at the restrictive temperature showed that scw1Δ cps1-191 cells were capable of making septa, unlike cps1-191 single mutant cells (Fig. 7B). Furthermore, the rescue of cps1-191 was not an indirect consequence of the cell separation defect in scw1Δ cells, since the sep1-1 cell separation mutant was unable to rescue the cps1-191 mutant strain (Fig. 7A).
FIG. 7.
scw1Δ rescues the cps1-191 mutant strain. (A) Strains of the indicated genotypes were tested by serial dilution patch test for growth. The dilutions shown were 10-fold, starting with 104 cells. Strains were pregrown in liquid YE at 25°C and then spotted onto YE plates at the indicated temperatures and incubated for 3 to 5 days before photography. (B) Cells of the indicated genotypes were grown in YE at 25°C to log phase and then shifted to 36°C for 4 h before being fixed and stained with DAPI. Septa are apparent as dark regions between nuclei (arrows).
DISCUSSION
In this study, we have identified the gene scw1+ in a genetic screen for potential regulators and effectors of the SIN pathway in S. pombe. An scw1 deletion mutation can suppress all of the mutations in the SIN pathway and shows a cell separation phenotype on its own. The suppression of the SIN seems to be specific, since the scw1Δ mutation does not suppress mutations in other cytokinesis genes, such as those required for actomyosin ring formation. How, then, does scw1 loss of function suppress SIN mutations? First, scw1Δ does not seem to bypass the SIN pathway, because scw1Δ does not rescue the strongest SIN mutations, such as sid4-A1 or sid1-125, at the highest restrictive temperature. In addition, the scw1Δ mutation is unable to suppress sid2-250 spg1-106 double mutants at 36°C, whereas it can suppress either single mutant at 36°C (data not shown). Together, these results indicate that scw1Δ cannot suppress a total loss of function in the SIN pathway. This suggests that scw1Δ either acts to enhance weak SIN signaling or removes an inhibitor downstream of the SIN. To study this, we examined cdc11-123 mutants, which have defects in localizing SIN components and activating Sid2p kinase activity. The scw1Δ mutation was unable to rescue the cdc11-123 defects in localization of SIN components or activation of Sid2p kinase, suggesting that scw1Δ does not directly enhance signaling through the SIN. In fact, scw1Δ single mutants had reduced Sid2p kinase activity. The reason for this is unclear. However, because the SIN seems to be down regulated once the septum has formed, the persistent presence of septa in scw1Δ cells could cause down regulation of Sid2p activity. Alternatively, increased septum-forming activity in scw1Δ mutants could inhibit the SIN through a feedback mechanism. Further study will be required to test these possibilities. Interestingly, SIN suppressors such as cdc16-116 (15) and par1/pbp1 (22, 25, 48), which are thought to suppress by enhancing signaling through the SIN, do not suppress cdc11-123, perhaps because the Cdc11-123p mutant protein does not localize properly to the SPB (24). Thus, the ability of scw1Δ to suppress cdc11-123 is consistent with a model in which it does not suppress by enhancing signaling through the SIN. Together, these results suggest that Scw1p may function as an inhibitor of septum formation, such that its loss of function allows weak SIN signaling to promote septum formation.
Consistent with this model are studies published during the course of this work showing that the scw1 mutant is resistant to cell wall-degrading enzymes, whereas SIN mutants are sensitive (23). The authors also found that scw1Δ rescued SIN mutants, and they proposed that it did so by restoring cell wall synthesis at the septum. Consistent with this model, we have also observed that scw1Δ mutants are resistant to Zymolyase treatment (data not shown), and in addition, we found that the scw1Δ mutation restored the septum synthesis defects of the cps1-191 1,3-β-glucan synthase mutant. 1,3-β-Glucan is the major component of the S. pombe division septum and cell wall, and previous studies have suggested that Cps1p may be a target of the SIN (29). Thus, one possible model for Scw1p function could be as a negative regulator of Cps1p, consistent with its loss of function rescuing weak activation of Cps1p by the SIN.
Given the effect of scw1Δ on the cell wall, it is interesting that scw1Δ mutants have defects in cell separation. It is not clear whether the cell separation defect is a representation of the SIN and cps1 suppression phenotype or a separate phenotype. It is possible that Scw1p promotes septum degradation leading to cell separation, and thus loss of this function in the scw1Δ mutant could rescue the septum synthesis defects of the SIN and cps1-191 mutants. Another suppressor of the SIN, the B′ regulatory subunit of protein phosphatase 2A called par1+/pbp1+, also has cell separation defects (22, 25, 48). This may be coincidental, since par1Δ mutations suppress only cdc7, cdc11, and spg1 mutations (22, 25), unlike scw1Δ mutations, which suppress all SIN mutations. Defects in cell separation alone are unlikely to suppress the SIN, since other mutants with cell separation defects, such as septin mutants (30) and sep1 mutants, do not suppress the SIN (44) (data not shown).
It is quite possible that Scw1p has multiple functions in the cell. We found that scw1Δ mutants could partially restore microtubules to the nda3-KM311 mutant strain. This effect is not simply from stabilization of the Nda3-KM311 mutant protein, because the scw1Δ mutation can partially stabilize microtubules in a wild-type background treated with the microtubule-destabilizing drug MBC. As with the effects of scw1Δ on cell separation, it is difficult to tell whether this phenotype is connected to the ability of the scw1Δ deletion to suppress the SIN. The SIN seems to be inhibited by microtubule defects, and thus it is possible that stabilization of microtubules could promote signaling through the SIN (20). However, this seems unlikely, since microtubule defects seem to inhibit SIN signaling, whereas scw1Δ deletion does not promote signaling through the SIN.
Understanding the relationship between the different phenotypes of the scw1Δ deletion mutant will likely depend on characterization of the targets of Scw1p action. Database comparisons revealed that Scw1p shows homology to Whi3p and Whi4p, two S. cerevisiae proteins containing RNA binding domains (39). Like Scw1p, Whi3p has also been implicated in cell cycle control. Whi3p specifically binds the G1 cyclin CLN3 mRNA and localizes the CLN3 mRNA into discrete cytoplasmic loci that may locally restrict Cln3p synthesis to modulate cell cycle progression (18). We find that Scw1p localizes to the cytoplasm; however, its localization is more diffuse than that observed for Whi3p. A similar localization pattern has been reported for another putative RNA binding protein, Sce3p, in S. pombe, which was isolated as a multicopy suppressor of certain alleles of cdc7, cdc11, and sid2 (11, 42). It is possible that Sce3p overproduction and scw1Δ deletion could rescue the SIN by affecting a common pathway; however, the genetics suggest that the wild-type gene products would be working in opposition to each other. It will be important in future studies to determine whether Scw1p, like Whi3p, binds specific RNAs and regulates their function. The use of DNA microarray technology may be a powerful approach to address this question.
Acknowledgments
We are grateful to Paul Young for communicating results prior to publication, to Kathy Gould for providing strains, to Susanne Trautmann and Ming-Chin Hou for comments on the manuscript, and to Jeff Salek for technical assistance.
This work was supported by National Institutes of Health grant GM58406 to D. McCollum.
REFERENCES
- 1.Bähler, J., A. B. Steever, S. Wheatley, Y. Wang, J. R. Pringle, K. L. Gould, and D. McCollum. 1998. Role of polo kinase and Mid1p in determining the site of cell division in fission yeast. J. Cell Biol. 143:1603-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bähler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. R. McKenzie, A. B. Steever, A. Wach, P. Philippsen, and J. R. Pringle. 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14:943-951. [DOI] [PubMed] [Google Scholar]
- 3.Balasubramanian, M. K., D. McCollum, and K. L. Gould. 1997. Cytokinesis in fission yeast Schizosaccharomyces pombe. Methods Enzymol. 283:494-506. [DOI] [PubMed] [Google Scholar]
- 4.Bardin, A. J., and A. Amon. 2001. Men and sin: what's the difference? Nat. Rev. Mol. Cell. Biol. 2:815-826. [DOI] [PubMed] [Google Scholar]
- 5.Basi, G., E. Schmid, and K. Maundrell. 1993. TATA bo mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123:131-136. [DOI] [PubMed] [Google Scholar]
- 6.Beltraminelli, N., M. Murone, and V. Simanis. 1999. The S. pombe zfs1 gene is required to prevent septation if mitotic progression is inhibited. J. Cell Sci. 112:3103-3114. [DOI] [PubMed] [Google Scholar]
- 7.Cerutti, L., and V. Simanis. 1999. Asymmetry of the spindle pole bodies and spg1p GAP segregation during mitosis in fission yeast. J. Cell Sci. 112:2313-2321. [DOI] [PubMed] [Google Scholar]
- 8.Chang, F., A. Wollard, and P. Nurse. 1996. Isolation and characterization of fission yeast mutants defective in the assembly and placement of the contractile actin ring. J. Cell Sci. 109:131-142. [DOI] [PubMed] [Google Scholar]
- 9.Chang, L., and K. L. Gould. 2000. Sid4p is required to localize components of the septation initiation pathway to the spindle pole body in fission yeast. Proc. Natl. Acad. Sci. USA 97:5249-5254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cortes, J. C., J. Ishiguro, A. Duran, and J. C. Ribas. 2002. Localization of the (1,3)beta-d-glucan synthase catalytic subunit homologue Bgs1p/Cps1p from fission yeast suggests that it is involved in septation, polarized growth, mating, spore wall formation and spore germination. J. Cell Sci. 115:4081-4096. [DOI] [PubMed] [Google Scholar]
- 11.Cullen, C. F., K. M. May, I. M. Hagan, D. M. Glover, and H. Ohkura. 2000. A new genetic method for isolating functionally interacting genes: high plo1(+)-dependent mutants and their suppressors define genes in mitotic and septation pathways in fission yeast. Genetics 155:1521-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fankhauser, C., J. Marks, A. Reymond, and V. Simanis. 1993. The S. pombe cdc16 gene is required both for maintenance of p34 Cdc2 kinase activity and regulation of septum formation: a link between mitosis and cytokinesis? EMBO J. 12:2697-2704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fankhauser, C., and V. Simanis. 1994. The cdc7 protein kinase is a dosage dependent regulator of septum formation in fission yeast. EMBO J. 13:3011-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fankhauser, C., and V. Simanis. 1993. The Schizosaccharomyces pombe cdc14 gene is required for septum formation and can also inhibit nuclear division. Mol. Biol. Cell 4:531-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fournier, N., L. Cerutti, N. Beltraminelli, E. Salimova, and V. Simanis. 2001. Bypass of the requirement for cdc16p GAP function in Schizosaccharomyces pombe by mutation of the septation initiation network genes. Arch. Microbiol. 175:62-69. [DOI] [PubMed] [Google Scholar]
- 16.Fujita, A., L. Vardy, M. A. Garcia, and T. Toda. 2002. A fourth component of the fission yeast gamma-tubulin complex, Alp16, is required for cytoplasmic microtubule integrity and becomes indispensable when gamma-tubulin function is compromised. Mol. Biol. Cell 13:2360-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furge, K. A., K. Wong, J. Armstrong, M. Balasubramanian, and C. F. Albright. 1998. Byr4 and Cdc16 form a two-component GTPase-activating protein for the Spg1 GTPase that controls septation in fission yeast. Curr. Biol. 8:947-954. [DOI] [PubMed] [Google Scholar]
- 18.Gari, E., T. Volpe, H. Wang, C. Gallego, B. Futcher, and M. Aldea. 2001. Whi3 binds the mRNA of the G1 cyclin CLN3 to modulate cell fate in budding yeast. Genes Dev. 15:2803-2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guertin, D. A., L. Chang, F. Irshad, K. L. Gould, and D. McCollum. 2000. The role of the sid1p kinase and cdc14p in regulating the onset of cytokinesis in fission yeast. EMBO J. 19:1803-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guertin, D. A., S. Venkatram, K. L. Gould, and D. McCollum. 2002. Dma1 prevents mitotic exit and cytokinesis by inhibiting the septation initiation network (SIN). Dev. Cell 3:779-790. [DOI] [PubMed] [Google Scholar]
- 21.Hou, M. C., J. Salek, and D. McCollum. 2000. Mob1p interacts with the Sid2p kinase and is required for cytokinesis in fission yeast. Curr. Biol. 10:619-622. [DOI] [PubMed] [Google Scholar]
- 22.Jiang, W., and R. L. Hallberg. 2001. Correct regulation of the septation initiation network in Schizosaccharomyces pombe requires the activities of par1 and par2. Genetics 158:1413-1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karagiannis, J., R. Oulton, and P. G. Young. 2002. The Scw1 RNA-binding domain protein regulates septation and cell-wall structure in fission yeast. Genetics 162:45-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krapp, A., S. Schmidt, E. Cano, and V. Simanis. 2001. S. pombe cdc11p, together with sid4p, provides an anchor for septation initiation network proteins on the spindle pole body. Curr. Biol. 11:1559-1568. [DOI] [PubMed] [Google Scholar]
- 25.Le Goff, X., S. Buvelot, E. Salimova, F. Guerry, S. Schmidt, N. Cueille, E. Cano, and V. Simanis. 2001. The protein phosphatase 2A B′-regulatory subunit par1p is implicated in regulation of the S. pombe septation initiation network. FEBS Lett. 508:136-142. [DOI] [PubMed] [Google Scholar]
- 26.Le Goff, X., A. Woollard, and V. Simanis. 1999. Analysis of the cps1 gene provides evidence for a septation checkpoint in Schizosaccharomyces pombe. Mol. Gen. Genet. 262:163-172. [DOI] [PubMed] [Google Scholar]
- 27.Li, C., K. A. Furge, Q. C. Cheng, and C. F. Albright. 2000. Byr4 localizes to spindle-pole bodies in a cell cycle-regulated manner to control Cdc7 localization and septation in fission yeast. J. Biol. Chem. 275:14381-14387. [DOI] [PubMed] [Google Scholar]
- 28.Liu, J., X. Tang, H. Wang, S. Oliferenko, and M. K. Balasubramanian. 2002. The localization of the integral membrane protein Cps1p to the cell division site is dependent on the actomyosin ring and the septation-inducing network in Schizosaccharomyces pombe. Mol. Biol. Cell 13:989-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, J., H. Wang, D. McCollum, and M. K. Balasubramanian. 1999. Drc1p/Cps1p, a 1,3-beta-glucan synthase subunit, is essential for division septum assembly in Schizosaccharomyces pombe. Genetics 153:1193-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Longtine, M. S., D. J. DeMarini, M. L. Valencik, O. S. Al-Awar, H. Fares, C. De Virgilio, and J. R. Pringle. 1996. The septins: roles in cytokinesis and other processes. Curr. Opin. Cell Biol. 8:106-119. [DOI] [PubMed] [Google Scholar]
- 31.Mata, J., and P. Nurse. 1997. tea1 and the microtubular cytoskeleton are important for generating global spatial order within the fission yeast cell. Cell 89:939-949. [DOI] [PubMed] [Google Scholar]
- 32.Maundrell, K. 1990. nmt1 of fission yeast. A highly transcribed gene completely repressed by thiamine. J. Biol. Chem. 265:10857-10864. [PubMed] [Google Scholar]
- 33.McCollum, D., and K. L. Gould. 2001. Timing is everything: regulation of mitotic exit and cytokinesis by the MEN and SIN. Trends Cell Biol. 11:89-95. [DOI] [PubMed] [Google Scholar]
- 34.Minet, M., P. Nurse, P. Thuriaux, and J. M. Mitchison. 1979. Uncontrolled septation in a cell division cycle mutant of the fission yeast Schizosaccharomyces pombe. J. Bacteriol. 137:440-446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moreno, S., J. Hayles, and P. Nurse. 1989. Regulation of p34cdc2 protein kinase during mitosis. Cell 58:361-372. [DOI] [PubMed] [Google Scholar]
- 36.Moreno, S., A. Klar, and P. Nurse. 1991. Molecular genetic analysis of fission yeast, Schizosaccharomyces pombe. Methods Enzymol. 194:795-823. [DOI] [PubMed] [Google Scholar]
- 37.Morgan, B. A., F. L. Conlon, M. Manzanares, J. B. Millar, N. Kanuga, J. Sharpe, R. Krumlauf, J. C. Smith, and S. G. Sedgwick. 1996. Transposon tools for recombinant DNA manipulation: characterization of transcriptional regulators from yeast, Xenopus, and mouse. Proc. Natl. Acad. Sci. USA 93:2801-2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Murone, M., and V. Simanis. 1996. The fission yeast dma1 gene is a component of the spindle assembly checkpoint, required to prevent septum formation and premature exit from mitosis if spindle function is compromised. EMBO J. 15:6605-6616. [PMC free article] [PubMed] [Google Scholar]
- 39.Nash, R. S., T. Volpe, and B. Futcher. 2001. Isolation and characterization of WHI3, a size-control gene of Saccharomyces cerevisiae. Genetics 157:1469-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ribar, B., A. Banrevi, and M. Sipiczki. 1997. sep1+ encodes a transcription-factor homologue of the HNF-3/forkhead DNA-binding-domain family in Schizosaccharomyces pombe. Gene 202:1-5. [DOI] [PubMed] [Google Scholar]
- 41.Salimova, E., M. Sohrmann, N. Fournier, and V. Simanis. 2000. The S. pombe orthologue of the S. cerevisiae mob1 gene is essential and functions in signalling the onset of septum formation. J. Cell Sci. 113:1695-1704. [DOI] [PubMed] [Google Scholar]
- 42.Schmidt, S., K. Hofmann, and V. Simanis. 1997. Sce3, a suppressor of the Schizosaccharomyces pombe septation mutant cdc11, encodes a putative RNA-binding protein. Nucleic Acids Res. 25:3433-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidt, S., M. Sohrmann, K. Hofmann, A. Woollard, and V. Simanis. 1997. The Spg1p GTPase is an essential, dosage-dependent inducer of septum formation in Schizosaccharomyces pombe. Genes Dev 11:1519-1534. [DOI] [PubMed] [Google Scholar]
- 44.Sipiczki, M., B. Grallert, and I. Miklos. 1993. Mycelial and syncytial growth in Schizosaccharomyces pombe induced by novel septation mutations. J. Cell Sci. 104:485-493. [DOI] [PubMed] [Google Scholar]
- 45.Sohrmann, M., S. Schmidt, I. Hagan, and V. Simanis. 1998. Asymmetric segregation on spindle poles of the Schizosaccharomyces pombe septum-inducing protein kinase Cdc7p. Genes Dev 12:84-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song, K., K. E. Mach, C. Y. Chen, T. Reynolds, and C. F. Albright. 1996. A novel suppressor of ras1 in fission yeast, byr4, is a dosage-dependent inhibitor of cytokinesis. J. Cell Biol. 133:1307-1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sparks, C. A., M. Morphew, and D. McCollum. 1999. Sid2p, a spindle pole body kinase that regulates the onset of cytokinesis. J. Cell Biol. 146:777-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanabe, O., D. Hirata, H. Usui, Y. Nishito, T. Miyakawa, K. Igarashi, and M. Takeda. 2001. Fission yeast homologues of the B′ subunit of protein phosphatase 2A: multiple roles in mitotic cell division and functional interaction with calcineurin. Genes Cells 6:455-473. [DOI] [PubMed] [Google Scholar]
- 49.Tomlin, G. C., J. L. Morrell, and K. L. Gould. 2002. The spindle pole body protein cdc11p links sid4p to the fission yeast septation initiation network. Mol. Biol. Cell 13:1203-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Umesono, K., T. Toda, S. Hayashi, and M. Yanagida. 1983. Cell division cycle genes nda2 and nda3 of the fission yeast Schizosaccharomyces pombe control microtubular organization and sensitivity to anti-mitotic benzimidazole compounds. J. Mol. Biol. 168:271-284. [DOI] [PubMed] [Google Scholar]
- 51.Vardy, L., and T. Toda. 2000. The fission yeast gamma-tubulin complex is required in G(1) phase and is a component of the spindle assembly checkpoint. EMBO J. 19:6098-6111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woods, A., T. Sherwin, R. Sasse, T. H. MacRae, A. J. Baines, and K. Gull. 1989. Definition of individual components within the cytoskeleton of Trypanosoma brucei by a library of monoclonal antibodies. J. Cell Sci. 93:491-500. [DOI] [PubMed] [Google Scholar]