Abstract
In the corn smut fungus Ustilago maydis, pathogenic development is initiated when two compatible haploid cells fuse and form the infectious dikaryon. Mating is dependent on pheromone recognition by compatible cells. In this report, we set out to evaluate the relationship between the cell cycle and the pheromone response in U. maydis. To achieve this, we designed a haploid pheromone-responsive strain that is able to faithfully reproduce the native mating response in nutrient-rich medium. Addition of synthetic pheromone to the responsive strain induces the formation of mating structures, and this response is abolished by mutations in genes encoding components of the pheromone signal transduction cascade. After recognition of pheromone, U. maydis cells arrest the cell cycle in a postreplicative stage. Visualization of the nucleus and microtubule organization indicates that the arrest takes place at the G2 phase. Chemical-induced cell cycle arrest and release in the presence of pheromone further support this conclusion.
Ustilago maydis, a basidiomycete fungus, is the agent responsible for corn smut, a disease with a worldwide distribution, which under some conditions may cause severe economic losses (1). In this fungus, pathogenesis and sexual development are intricately interconnected. A prerequisite for generating the infectious stage is the mating of two compatible haploid cells and the generation of an infective dikaryotic filament after cell fusion (3, 14). Because of this prerequisite, elucidation of the steps involved in mating has attracted considerable attention. Mating in U. maydis is initiated by a pheromone-based cell recognition system encoded by the biallelic a mating type locus. Each allele encodes a lipopeptide pheromone precursor (mfa) and a receptor (pra) that recognizes pheromone secreted by cells of the opposite mating type (6). In order to induce the mating process, the binding of pheromones to their cognate receptors is required. The pheromone signal has been proposed to be transmitted by a mitogen-activated protein kinase (MAPK) cascade, which is composed of a MAPK kinase kinase (Ubc4) (2), a MAPK kinase (Fuz7/Ubc5) (2, 5), and a MAPK (Ubc3/Kpp2) (16, 17). The MAPK cascade is believed to feed in the transcription factor Prf1, as well as additional unknown targets (12, 14, 17).
The first gross morphological alteration observed in response to pheromone is the production of the conjugation tubes, which are tail-like structures that are formed preferentially at one tip of the cell (21, 22). Conjugation tubes of compatible cells grow towards each other, resulting in cell fusion. U. maydis cells that develop conjugation tubes are rarely in the process of budding, suggesting that either the mating structures are induced in a specific stage of the cell cycle or, alternatively, there is a cell cycle arrest induced by pheromone. In ascomycete fungi, such as Saccharomyces cerevisiae and Schizosaccharomyces pombe, cell fusion requires a previous G1 cell cycle arrest in response to pheromone recognition (10, 24). To date, there have been no studies carried out to evaluate the pheromone-associated events with respect to the cell cycle in U. maydis or in other basidiomycete fungi.
In this study the aim was to explore the effects of pheromone addition on cell cycle progression in U. maydis. Here we describe the construction and use of a pheromone-responsive strain to approach this issue. By using this strain, we have found that when exposed to pheromone, cells of U. maydis undergo a cell cycle arrest in G2 phase. The results of the study lead to the conclusion that in U. maydis the response to pheromone produces a cell cycle arrest different from the well-known G1 cell cycle arrest induced by pheromone recognition in ascomycete fungi (10, 24), and they raise the question of how the pheromone recognition cascade is linked to the cell cycle.
MATERIALS AND METHODS
Strains and growth conditions.
For cloning purposes, the Escherichia coli K-12 derivative DH5α (Bethesda Research Laboratories) was used. The U. maydis strains employed in this study are listed in Table 1. Strains were grown at 28°C in yeast extract-peptone-dextrose (YPD) (9), YEPS (28), or complete medium (CM) (13). Hygromycin B was purchased from Roche, and carboxin was purchased from Riedel de Haen. All chemicals used were of analytical grade and were obtained from Sigma or Merck. The synthetic a1 pheromone (a1-Ahds-Ome) was a generous gift of H. Kessler (Technical University of Munich, Munich, Germany) (15).
TABLE 1.
U. maydis strains
| Strain | Relevant genotype | Parent strain | Reference |
|---|---|---|---|
| FB1 | a1 b1 | 4 | |
| FB2 | a2 b2 | 4 | |
| FB1c | a1 b1 cbx::Ptef-gfp | FB1 | This work |
| TAU3 | a1 b1 cbx::Ptef-pra2 | FB1 | This work |
| TAU21 | a1 b1 cbx::Ptef-pra2 Δprf1-1 | TAU3 | This work |
| TAU38 | a1 b1 cbx::Ptef-pra2 Δubc3-1 | TAU3 | This work |
| TAU50 | a1 b1 cbx::Ptef-pra2 Potef-cfp-tub1 | TAU3 | This work |
Strain constructions.
To produce the pheromone-responsive strain, TAU3, a construction carrying the cDNA encoding the Pra2 receptor (a generous gift of R. Kahmann, MPI, Marburg, Germany) under the control of the constitutive promoter Ptef1 (23) was integrated into the succinate dehydrogenase (cbx) locus from the wild-type a1 mating type strain FB1. The control strain, FB1c, was constructed by integration of a DNA fragment carrying a Ptef1-gfp fusion in the cbx locus of wild-type FB1 cells. To construct strain TAU50, we integrated ectopically the plasmid pCFPtub1 (30). In brief, this plasmid contains a cfp-tub1 fusion construct behind the otef promoter and a hygromycin resistance cassette. The integration of the cfp-tub1 fusion in the pheromone-responsive strain allows visualization of the microtubule cytoskeleton by epifluorescence. To construct the TAU38 strain, in which the ubc3 gene was removed, the construction carrying the Δubc3-1 allele was utilized as described by Garrido and Pérez-Martín (11). To produce the TAU21 strain, in which the prf1 gene was removed, a disruption cassette carrying the Δprf1-1 allele was constructed by ligation of a pair of DNA fragments flanking the prf1 open reading frame into pNEBHyg(+)ΔEcoRI, a U. maydis integration vector containing a hygromycin B resistance cassette (8). The 5′ fragment spans the sequence from nucleotide −355 to −1 (considering the adenine in the ATG to be nucleotide +1), and it was produced by PCR amplification with primers PRF1-A (5′CGGAATTCCCAAACCAGCTTCGGTCTCTTT3′) and PRF1-B (5′CCGCTCGAGGGTGAGCACGACTTTGCCTTAGAA3′). The 3′ fragment spans the sequence from nucleotide +2551 to +2896, and it was produced by PCR amplification with primers PRF1-C (5′CGGGATCCATGCTTTTTCAATCTCTTTGCAAC3′) and PRF1-D (5′ACATGCATGCTGATTTTACTTTTTGTTTCAGCGG3′). The resulting plasmid, pPRF1-KO, was digested with EcoRI and SphI and transformed into the appropriate U. maydis strains. Transformants were screened for the loss of the wild-type copy by PCR analysis, and this was confirmed by Southern analysis.
Induction of the pheromone response.
Tester and control strains were grown in YPD to an optical density at 600 nm (OD600) of 0.2. The cells were then washed three times with CM and resuspended in a similar volume of prewarmed CM amended with 3 mM cyclic AMP and a1 synthetic pheromone (a1-Ahds-Ome) (0.1 ng/ml). The addition of cyclic AMP had no effect by itself but decreased the time of response to pheromone to less than 1 h and increased the reproducibility of the experiments (unpublished observations). Cells were incubated at 28°C, and samples were removed at the indicated times.
Cell cycle arrest and release experiments.
To induce cell cycle arrest in S phase, liquid cultures were grown in YPD at 28°C to an OD600 of 0.2 and then incubated for 90 min in the presence of 1 mg of hydroxyurea (HU) per ml. After this incubation, cells were washed three times with sterile water and resuspended in the same volume of prewarmed CM with synthetic a1 pheromone (0.1 ng/ml). Arrest and release from the G2/M transition were performed with benomyl. As before, liquid cultures were grown in YPD to an OD600 of 0.2, and they were then incubated for 60 min in the presence of 10 μM benomyl. After this incubation, the cells were washed three times with sterile water and resuspended in the same volume of prewarmed CM amended with synthetic a1 pheromone.
RNA analysis.
U. maydis cells for RNA isolation were prepared by collecting the cells by centrifugation at 4°C, washed once with ice-cold water, and stored at −70°C. Total RNA was isolated (19), separated on formaldehyde-containing agarose gels, transferred to Zeta-Probe blotting membranes (Bio-Rad), and hybridized with 32P-labeled probes. Radioactive labeling was performed with the Ready-To-Go labeling kit (Amersham Pharmacia Biotech Inc.). A 675-bp EcoRV fragment spanning the sequence of the mfa1 gene was used as a probe (29). A 5′-end-labeled oligonucleotide complementary to the U. maydis 18S rRNA (7) was used as a loading control in Northern analyses. A phosphorimager (Molecular Imager FX; Bio-Rad) and a suitable program (Quantity One; Bio-Rad) were used for visualization and quantification of radioactive signals.
Flow cytometry.
DNA content was measured by flow cytometry. Cells (107) were harvested, washed twice with cold water, fixed in 70% ethanol overnight, and resuspended in 50 mM sodium citrate, pH 7.5. Cellular RNA was destroyed by incubation with RNase A (0.25 mg/ml) at 50°C for 1 h, and then, proteinase K (1 mg/ml) was added and the cells were incubated for another hour at 50°C. Cells were stained at 4°C with propidium iodide (16 μg/ml) and analyzed with a Coulter Epics XL-MLC instrument. For each acquisition, 104 events were measured at a flow rate of 60 to 100 events per s for both DNA content (FL3) and cell size (FSC). In the DNA histograms, relative fluorescence intensities are given on the horizontal axes and cell numbers are given on the vertical axes.
Light microscopy and image processing.
Nuclear staining was done by using DAPI (4′,6′-diamidino-2-phenylindole) staining. To prepare cells for staining, 1 ml of each culture was washed twice with 1 ml of phosphate-buffered saline and concentrated 10 times in phosphate-buffered saline. Ten microliters of this cell concentrate was applied to a coverslip and dried with a hair dryer. Two microliters of mounting medium (10 μl of DAPI plus 90 μl of elvanol) was added to a glass microscope slide, and the coverslip carrying the cells was inverted on the small drop. Microscopy analysis was performed with a Zeiss Axiophot microscope. Frames were taken with a cooled charge-coupled device camera (Hamamatsu C4742-95). Epifluorescence was observed with a standard DAPI filter set. CFP was analyzed with a specific filter set (BP436, FT455, and BP480-500). Image processing was performed with Image Pro Plus (Media Cybernetics) and Photoshop (Adobe).
RESULTS
Design of a haploid pheromone-responsive strain.
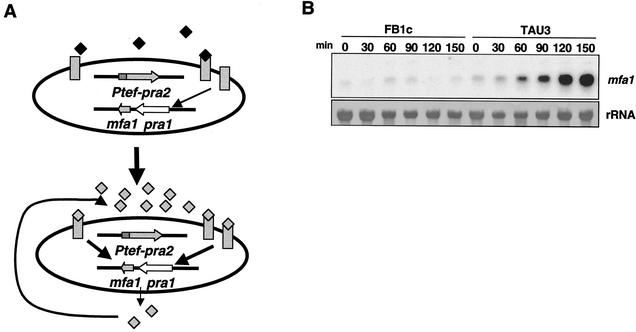
Expression of the mating locus in U. maydis requires a nutritional starvation signal to enhance the transcription of the mating genes (22). The effects on the cell cycle regulation that impose this requirement could mask the analysis of putative interactions between the mating response and the cell cycle. To overcome this problem, we designed a haploid pheromone-responsive strain, TAU3, that is able to acquire mating competence under conditions of growth in nutrient-rich medium (Fig. 1A). The rationale of the design relies on the previous observation that receptor and pheromone genes are up regulated through pheromone stimulation (22). To produce the TAU3 strain, we constitutively expressed the pra2 pheromone receptor gene in the wild-type strain FB1, which is able to produce the compatible pheromone a1. For a control strain, we constructed strain FB1c, which is FB1 containing a construction constitutively expressing the green fluorescent protein gene integrated at the same locus where the pra2 gene was integrated in the TAU3 strain. TAU3 cells were unable to express the compatible a1 pheromone gene (mfa1) to high levels while they were growing in nutrient-rich medium (not shown). However, because of the continuous expression of the compatible Pra2 receptor, these cells were able to detect minute amounts of a1 pheromone under any given nutritional condition. Therefore, addition of a trace amount of a synthetic analog of pheromone (a1-Ahds-Ome [15]) triggered mfa1 gene expression in nutrient-rich medium, bypassing the nutritional repression (Fig. 1B). After 4 h of incubation, nearly 100% of the responsive cells treated with pheromone developed structures morphologically indistinguishable from conjugation tubes (Fig. 1C). The response observed in TAU3 is dependent on the addition of exogenous pheromone; neither conjugation tube formation (Fig. 1C) nor an increase in mfa1 expression (not shown) was detected in strain TAU3 without pheromone addition. In a similar way, the nonresponsive FB1c control strain showed neither induction of mfa1 expression nor conjugation tube formation in the presence of exogenous pheromone (Fig. 1B and C).
FIG. 1.
Design of a haploid pheromone-responsive strain. (A) Experimental system. TAU3 cells (a1 mating type) express constitutively an ectopic copy of the pra2 gene, which encodes the a1 pheromone receptor (gray rectangles). Addition of minute amounts of a synthetic pheromone (black diamonds) induce the secretion of endogenous a1 pheromone (gray diamonds) that feeds back into the system in an autocrine manner and induces the mating program. (B) Induction of expression of the mfa1 gene, encoding a1 pheromone precursor. FB1c (control strain) and TAU3 (pheromone-responsive strain) cells were incubated at 28°C in the presence of pheromone at time zero. Aliquots were extracted at the indicated times, and total RNA was isolated and subjected to Northern analysis after loading 10 μg of total RNA per lane. The same filter was hybridized in succession with probes for mfa1 and 18S rRNA. (C) Mating structures induced by pheromone. FB1c (control strain) and TAU3 (pheromone-responsive strain) cells were incubated with (+a1) or without pheromone for 4 h. Conjugative tubes are visible only in pheromone-treated TAU3 cells. Bars, 5 μm.
The pheromone-responsive strain reproduces the mating response of U. maydis.
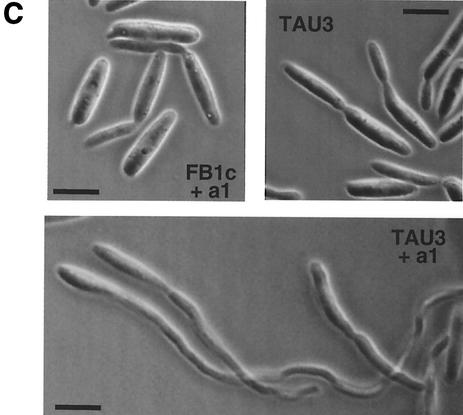
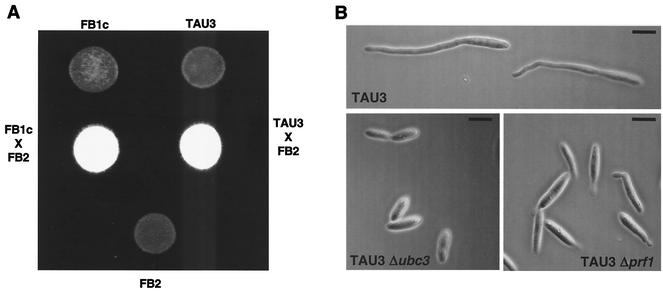
To further characterize the pheromone-responsive strain, we evaluated the ability of TAU3 cells to mate with a wild-type compatible partner. To this end, we cospotted either FB1c or TAU3 cells along with FB2 (a2 b2) cells on charcoal mating plates. Under these conditions, white colonies that are indicative of the dikaryon formation developed, and no difference between TAU3 and control crosses was observed (Fig. 2A). In addition, we analyzed whether mutations in genes encoding proteins known to be required for the transmission of the pheromone signal abolished the ability to respond to pheromone in our system. For this purpose, we deleted the genes encoding either the MAPK protein Ubc3/Kpp2 (16, 17) or the transcription factor Prf1 (12) in TAU3 cells. The resulting mutant strains were unable to develop conjugation tubes in response to the addition of pheromone (Fig. 2B). They were also unable to induce the expression of the mfa1 gene in response to pheromone addition (not shown). Taken together, these results indicate that our tester strain faithfully reproduces the native mating response of U. maydis.
FIG. 2.
TAU3 faithfully reproduces the mating response of U. maydis. (A) Mating ability of TAU3 cells. Around 102 cells from strains FB1c (a1 b1 Ptef::gfp) and TAU3 (a1 b1 Ptef::pra2) were spotted alone or in combination with a similar number of cells from wild-type strain FB2 (a2 b2) on charcoal-containing PD plates and incubated at 24°C for 48 h. Dikaryotic filaments render colonies white and fuzzy looking. (B) Components of the pheromone transduction cascade are required for the pheromone response in TAU3 cells. The TAU3 strain and its derivatives TAU38 (TAU3 Δubc3) and TAU21 (TAU3 Δprf1), which are defective in the Ubc3/Kpp2 MAPK and the Prf1 transcription factor, respectively, were incubated for 4 h in the presence of pheromone as described for Fig. 1. Bars, 5 μm.
Response to pheromone causes a G2 cell cycle arrest in U. maydis.
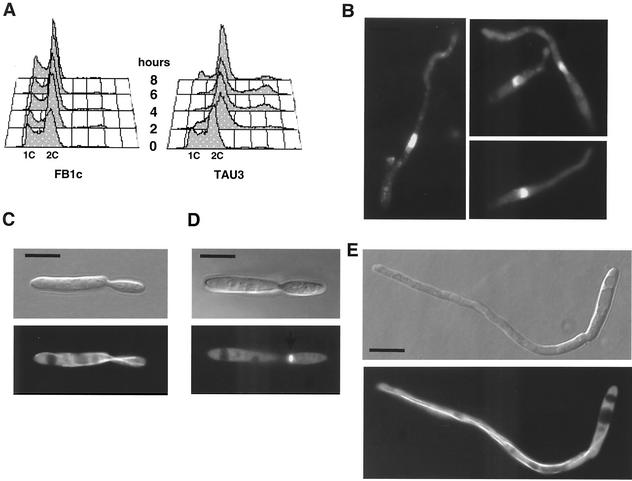
To investigate whether the response to pheromone in U. maydis correlated with specific cell cycle changes, we added the pheromone to TAU3 cultures and monitored the DNA content profile by fluorescence-activated cell sorter (FACS) analysis (Fig. 3A). Addition of pheromone resulted in the accumulation of cells with a 2C DNA content. We have found that this postreplicative arrest is transient and that a certain fraction of cells exposed to pheromone resumed growth thereafter, as evidenced by the steadily increasing 1C DNA peak that can be observed in responsive cells after 8 h of pheromone treatment (Fig. 3A). In contrast to the cell cycle arrest observed in TAU3 cells, the DNA content profile was not affected after pheromone addition in nonresponsive FB1c cells (Fig. 3A). TAU3 cells not treated with pheromone showed a FACS profile similar to that of the nonresponsive control strain (not shown).
FIG. 3.
Response to pheromone causes a G2 cell cycle arrest. (A) DNA content of pheromone-responding cells. The control FB1c and tester TAU3 cells were incubated in the presence of a1 synthetic pheromone as described in the text. Samples were removed at 0, 2, 4, 6, and 8 h, and the DNA content was measured by FACS analysis. Relative fluorescence intensities are given on the horizontal axes, indicating the 1C and 2C DNA contents. The adhesiveness of cells forming conjugative tubes produces a small peak corresponding to 4C DNA content. (B) DAPI staining of TAU3 cells producing conjugation tubes after incubation in the presence of pheromone. Note the presence of a single uncondensed nucleus per cell, which is typical of G2 phase. (C) Nomarski (top) and epifluorescence (bottom) images of cells of TAU50, a pheromone-responsive strain carrying a CFP-α-tubulin fusion, grown in the absence of pheromone, showing the typical morphology and microtubule organization at G2 phase. (D) Same cell as in panel C but at the onset of mitosis, where the spindle is present near the neck area. (E) Pheromone-treated TAU50 cell developing a conjugative tube. Note that the microtubule organization of the cell is similar to that present at G2 phase. Bars, 5 μm.
Our FACS analysis demonstrated that pheromone-arrested cells had a 2C DNA content, suggesting that they are either in G2 or in mitosis. To distinguish between these stages, we visualized the nuclei by DAPI staining. We detected one large nucleus per cell, in agreement with an arrest in G2 (Fig. 3B). This is in contrast to early mitotic nuclei, which are small and condensed (25). To further support this interpretation we examined the microtubule cytoskeleton, since U. maydis cells undergoing G2 phase assemble long microtubules towards the growth region, while at the onset of mitosis, cytoplasmic microtubules disappear and short spindles are formed (25). For this, we constructed a pheromone-responsive strain (TAU50) carrying a CFP-Tub1 fusion protein (30). In the absence of pheromone, TAU50 cells in G2 phase formed polar buds and long microtubules reaching the growth region, supporting bud formation (Fig. 3C). As soon these cells entered mitosis, a spindle appeared at the neck region and the long microtubules disassembled (Fig. 3D). Pheromone-treated TAU50 cells contained long microtubules that reached the tip of the growing conjugation tube (Fig. 3E), which argues in favor of an arrest in G2 rather than in mitosis. Furthermore, pheromone-treated cells contained paired tubulin structures (not shown) that are characteristic of G2 phase (25) and most likely participate in microtubule organization at the growing bud (27). Taken together, these results are in accordance with a G2 arrest in response to pheromone in U. maydis.
Experiments involving chemical-induced cell cycle arrest and release in the presence of pheromone support a G2 cell cycle arrest.
To reinforce the conclusion that pheromone-responding cells are arrested at G2 phase, we carried out experiments in which cells were arrested by chemical treatment at specific cell cycle points and then released from this arrest in the presence of pheromone. We reasoned that cells arrested before G2 phase and released in the presence of pheromone would arrest the cell cycle in response to pheromone immediately, as soon as responding cells entered G2. However, if the cells were arrested after G2 phase and released in the presence of pheromone, then the pheromone-induced cell cycle arrest should take place in the succeeding cell cycle.
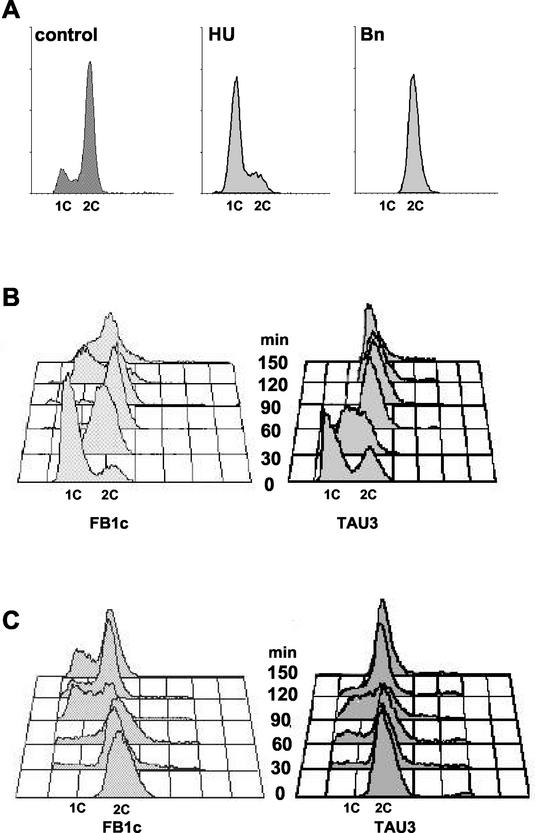
First, we determined the conditions to arrest the cell cycle before G2 phase. HU, an inhibitor of the enzyme ribonucleotide reductase, has been previously used to arrest yeast cells in early stages of S phase (9). We found that incubation of U. maydis wild-type cells in the presence of 1 mg of HU per ml for 90 min resulted in the accumulation of cells with DNA content ranging from 1C to 2C (Fig. 4A), consistent with a blockage of the progression through the S phase in response to unreplicated DNA. Removal of HU from the incubation medium resulted in a synchronous release from cell cycle arrest (not shown). TAU3 and FB1c cells were arrested with HU and released in medium amended with pheromone. While the control strain resumed its growth and passed through G2/M, entering G1 phase after 120 min, the pheromone-responsive strain arrested in G2 as expected (Fig. 4B) and started to produce conjugative tubes (not shown).
FIG.4.
Chemical-induced cell cycle arrest and release in the presence of pheromone. (A) DNA content of cells arrested by chemical treatment. Wild-type FB1 cells were incubated in YPD (control), YPD with 1 mg of HU per ml, or YPD with 10 μM benomyl (Bn). Samples were removed after 90 min, and the DNA content was measured by FACS analysis. (B) Release of HU-arrested cells in the presence of pheromone. FB1c and TAU3 cells were grown in YPD to an OD600 of 0.2 and then incubated for 90 min in the presence of 1 mg of HU per ml. After this incubation, cells were washed three times and then released in CM with synthetic a1 pheromone (0.1 ng/ml). Numbers indicate times after transfer to fresh medium without HU and pheromone. (C) Release of benomyl-arrested cells in the presence of pheromone. FB1c and TAU3 cells were grown in YPD to an OD600 of 0.2 and then incubated for 60 min in the presence of 10 μM benomyl. After this incubation, cells were washed three times and then released in CM with synthetic a1 pheromone (0.1 ng/ml). Numbers indicate times after transfer to fresh medium without benomyl and pheromone.
To arrest cells after G2 phase, we took advantage of previous work (18) indicating that incubation of U. maydis cells in medium containing methyl-1-(butylcarbamoyl)-2-benzimidazole-carbamate, a microtubule inhibitor that prevents tubulin polymerization and blocks chromosome segregation by inhibiting spindle assembly, holds cells in G2/M phase. We reproduced those experiments, but we found that the cell cycle release after removal of methyl-1-(butylcarbamoyl)-2-benzimidazole-carbamate was quite unpredictable (not shown). In contrast, the chemical analog benomyl, which also prevents tubulin polymerization, produces a clear cell cycle arrest in G2/M (Fig. 4A) and a better release once the drug is removed (not shown). TAU3 and FB1c cells were arrested with benomyl and released in medium amended with pheromone. Responsive and control cells completed division and entered a new cell cycle. However, responsive cells then arrested postreplication in the succeeding cell cycle, while control cells were not affected (Fig, 4C). Taken together, these results support a cell cycle arrest occurring after S phase and before the G2/M transition (i.e., in G2 phase).
DISCUSSION
From the data presented in this work, we conclude that in the phytopathogenic fungus U. maydis, the response to pheromone induces a specific G2 cell cycle arrest. This conclusion contrasts with the previously characterized pheromone-induced G1 cell cycle arrest in ascomycete yeasts such as S. cerevisiae and S. pombe (10, 24). We believe that this specific cell cycle arrest in U. maydis has a mechanistic reason. In U. maydis, the G2 phase is characterized by polar formation of a bud (20), which requires the rearrangement of the cytoskeleton (25) and involves a specialized set of motors, such as cytoplasmic dynein (26, 30), which support the polar extension of the cell. In other words, in the G2 phase, the cytoskeletal growth machinery is set up to support polar growth. Assuming that the formation of a conjugation tube is based on a mechanism similar to that for polar bud growth, a prolonged G2 phase is best suited to support tip growth during tube formation. We could then postulate that in U. maydis, a point of decision exists once DNA has been replicated, in such a way that in response to external stimuli, the cell decides either to bud or to enter a mating program. A decision-making point, in which the cell must choose between alternative developmental fates, is consistent with the previous observation that cells responding to pheromone do not form buds (22).
In summary, we have shown in this study that recognition of pheromone by the basidiomycete U. maydis induces a G2 cell cycle arrest. It could be interesting to address whether this particular cell cycle arrest is a consequence of the particular lifestyle of U. maydis or whether it is a more general mechanism in other basidiomycete fungi, such as Cryptococcus neoformans, for instance. In addition, the differences in cell cycle arrest between U. maydis and ascomycete yeasts suggest the existence of alternative mechanisms linking the pheromone response and the cell cycle, which will be an exciting challenge to analyze.
Acknowledgments
We thank Regine Kahmann and the members of her laboratory for their generous help in sharing with us techniques, material, and information; H. Kessler for providing the synthetic pheromone; and K. M. Snetselaar for critical reading of the manuscript.
This work was supported by grants BIO99-0906 and BIO2002-03503 from MCyT to J. Pérez-Martín and grant SP1111 from DFG to G. Steinberg.
REFERENCES
- 1.Agrios, G. N. 1988. Plant pathology. Academic Press, Inc., San Diego, Calif.
- 2.Andrews, D. L., J. D. Egan, M. E. Mayorga, and S. E. Gold. 2000. The Ustilago maydis ubc4 and ubc5 genes encode members of a MAP kinase cascade required for filamentous growth. Mol. Plant-Microbe Interact. 13:781-786. [DOI] [PubMed] [Google Scholar]
- 3.Banuett, F. 1995. Genetics of Ustilago maydis, a fungal pathogen that induces tumors in maize. Annu. Rev. Genet. 29:179-208. [DOI] [PubMed] [Google Scholar]
- 4.Banuett, F., and I. Herskowitz. 1989. Different a alleles are necessary for maintenance of filamentous growth but not for meiosis. Proc. Natl. Acad. Sci. USA 86:5878-5882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banuett, F., and I. Herskowitz. 1994. Identification of fuz7, a Ustilago maydis MEK/MAPKK homolog required for a-locus-dependent and -independent steps in the fungal life cycle. Genes Dev. 8:1367-1378. [DOI] [PubMed] [Google Scholar]
- 6.Bölker, M., M. Urban, and R. Kahmann. 1992. The a mating type locus of U. maydis specifies cell signaling components. Cell 68:441-450. [DOI] [PubMed] [Google Scholar]
- 7.Bottin, A., J. Kämper, and R. Kahmann. 1996. Isolation of a carbon source-regulated gene from Ustilago maydis. Mol. Gen. Genet. 253:342-352. [DOI] [PubMed] [Google Scholar]
- 8.Brachmann, A., G. Weinzierl, J. Kämper, and R. Kahmann. 2001. Identification of genes in the bW/bE regulatory cascade in Ustilago maydis. Mol. Microbiol. 42:1047-1063. [DOI] [PubMed] [Google Scholar]
- 9.Burke, D., D. Dawson, and T. Stearns. 2000. Methods in yeast genetics. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 10.Davey, J. 1998. Fusion of a fission yeast. Yeast 14:1529-1566. [DOI] [PubMed] [Google Scholar]
- 11.Garrido, E., and J. Pérez-Martín. 2003. The crk1 gene encodes an Ime2-related protein that is required for morphogenesis in the plant pathogen Ustilago maydis. Mol. Microbiol. 47:729-743. [DOI] [PubMed]
- 12.Hartmann, H. A., R. Kahmann, and M. Bölker. 1996. The pheromone response factor coordinates filamentous growth and pathogenic development in Ustilago maydis. EMBO J. 15:1632-1641. [PMC free article] [PubMed] [Google Scholar]
- 13.Holliday, R. 1974. Ustilago maydis, p. 575-595. In R. C. King (ed.), Handbook of genetics, vol. 1. Plenum Press, New York, N.Y.
- 14.Kahmann, R., G. Steinberg, C. Basse, M. Feldbrügge, and J. Kämper. 2000. Ustilago maydis, the causative agent of corn smut disease, p. 347-371. In J. W. Kronstad (ed.), Fungal pathology. Kluwer Academic Publishers, Dodrecht, The Netherlands.
- 15.Koppitz, M., T. Spellig, R. Kahmann, and H. Kessler. 1996. Lipoconjugates: structure-activity studies for pheromone analogues of Ustilago maydis with varied lipophilicity. Int. J. Peptide Res. 48:377-390. [DOI] [PubMed] [Google Scholar]
- 16.Mayorga, M. E., and S. E. Gold. 1999. A MAP kinase encoded by the ubc3 gene of Ustilago maydis is required for filamentous growth and full virulence. Mol. Microbiol. 34:485-497. [DOI] [PubMed] [Google Scholar]
- 17.Müller, P., C. Aichinger, M. Feldbrugge, and R. Kahmann. 1999. The MAP kinase kpp2 regulates mating and pathogenic development in Ustilago maydis. Mol. Microbiol. 34:1007-1017. [DOI] [PubMed] [Google Scholar]
- 18.Onel, K., A. Koff, R. L. Bennett, P. Unrau, and W. K. Holloman. 1996. The REC1 gene of Ustilago maydis, which encodes a 3′5′ exonuclease, couples DNA repair and completion of DNA synthesis to a mitotic checkpoint. Genetics 143:165-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmitt, M. E., T. A. Brown, and B. L. Trumpower. 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18:3091-3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snetselaar, K. M. 1993. Microscopic observation of Ustilago maydis mating interactions. Exp. Mycol. 17:345-355. [Google Scholar]
- 21.Snetselaar, K. M., and C. W. Mims. 1992. Sporidial fusion and infection of maize seedlings by the smut fungus Ustilago maydis. Mycologia 84:193-203. [Google Scholar]
- 22.Spellig, T., M. Bölker, F. Lottspeich, R. W. Frank, and R. Kahmann. 1994. Pheromone trigger filamentous growth in Ustilago maydis. EMBO J. 13:1620-1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spellig, T., A. Bottin, and R. Kahmann. 1996. Green fluorescent protein (GFP) as a new vital marker in the phytopathogenic fungus Ustilago maydis. Mol. Gen. Genet. 252:503-509. [DOI] [PubMed] [Google Scholar]
- 24.Sprague, G. F., and J. W. Thorner. 1992. Pheromone response and signal transduction during the mating process of Saccharomyces cerevisiae, p. 657-744. In J. R. Broach, J. R. Pringle, and E. W. Jones (ed.), The molecular and cellular biology of the yeast Saccharomyces: gene expression. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 25.Steinberg, G., R. Wedlich-Söldner, M. Brill, and I. Schulz. 2001. Microtubules in the fungal pathogen Ustilago maydis are highly dynamic and determine cell polarity. J. Cell Sci. 114:609-622. [DOI] [PubMed] [Google Scholar]
- 26.Straube, A., W. Enard, A. Berner, R. Wedlich-Söldner, R. Kahmann, and G. Steinberg. 2001. A split motor domain in a cytoplasmic dynein. EMBO J. 20:5091-5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Straube, A., M. Brill, B. R. Oakley, T. Horio, and G. Steinberg. 2003. Microtubule organization requires cell cycle dependent nucleation at dispersed cytoplasmic sites, polar and perinuclear MTOCs in the plant pathogen Ustilago maydis. Mol. Biol. Cell 14:642-657. [DOI] [PMC free article] [PubMed]
- 28.Tsukuda, T., S. Carleton, S. Fotheringham, and W. K. Holloman. 1988. Isolation and characterization of an autonomously replicating sequence from Ustilago maydis. Mol. Cell. Biol. 8:3703-3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Urban, M., R. Kahmann, and M. Bölker. 1996. Identification of the pheromone response element in Ustilago maydis. Mol. Gen. Genet. 251:31-37. [DOI] [PubMed] [Google Scholar]
- 30.Wedlich-Söldner, R., A. Straube, M. W. Friedrich, and G. Steinberg. 2002. A balance of KIF1A-like kinesin and dynein organizes early endosomes in the fungus Ustilago maydis. EMBO J. 21:2946-2957. [DOI] [PMC free article] [PubMed] [Google Scholar]