Abstract
A detailed characterization of the central metabolic network of Saccharomyces cerevisiae CEN.PK 113-7D was carried out during cometabolism of different mixtures of glucose and acetate, using aerobic C-limited chemostats in which one of these two substrates was labeled with 13C. To confirm the role of malic enzyme, an isogenic strain with the corresponding gene deleted was grown under the same conditions. The labeling patterns of proteinogenic amino acids were analyzed and used to estimate metabolic fluxes and/or make inferences about the in vivo activities of enzymes of the central carbon metabolism and amino acid biosynthesis. Malic enzyme flux increased linearly with increasing acetate fraction. During growth on a very-high-acetate fraction, the activity of malic enzyme satisfied the biosynthetic needs of pyruvate in the mitochondria, while in the cytosol pyruvate was supplied via pyruvate kinase. In several cases enzyme activities were unexpectedly detected, e.g., the glyoxylate shunt for a very-low-acetate fraction, phosphoenolpyruvate carboxykinase for an acetate fraction of 0.46 C-mol of acetate/C-mol of substrate, and glucose catabolism to CO2 via the tricarboxylic acid cycle for a very-high-acetate fraction. Cytoplasmic alanine aminotransferase activity was detected, and evidence was found that α-isopropylmalate synthase has two active forms in vivo, one mitochondrial and the other a short cytoplasmic form.
Isotope labeling experiments have been used extensively for identification of metabolic pathways. They have also been used to characterize the structures of metabolic networks and to detect which pathways are active in vivo, both under different growth conditions and for different mutants (4, 7). Isotope labeling experiments can be used for functional analysis, for example, to detect the in vivo activity of an enzyme when a deletion mutant for the corresponding gene has no discernible phenotype, as illustrated by this work. To understand the metabolic network in detail, it is often important to quantify the activities of different pathways or enzymes leading to the same product. Isotope labeling experiments may allow for this quantification, as long as there is a difference in the labeling of the products originating from the different pathways (4, 36). Quantification of bidirectional fluxes is also possible (4, 24, 27).
Analysis of the metabolism of eukaryotes encompasses one extra degree of complexity due to the existence of physically separated compartments in the cell. Information on compartmentalization of metabolic reactions can be derived from labeling experiments (4, 5). Compared with enzyme activity assays and compartment fractionation, labeling experiments have the advantage of being rapid and can give information about both the intracellular locations of specific reactions and the in vivo fluxes.
Saccharomyces cerevisiae not only is a model eukaryotic organism for numerous biological phenomena but also is the most widely used organism in biotechnology. Understanding its metabolism in detail is therefore of utmost importance, and consequently, it has been studied extensively. Its biochemical reaction network is, in general, very well described. One exception to this is malic enzyme, whose role is not well understood. Activity of this enzyme is detected in cell extracts of cultures grown under different conditions; however, a deletion mutant for the corresponding gene has no discernible growth phenotype under the same conditions (2).
Malic enzyme is located in the middle of the metabolic network of S. cerevisiae, converting malate, an intermediate of the tricarboxylic acid (TCA) cycle, into pyruvate, a key metabolite for yeast in the split of respiration and fermentation, with production of one NADPH molecule. This mitochondrial enzyme (2) could as a result be involved in a number of different metabolic functions.
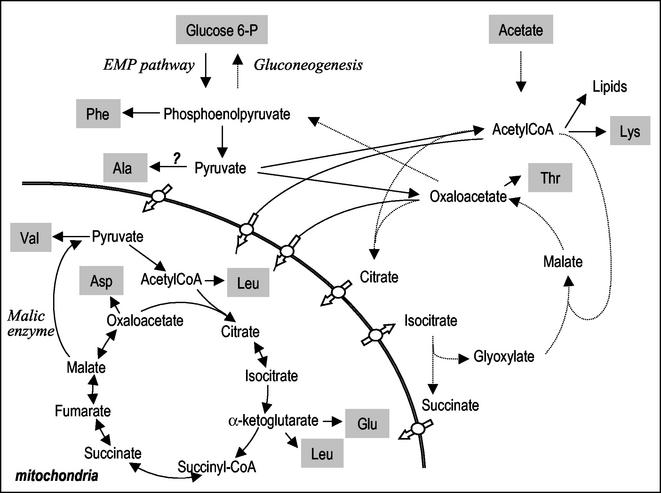
During aerobic growth in chemostats run at low specific growth rates, glucose is completely converted to CO2 and biomass, via the Embden-Meyerhof-Parnas pathway, the TCA cycle, and activity of cytosolic pyruvate carboxylase. During growth on acetate, on the other hand, this substrate is metabolized via acetyl coenzyme A (acetyl-CoA) formed by one of the two acetyl-CoA synthethases: the peroxisomal Acs1p and the cytosolic Acs2p (30). Acetyl-CoA can then be consumed in the glyoxylate shunt (by isocitrate lyase and malate synthase, outside the mitochondria) or enter the mitochondria to be metabolized in the TCA cycle. Gluconeogenesis also needs to be active (Fig. 1).
FIG. 1.
Aerobic glucose and acetate metabolism. Dashed arrows represent reactions that do not operate during growth on glucose. 6-P, 6-phosphate; EMP, Embden-Meyerhof-Parnas.
Acetate metabolism, similarly to almost all other yeast carbon sources, is subjected to glucose repression. However, during chemostat cultivations the glucose concentration inside the fermentor is so low that cells are no longer repressed and can metabolize acetate concomitantly with glucose. This experimental setup therefore provides a unique opportunity to study cometabolism of substrates.
In this work, S. cerevisiae was grown on either a glycolytic substrate (glucose), a gluconeogenic substrate (acetate), or mixtures of both. The purpose of the study was to detect the in vivo activities of different enzymes of the central metabolism for conditions intermediate to the well-studied glucose catabolism and gluconeogenesis. Special emphasis was put on the enzymes around the pyruvate node, and in particular on the biosynthesis of pyruvate via either pyruvate kinase or malic enzyme, which illustrates the quantification of alternative pathways leading to the same product. Information on the cellular compartment where enzyme activity is present and on amino acid biosynthesis was derived whenever possible. Finally, a strain with malic enzyme deleted was investigated under the same conditions to gain further insight into the function of this enzyme.
MATERIALS AND METHODS
Strains and maintenance.
The genetic reference strain used in this study was strain CEN.PK113-7D (MATa MAL2-8c SUC2). The isogenic mae1Δ strain was kindly provided by E. Boles (2). Working stocks of both strains were maintained at 4°C on YPD agar plates (containing, per liter, 10 g of Difco yeast extract, 20 g of Difco peptone, 20 g of glucose, and 20 g of agar), which were prepared monthly. Single colonies were used to inoculate precultures.
Chemostat cultivations.
Aerobic carbon-limited chemostats were operated at 30°C with a dilution rate (which equals the specific growth rate in steady-state cultures) of 0.1 h−1, using homemade fermentors with a working volume of approximately 170 ml. The volume was kept constant by withdrawing liquid with a continuously operating peristaltic pump. The pH was controlled to 5 with NaOH, except for cultivations with an acetic acid fraction in the feed of 0.95 C-mol/C-mol of substrate, for which a pH of 6 was kept constant by addition of HCl (pH 6 was used because acetic acid inhibition is lower at this pH than at pH 5). In all cases, the dissolved oxygen concentration was kept above 70% saturation by sparging sterile air and using a magnetic stirrer at 800 rpm.
Cells were cultivated with mineral medium supplemented with vitamins, prepared as described by Verduyn et al. (32). The antifoam used was 289 from Sigma. The filter-sterilized carbon source was added at a final concentration of 2 g/liter as a mixture of glucose and acetic acid (added as the sodium salt when unlabeled). A feed with unlabeled substrates was used until steady state was reached. Steady state was defined as the situation in which at least five residence times (the inverse of the dilution rate) had passed since the last change in growth conditions and the biomass concentration, monitored by optical density at 600 nm, had remained constant during at least two volume changes. Thereafter, the feed was replaced by a feed of similar composition, except for the carbon source, which was labeled in the following way: [1-13C]glucose when it was the sole carbon source, unlabeled glucose and [1,2-13C]acetic acid for acetic acid fractions in the feed from 0.05 to 0.5 C-mol/C-mol of substrate (for the acetic acid fraction of approximately 0.5, only 20% of the added acetate was labeled), and [13C6]glucose and unlabeled acetic acid for an acetic acid fraction in the feed of 0.95 C-mol/C-mol of substrate. In all experiments, the chemostats were fed with labeled substrates until an isotopic steady state was obtained (i.e., when the labeling patterns of the biomass components did not change over time).
Precultures were grown in baffled shake flasks at 30°C and 150 rpm for 24 to 30 h. The preculture medium had a composition similar to that of the chemostat medium, except that a higher concentration was used in order to obtain a high biomass concentration for the inoculation. The composition of the medium was as follows: 10 g of glucose per liter, 7.5 g of (NH4)2SO4 per liter, 14.4 g of KH2PO4 per liter, and trace metal solution (2× concentrated). The pH was adjusted to 6.5. The bioreactor was inoculated to an initial biomass concentration of 20 mg (dry weight) per liter and run as a batch with 2 g of glucose per liter until depletion of the substrate (for approximately 12 h). At this time, the feed was started and the fermentation was run in a continuous mode.
Analysis of glucose and acetate.
Glucose and acetate levels were determined by high-performance liquid chromatography with an Aminex HPX-87H ion exclusion column (Bio-Rad, Hercules, Calif.) coupled to two detectors connected in series, a Waters 410 differential refractometer detector for glucose determination and a Waters 486 tunable absorbance detector set at 210 nm for acetate determination. At steady state, no substrate was detected inside the fermentor except otherwise stated; excretion of metabolites was never detected.
Preparation of biomass hydrolysates.
After feeding of the chemostats with labeled substrate for at least four residence times (volume changes in the fermentor), the biomass was harvested by filtration, washed with distilled water, and frozen at −80°C. The biomass was hydrolyzed in 6 M HCl at 105°C for 0.5 and 16 h for analysis of glucose and amino acids, respectively. The hydrolysates were filtered to remove the cell debris and dried under a current of air in test tubes that contained the hydrolysis product of approximately 4 mg (wet weight) of biomass.
GC-MS.
The labeling patterns of amino acids and glucose incorporated in the biomass were determined by gas chromatography-mass spectrometry (GC-MS) and taken as an indirect measure of the labeling patterns of the intermediates of the central carbon metabolism that are precursors for the respective compounds. Derivatization procedures that render these compounds volatile were performed directly on the crude hydrolysates as described by Christensen and Nielsen (5); amino acids were analyzed as N-ethoxycarbonyl amino acid ethylesters and/or N-(N,N-dimethylaminomethylene)-amino acid methylesters, and glucose was analyzed as glucose pentaacetate. The GC-MS analysis was done as previously described (6).
When a compound of interest, separated by GC, enters the mass spectrometer, positively charged ions with the same mass as the original molecule are generated and detected. The masses of the ions depend on the number of 13C isotopes in the compound, and this distribution of masses gives rise to a cluster of signals that correspond to ions with identical elemental compositions but, due to the presence of 13C isotopes, with different masses, i.e., the mass isotopomers. Since the ions are unstable, they usually fragment into smaller pieces, leading to ions that contain only a subset of the carbon atoms found in the original molecule. Frequently, only such fragments are observed. Comparison of the mass distribution of the intact ion with that of the fragments arising from it can be used to deduce to what extent the various carbon positions in the molecule are labeled. Spectra were corrected for the background contribution to the signals. The signals were subsequently corrected for the influence of naturally occurring isotopes found in, e.g., the atoms introduced by the derivatization (17, 18). This modified mass spectrum was used to calculate the sum of the fractional labelings (in percent) according to the formula
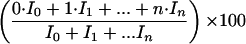 |
where I0 is the intensity of the peak corresponding to the mass isotopomer without 13C and In is the intensity of the peak corresponding to the mass isotopomer where all carbon atoms of the fragment are labeled with 13C. This value equals the number of 13C atoms per 100 fragment ions. Thus, for example, the sum of fractional labelings of a naturally labeled compound with n carbon atoms will be n.n%, whereas for the [1-13C]glucose used in this work it will be 104.5% (99% from C-1 and 1.1% from each of the other five carbon atoms).
For every biomass sample the number of ions analyzed was 26, involving quantification of approximately 104 mass isotopomers. These 26 ions were selected through an evaluation of the individual ion clusters (6). The reproducibility of the sum of fractional labelings has been previously determined, and therefore a statistical analysis was not performed here, but the standard deviation was 0.6% or lower for 19 of the 26 fragments analyzed, and only for Phe143 and Glc331 was it above 1% (15).
Chemicals.
[1,2-13C]acetate was purchased from Euriso-Top (Saint-Aulin, France), d-[1-13C]glucose was from Omicron (South Bend, Ind.), and [13C6]glucose was from Campro Scientific (Veenendaal, The Netherlands). All three compounds had 99% enrichment.
Flux determination.
Estimation of the metabolic flux distribution for the mae1Δ strain growing on glucose as the sole carbon source, using the determined sums of fractional labelings, was performed exactly as described by Gombert et al. (15). The biochemical model is given in the appendix; malic enzyme was omitted from the model for the deletion mutant. In short, the numeric method employed (described in more detail in reference 5) consisted of an iterative process of error minimization where the measured labeling patterns were compared with the ones calculated from a set of guessed fluxes and a mathematical model, thereby generating one error. Another error was generated by comparing the guessed fluxes with the measured ones (fluxes leading to biomass formation were not directly measured but were obtained from literature data on biomass composition and the measured biomass yield on glucose), and an optimization algorithm was used to minimize the sum of these two errors. The mathematical model based on the stoichiometry of the central metabolic network included bidirectional reaction rates as exchange fluxes (35).
RESULTS AND DISCUSSION
The metabolic network of S. cerevisiae CEN.PK113-7D (reference strain) and the influence of the carbon source composition on the active network were studied in chemostats operated with different mixtures of glucose and acetate (Table 1). The same cultivations were performed with an isogenic malic enzyme deletion strain, with the purpose of understanding the physiological role of this enzyme. The central metabolism was furthermore analyzed in detail by investigating acetate metabolism (including activity of the glyoxylate shunt and gluconeogenesis) and amino acid biosynthesis.
TABLE 1.
Biomass yields determined at dilution rate of 0.1 h−1 for different mixtures of glucose and acetate and the different strains studied
| Acetate fraction (C-mol/C-mol of substrate) | Biomass yield (g [dry wt]/g of substrate) for strain:
|
Labeled substrate used | |
|---|---|---|---|
| Reference | mae1Δ | ||
| 0.00 | 0.52a | 0.50 | [1-13C]glucose |
| 0.04 | 0.53 | [1,2-13C]acetate | |
| 0.05 | 0.44 | [1,2-13C]acetate | |
| 0.46 | 0.49 | [1,2-13C]acetate | |
| 0.90 | 0.45b,c | [13C6]glucose | |
| 0.94 | 0.36b | [13C6]glucose | |
Data are from reference 15.
pH 6.
There was a considerable amount of residual acetate in the fermentation medium. Only the acetate consumed by the cells was considered for the calculation of both the biomass yield and the acetate fraction.
For every cultivation, a substrate labeled with 13C was fed to the cells. The choice of labeled substrate was considered carefully for each feed composition (Table 1). The substrate present in smaller amounts was selected to be the substrate added as fully labeled. This was done in order to allow detection of very low fluxes. Since a key point of this study was to gain insight into acetate metabolism, this compound was used as the source of label for the feed that was composed of approximately equal amounts of glucose and acetate. In the situation where glucose was used as the sole substrate, [1-13C]glucose was chosen because it allows for distribution of the labeled C asymmetrically throughout the whole metabolic network, as required for flux estimation.
The labeling patterns of the intermediates of the central metabolism were not measured directly; instead, the labeling patterns of glucose and amino acids incorporated in the biomass, measured by GC-MS, were taken as an indirect measure of the labeling patterns of the intermediates of the central carbon metabolism that are precursors for the respective compounds, using the present knowledge on the routes for amino acid biosynthesis. Conversely, it was in some cases possible to derive information on amino acid biosynthesis from this analysis (see below). This approach has been extensively used with success (for examples, see references 5, 20, 27, and 28). The isotopic labelings measured for the different cultivations are summarized in Table 2 (for the reference strain) and Table 3 (for the mae1Δ strain) and are the basis of the presentation of results below.
TABLE 2.
Summed fractional labelings obtained with the reference strain in continuous cultivations at dilution rate of 0.1 h−1 for different acetate fractionsa
| Compound analyzed | Mol wt of unlabeled isotopomer | Biosynthetic precursor(s) | C atoms from biosynthetic precursor(s) present in fragment | Sum of fractional labelings (%) of C atoms in a fragment with the following acetate fraction (C-mol/C-mol of substrate) and labeled substrate used:
|
|||
|---|---|---|---|---|---|---|---|
| 0b [1-13C]glucose | 0.04, [1,2-13C]acetate | 0.46c, [1,2-13C]acetate | 0.90d, [13C6]glucose | ||||
| Glc | 331 | Glc 6P | 1, 2, 3, 4, 5, 6 | 91.0 | 6.9 | 277.9 | |
| Gly | 175e | 3-PG | 1, 2 | 5.9 | 3.2 | 10.2 | 21.3 |
| 144f | 3-PG | 1, 2 | 6.2 | 3.1 | 11.4 | ||
| 85f | 3-PG | 2 | 3.7 | 1.3 | 5.1 | 9.6 | |
| Ser | 175e | 3-PG | 1, 2 | 3.1 | 3.1 | 5.9 | 34.0 |
| 132e | 3-PG | 2, 3 | 33.8 | 2.4 | 5.6 | 31.5 | |
| Phe | 143f | PEP | 1, 2 | 3.0 | 2.2 | 4.3 | 25.3 |
| 192e | PEP + E4P | 2, 2, 3, 3 + 1, 2, 3, 4 | 90.6 | 9.4 | 14.9 | 212.1 | |
| Ala | 116e | Pyr | 2, 3 | 36.0 | 3.0 | 12.5 | 14.9 |
| 99f | Pyr | 2, 3 | 36.0 | 3.2 | 12.5 | 15.1 | |
| 158f | Pyr | 1, 2, 3 | 38.5 | 4.7 | 18.3 | 21.1 | |
| Val | 144e | Pyr | 2, 2, 3, 3 | 73.3 | 6.5 | 27.3 | 21.2 |
| 143f | Pyr | 1, 2 | 6.9 | 2.9 | 13.8 | 10.2 | |
| 127f | Pyr | 2, 2, 3, 3 | 73.1 | 6.3 | 26.8 | 20.1 | |
| 186f | Pyr | 1, 2, 2, 3, 3 | 74.9 | 7.9 | 33.3 | 26.5 | |
| Leu | 158e | Pyr + AcCoA | 2, 2, 3, 3 + 2 | 106.1 | 15.6 | 42.3 | 19.8 |
| Asp | 188e | OAA | 2, 3, 4 | 57.1 | 11.4 | 39.8 | 7.2 |
| 115f | OAA | 2 | 12.0 | 3.4 | 11.7 | 2.5 | |
| 216f | OAA | 1, 2, 3, 4 | 64.4 | 14.3 | 52.8 | 10.0 | |
| Thr | 175e | OAA | 1, 2 | 17.8 | 6.8 | 25.0 | 5.0 |
| 146e | OAA | 2, 3, 4 | 54.5 | 12.1 | 35.7 | 8.2 | |
| Ile | 158e | OAA + Pyr | 2, 3, 4 + 2, 3 | 93.7 | 14.5 | 53.4 | 16.9 |
| Pro | 142e | α-KG | 2, 3, 4, 5 | 85.5 | 18.8 | 58.3 | 6.9 |
| Glu | 143f | α-KG | 1, 2 | 39.9 | 10.5 | 28.4 | 6.5 |
| 230f | α-KG | 1, 2, 3, 4, 5 | 100.1 | 23.7 | 72.1 | 11.6 | |
| Lys | 156e | α-KG + AcCoA | 2, 3, 4, 5 + 2 | 117.2 | 48.5 | 75.6 | 8.4 |
A naturally labeled fragment with n carbon atoms will be labeled n.n%. The biosynthetic precursors indicated are for growth on glucose and ammonia; for some amino acids, the biosynthetic pathways may change for growth on acetate. Abbreviations: 3-PG, 3-phosphoglycerate; Pyr, pyruvate; AcCoA, acetyl-CoA; OAA, oxaloacetate; α-KG, α-ketoglutarate; PEP, phosphoenolpyruvate; E4P, erythrose-4-phosphate; Glc, glucose; Glc 6P, glucose-6-phosphate.
Data are from reference 15.
Only 20% of the acetate fed to the culture was [1,2-13C] acetate.
pH6.
Amino acid analyzed as N-ethoxycarbonyl amino acid ethylester.
Amino acid analyzed as N-(N,N-dimethylaminomethylene) amino acid methylester.
TABLE 3.
Summed fractional labelings obtained with the mae1Δ strain in continuous cultivations at dilution rate of 0.1 h−1 for different acetate fractionsa
| Compound analyzed | Mol wt of unlabeled isotopomer | Biosynthetic precursor(s)b | C atoms from biosynthetic precursor(s) present in fragment | Sum of fractional labelings (%) of C atoms in a fragment with the following acetate fraction (C-mol/C-mol of substrate) and labeled substrate used:
|
||
|---|---|---|---|---|---|---|
| 0, [1-13C]glucose | 0.05, [1,2-13C]acetate | 0.94c, [13C6]glucose | ||||
| Glc | 331 | Glc 6P | 1, 2, 3, 4, 5, 6 | 91.7 | 133.0 | |
| Gly | 175d | 3-PG | 1, 2 | 6.8 | 3.1 | 9.3 |
| 144e | 3-PG | 1, 2 | 6.4 | 2.5 | 8.7 | |
| 85e | 3-PG | 2 | 3.7 | 1.3 | 5.1 | |
| Ser | 175d | 3-PG | 1, 2 | 3.4 | 11.2 | |
| 132d | 3-PG | 2, 3 | 36.3 | 2.0 | 13.5 | |
| Phe | 143e | PEP | 1, 2 | 31.1 | 2.2 | 12.2 |
| 192d | PEP + E4P | 2, 2, 3, 3 + 1, 2, 3, 4 | 80.4 | 10.0 | 101.7 | |
| Ala | 116d | Pyr | 2, 3 | 35.6 | 2.2 | 12.6 |
| 99e | Pyr | 2, 3 | 35.6 | 2.3 | 12.8 | |
| 158e | Pyr | 1, 2, 3 | 37.5 | 3.6 | 18.3 | |
| Val | 144d | Pyr | 2, 2, 3, 3 | 72.0 | 5.0 | 24.6 |
| 143e | Pyr | 1, 2 | 4.6 | 2.2 | 12.7 | |
| 127e | Pyr | 2, 2, 3, 3 | 71.9 | 5.1 | 25.0 | |
| 186e | Pyr | 1, 2, 2, 3, 3 | 69.6 | 6.2 | 29.1 | |
| Leu | 158d | Pyr + AcCoA | 2, 2, 3, 3 + 2 | 106.0 | 17.0 | 25.6 |
| Asp | 188d | OAA | 2, 3, 4 | 60.5 | 10.1 | 4.5 |
| 115e | OAA | 2 | 16.6 | 2.1 | 1.4 | |
| 216e | OAA | 1, 2, 3, 4 | 70.6 | 13.4 | 6.9 | |
| Thr | 175d | OAA | 1, 2 | 23.5 | 5.5 | 3.1 |
| 146d | OAA | 2, 3, 4 | 58.2 | 9.6 | 4.3 | |
| Ile | 158d | OAA + Pyr | 2, 3, 4 + 2, 3 | 96.3 | 12.5 | 16.8 |
| Pro | 142d | α-KG | 2, 3, 4, 5 | 90.2 | 15.6 | 6.1 |
| Glu | 143e | α-KG | 1, 2 | 40.9 | 8.5 | 4.2 |
| 230e | α-KG | 1, 2, 3, 4, 5 | 103.7 | 19.7 | 8.5 | |
| Lys | 156d | α-KG + AcCoA | 2, 3, 4, 5 + 2 | 120.2 | 65.4 | 7.7 |
A naturally labeled fragment with n carbon atoms will be labeled n.n%. Abbreviations are as for Table 2.
For growth on glucose.
pH 6.
Amino acid analyzed as N-ethoxycarbonyl amino acid ethylester.
Amino acid analyzed as N-(N,N-dimethylaminomethylene) amino acid methylester.
Biomass yields.
For both the reference and the mae1Δ strains, the biomass yield decreased with increased acetate fraction in the feed (Table 1). This result was expected in view of the fact that the biomass yield on acetate is lower than that on glucose (11, 33). For all acetate fractions tested, the biomass yield was lower for the deletion mutant.
Malic enzyme activity.
A continuous culture was operated with a mixture of unlabeled glucose and [1,2-13C]acetate. The acetate fraction in the feed was 0.04 C-mol/C-mol of substrate. At steady state, all TCA cycle intermediates were labeled (Table 2). Phosphoenolpyruvate was unlabeled (Table 2) (Phe [C-1,2] is naturally labeled), indicating that the enzyme phosphoenolpyruvate carboxykinase (converting oxaloacetate into phosphoenolpyruvate) was not active at this low acetate ratio. Pyruvate, however, was labeled to some extent (Table 2), which is considerable taking into account that the net flow of carbon is in the opposite direction and that this 13C enrichment must originate from a C4 intermediate (as opposed to a C2 intermediate, since acetate cannot be directly converted to pyruvate). Malic enzyme was found to be the most likely candidate for the generation of labeled pyruvate. Similar fermentations performed with a mae1Δ strain supported this hypothesis. The main difference between the two strains was that for the deletion mutant there was no labeling of pyruvate (Table 3). This was taken as evidence of malic enzyme being active in the reference strain. Except for this, the results obtained for the mae1Δ strain grown with this acetate fraction were similar to those for the reference strain.
For an acetate fraction of 0.46 C-mol/C-mol of substrate in the feed, pyruvate also was labeled to a considerably higher extent than phosphoenolpyruvate (Table 2), again indicating in vivo activity of malic enzyme.
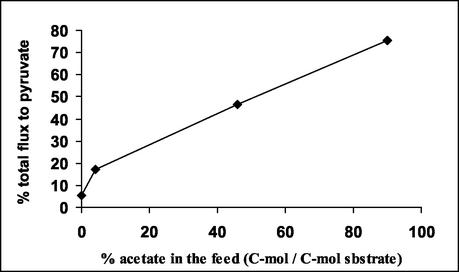
For each of the acetate fractions in the feed tested, a labeling balance was used to calculate the percentage of pyruvate in the cell synthesized via the malic enzyme (x in the following equation). The calculations were performed by using a labeling balance according to the following equation: Phe(C-1,2) · (100 − x) + Asp(C-1,2) · x = Val(C-1,2) · 100. This labeling balance is based on the assumption that phosphoenolpyruvate is the precursor of cytosolic pyruvate via pyruvate kinase and thereby indirectly a precursor of mitochondrial pyruvate; malate (in equilibrium with oxaloacetate) is another precursor of mitochondrial pyruvate via malic enzyme, and valine directly measures the labeling in mitochondrial pyruvate (Fig. 1). The results are presented in Fig. 2. It is very clear that the relative importance of malic enzyme for pyruvate synthesis increased with increased acetate fraction in the feed. However, despite the high flux through malic enzyme for high-acetate fractions, this enzyme does not seem to be essential for metabolizing acetate. Very striking is the fact that malic enzyme was also active during growth on glucose.
FIG. 2.
Percentage of pyruvate synthesized via the malic enzyme for different mixtures of glucose and acetate. The value for 0% acetate was obtained by using a flux estimation procedure for the whole metabolic network (Fig. 3).
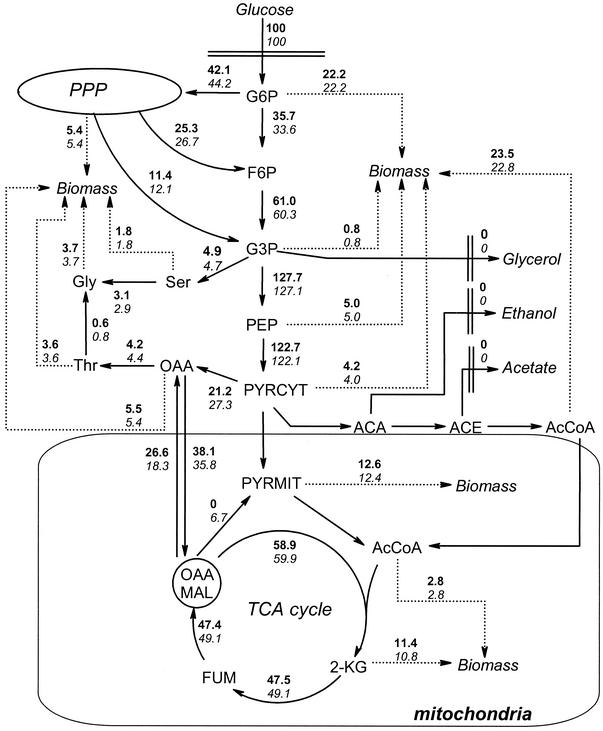
Malic enzyme deletion was also investigated during growth on [1-13C]glucose. The metabolic fluxes of the mae1Δ strain were estimated with the data from Table 3, using the biochemical model shown in the appendix, with the one exception that malic enzyme was removed. Selected fluxes are presented in Fig. 3. It can be seen that deletion of the gene coding for malic enzyme had a significant impact on the flux through pyruvate carboxylase but only a minor impact on the other fluxes of the metabolic network.
FIG. 3.
Distribution of metabolic fluxes for a malic enzyme deletion strain (values in boldface) and the reference strain (values in italics) during chemostat cultivation with [1-13C]glucose. Relative fluxes through pyruvate dehydrogenase and the pyruvate dehydrogenase bypass cannot be discriminated, but their sum was 61.6 for the deletion mutant and 62.7 for the reference strain. All of the net metabolic fluxes are given in millimoles per 100 mmol of glucose. For abbreviations, see Table A1.
Acetate metabolism.
The labeling data obtained for the cultivation with an acetate fraction in the feed of 0.04 C-mol/C-mol of substrate indicated that acetate was consumed primarily outside the mitochondria. This is supported by comparing the labeling of the lysine carbon atom that derives from C-2 of acetyl-CoA (acetyl-CoA condensation in the first step of lysine synthesis by homocitrate synthase takes place in the nucleus [3]) with the labeling of the C-4 of α-ketoglutarate, which derives from C-2 of acetyl-CoA (in the mitochondria): the first value was 29.7% (Table 2, Lys − Pro), and the second was around 4% (Table 2, Glu; the average for C-3, C-4, and C-5 is 4.3). Cytosolic acetyl-CoA is not 100% labeled due to activities of pyruvate decarboxylase, acetaldehyde dehydrogenase, and acetyl-CoA synthetase, and the labeling of acetyl-CoA in mitochondria indicates transfer of this compound from the cytosol to this compartment.
It is well established that acetyl-CoA condensing with oxaloacetate in the first step of the TCA cycle is converted to carbon atoms 4 and 5 of α-ketoglutarate. If the acetyl-CoA is labeled, scrambling at the level of succinate dehydrogenase and fumarase further on in the TCA cycle will make carbon atoms 1 and 2 of the regenerated oxaloacetate molecules labeled to the same extent as carbon atoms 3 and 4. All of them will, of course, be labeled to a lower extent than the labeled acetyl-CoA. Therefore, when labeled acetyl-CoA is the origin of the labeling of the TCA cycle intermediates, C-4 and C-5 of α-ketoglutarate will be more labeled than the other carbon atoms. However, for an acetate fraction in the feed of 0.04 C-mol/C-mol of substrate, this was not the case, according to the labeling pattern obtained for α-ketoglutarate in this experiment (Table 2, Glu; the average for C-1 and C-2 is 5.3, and the average for C-3, C-4, and C-5 is 4.3). Also, the isotopic enrichment of C-1 and C-2 of oxaloacetate was lower than that of C-3 and C-4, again indicating that the pathway for incorporation of 13C into oxaloacetate was not the TCA cycle (Table 2; for Asp the labeling of C-3 plus C-4 is 11.4 − 3.4 and therefore that of C-1 plus C-2 is 14.3 − 8, and for Thr the average for C-1 and C-2 is 3.4 and so the average for C-3 and C-4 is 4.4). A possible pathway for the generation of labeled oxaloacetate is the glyoxylate shunt. A flux analysis including the glyoxylate shunt in a simplified biochemical model was performed (not shown), and it was possible to minimize the error in the procedure for flux estimation, as opposed to the case for a model where the reactions for the glyoxylate shunt were omitted. The glyoxylate shunt therefore seemed to be active even with this low-acetate fraction in the feed. Nevertheless, pyruvate carboxylase was certainly the main anaplerotic reaction under these growth conditions.
Phosphoenolpyruvate was labeled for an acetate fraction in the feed of 0.46 C-mol/C-mol of substrate (Table 2), indicating that for this acetate fraction in the feed the enzyme phosphoenolpyruvate carboxykinase was active (as opposed to growth on the 0.04 acetate fraction). During this intermediate condition of glucose catabolism and gluconeogenic growth on acetate, the isotopic labeling of the lysine C atom that derives from C-2 of acetyl-CoA (in the nucleus) was 17.3% (Table 2, Lys − Pro), and because this is lower than the labeling of the added acetate (20%), it appears that pyruvate decarboxylase was active. For leucine, the isotopic labeling of the C atom that derives from C-2 of acetyl-CoA was 15%, indicating that pyruvate dehydrogenase was active. Fructose 1,6-bisphosphatase was not active (Table 2) (glucose-6-phosphate has natural labeling).
A continuous cultivation was performed with an acetate fraction of 0.90 C-mol/C-mol of substrate, where glucose was added as [13C6]glucose. The results obtained for the reference strain show that all of the intermediates in the central metabolism were labeled (Table 2), indicating that glucose was catabolized concomitantly with gluconeogenesis, the main pathway active during growth on a high-acetate fraction. It should be noted that glucose degradation down to pyruvate, and further down to intermediates of the TCA cycle, occurred even though the amount of glucose in the feed was not enough to supply all of the glucose needed for synthesis of polysaccharides (22, 34). This means that labeled carbon atoms from small amounts of glucose were distributed all over the metabolic network at the same time as the cell was spending energy synthesizing glucose-6-phosphate from C2 units. It is likely that glucose degradation occurred via the pentose phosphate pathway, since it has been shown by in vivo nuclear magnetic resonance that during growth on acetate, glucose synthesized via gluconeogenesis is shunted through this pathway (10). It is not clear if this is due to a need for NAPDH generation, but some evidence for this comes from the following observation. A major difference between the reference strain and the mae1Δ strain for this acetate fraction was the isotopic enrichment of glucose-6-phosphate. It was much lower for the mutant, indicating a higher flux through the pentose phosphate pathway, perhaps to compensate for the lack of NADPH synthesized by the malic enzyme.
Amino acid biosynthesis.
It is well established that pyruvate is the precursor for both valine in the mitochondria (12) and alanine in an unknown compartment. In the results obtained for the reference strain growing on the 0.90 acetate fraction, the labeling patterns of these two amino acids were considerably different (Table 2). This could be explained by the existence of two different pyruvate pools, one mitochondrial and the other cytosolic: pyruvate from the mitochondrial pool would be synthesized via the malic enzyme from malate (the carbon atoms of this compound deriving mainly from unlabeled acetate assimilated by the glyoxylate shunt) and used for valine biosynthesis, and the cytosolic pool would originate from phosphoenolpyruvate (partly derived from the labeled glucose) through pyruvate kinase and would be used for alanine biosynthesis. It can hence be inferred that alanine synthesis occurred from cytosolic pyruvate. This interpretation of the results is furthermore strengthened by the observation that the most striking difference between the reference strain and the mae1Δ strain for this acetate fraction was that the labeling patterns of alanine and valine (both derived from pyruvate) were the same for the deletion mutant.
Even though there are two pyruvate pools, it can be seen from the labeling of valine that pyruvate was transported into the mitochondria, since this metabolite was more labeled than any intermediate of the TCA cycle (Table 2).
It has been reported that α-isopropylmalate synthase, the mitochondrial enzyme utilizing acetyl-CoA for leucine biosynthesis, has a short form targeted to the cytosol (1). The results obtained in the present work for an acetate fraction in the feed of 0.04 C-mol/C-mol of substrate seem to indicate that both forms of this enzyme were active in vivo, since the labeling of the carbon atom in leucine that derives from C-2 of acetyl-CoA (Table 2, Leu − 2 × Pyr[C-2,3] ∼ 9.2) was intermediate to those referred to above for lysine (30%) and α-ketoglutarate (around 4%), which were taken as an indication of the isotopic labeling of acetyl-CoA in the cytoplasm-nucleus and in the mitochondria, respectively.
For this acetate fraction in the feed, glycine and serine do not reflect the labeling pattern of 3-phosphoglycerate (they have a higher labeling than phosphoenolpyruvate). This is probably due to the fact that during growth on acetate, another pathway for the biosynthesis of these amino acids is occurring, where glycine is formed by transamination of glyoxylate. Part of it is then decarboxylated to form 5,10-methylene tetrahydrofolate, which may be used to methylate another glycine molecule, generating enriched serine in the reaction catalyzed by serine hydroxymethyltransferase (16). Furthermore, we cannot exclude the possibility of threonine aldolase also being active, as seen for growth on glucose (Fig. 3).
Overall physiological conclusions.
In vivo activity of malic enzyme was found for all mixtures of glucose and acetate tested, and its relative importance increased with increased acetate fraction in the feed. It is not clear what role this enzyme plays during growth on glucose, since the combined activities of pyruvate carboxylase, malate dehydrogenase, and malic enzyme result in net ATP consumption and can therefore be regarded as a futile cycle. It is generally accepted that malic enzyme produces NADPH, so it is possible that this enzyme plays a role in NADPH production in the mitochondria during growth on glucose.
When cells grow on acetate, the picture is quite different. Synthesis of pyruvate can occur in the mitochondria via the malic enzyme from malate or in the cytoplasm from oxaloacetate via phosphoenolpyruvate carboxykinase and pyruvate kinase. The energetic costs for these alternative pathways are the same. With this carbon source, it seems that pyruvate synthesis in both compartments of the cell is preferred over pyruvate transport across the mitochondrial membrane, even if, presumably, it can do so in both directions (a pyruvate kinase 1 and 2 double mutant is viable [2]). In fact, it is possible to calculate what percentage of the total amount of pyruvate needed for biosynthesis in the cell is consumed in the mitochondria. Using the data from reference 23 and considering that the first step in alanine biosynthesis takes place in the cytosol (need for pyruvate, 0.452 mmol/g [dry weight] of cells) and that valine, isoleucine, and leucine biosyntheses take place in the mitochondria (need for pyruvate, 1.38 mmol/g [dry weight] of cells), we obtain the result that the percentage of pyruvate consumed in the mitochondria is 75.3% of the total pyruvate requirement, and this fits very well with the experimental value obtained of 75.7% pyruvate in the cell synthesized via the malic enzyme (Fig. 2).
Malic enzyme has been assigned to have a role in lipid accumulation of oleaginous fungi, which occurs under conditions of growth limitation, e.g., nitrogen limitation (26). The role of malic enzyme in S. cerevisiae has not yet been tested under these conditions, and it may very well be that this enzyme has a function in lipid metabolism.
At the low-acetate fraction of 0.04 C-mol/C-mol of substrate, acetate was expected to be consumed via the TCA cycle, but this was not the case. There was evidence not only that most of it was consumed outside the mitochondria but also that the glyoxylate shunt was active. This result was surprising, even though low activities of both malate synthase and isocitrate lyase have been detected in extracts of yeast cells of the strain used in the present study grown in glucose-limited chemostats (8). A possible explanation could be the fact reported in the literature that acetate induces expression of both enzymes of the glyoxylate shunt, particularly under the glucose-derepressed conditions prevailing in the chemostat cultures applied in this study (13, 31).
Although in previous studies considerable activity of phosphoenolpyruvate carboxykinase has been detected in cell extracts of glucose-grown cells (8), it was clearly seen here that this enzyme was not active in vivo during growth on an acetate fraction of 0.04 C-mol/C-mol of substrate. For a fraction of 0.46 there was considerable activity of this enzyme, even though it has been calculated that its activity becomes necessary for the synthesis of biomass precursors only above an acetate fraction in the feed of 0.71 (9).
Activities of glyoxylate shunt enzymes and phosphoenolpyruvate carboxykinase were detected under conditions not expected from considerations of cellular requirements for growth. This was also the case for pyruvate decarboxylase, which was active although the acetate fed to the cells in this work (even for the lowest acetate fraction used) was above the cytoplasmic need for acetyl-CoA, which was determined to be 1.04 to 1.05 mmol/g of biomass both theoretically and experimentally (14). Glucose degradation occurred during growth under gluconeogenic conditions, and even TCA cycle intermediates were labeled, in a way presumably due to activity of pyruvate carboxylase, which is constitutively expressed (8, 25).
In the present work, information not covered in the literature regarding the localization and activity of enzymes involved in amino acid biosynthesis was obtained. For alanine aminotransferase, this is, to our knowledge, the first report providing evidence for its activity in the cytosol. While this was deduced from data obtained for growth under gluconeogenic conditions, other authors have interpreted their own results for growth on glucose in a conflicting way (19). In this context, it should be said that two putative alanine aminotransferase genes have been found in the genome of S. cerevisiae, one coding for a putative mitochondrial enzyme and the other coding for a putative cytoplasmic isoenzyme (sequence from Incyte Genomics). Another possible route for biosynthesis of alanine in the cytosol could be the enzyme alanine glyoxylate aminotransferase, for which the location is unknown. However, this enzyme has been identified as the main enzyme responsible for the production of glycine during growth on nonfermentable carbon sources (and is glucose repressed) (21, 29); it is therefore unlikely to be involved in biosynthesis of alanine.
For α-isopropylmalate synthase, a mitochondrial enzyme for which the existence of a short cytoplasmic form has been reported, evidence was found that both forms are active.
Acknowledgments
Margarida Moreira dos Santos acknowledges Fundação para a Ciência e Tecnologia, Portugal, for the award of a research fellowship. Andreas Karoly Gombert gratefully acknowledges financial support by CAPES (Brasília, Brazil) (grant BEX1098/98-5).
APPENDIX
The model used for flux calculations is presented in Table A1. The C atom transitions, which are used by the flux estimation routine, are also indicated.
TABLE A1.
Model used for flux calculationsa
| Reaction |
|---|
| % Glucose uptake |
| GLC (abcdef) → G6P (abcdef) |
| % EMP pathway |
| G6P (abcdef) → F6P (abcdef) |
| F6P (abcdef) + G6P (ghijkl) → G6P (abcdef) + F6P (ghijkl) |
| F6P (abcdef) → G3P (cba) + G3P (def) |
| G3P (abc) → PEP (abc) |
| PEP (abc) → PYRCYT (abc) |
| % PP pathway |
| G6P (abcdef) → CO (a) + P5P (bcdef) |
| P5P (abcde) + P5P (fghij) → S7P (fgabcde) + G3P (hij) |
| S7P (fgabcde) + G3P (hij) + P5P (klmno) + P5P (pqrst) → P5P (abcde) + P5P (fghij) + S7P (klpqrst) + G3P (mno) |
| S7P (abcdefg) + G3P (hij) → F6P (abchij) + E4P (defg) |
| F6P (abchij) + E4P (defg) + S7P (klmnopq) + G3P (rst) → S7P (abcdefg) + G3P (hij) + F6P (klmrst) + E4P (nopq) |
| P5P (abcde) + E4P (fghi) → F6P (abfghi) + G3P (cde) |
| F6P (abfghi) + G3P (cde) + P5P (jklmn) + E4P (opqr) → P5P (abcde) + E4P (fghi) + F6P (jkopqr) + G3P (lmn) |
| % Ethanol, acetate, and glycerol formation |
| PYRCYT (abc) → ACA (bc) + CO (a) |
| ACA (ab) → ETH (ab) |
| ACA (ab) → ACE (ab) |
| G3P (abc) → GLYC (abc) |
| % Formation of acetyl-CoA in the cytosol |
| ACE (ab) → ACCOACYT (ab) |
| % Anaplerotic reaction (cytosolic) |
| PYRCYT (abc) + CO (d) → OAACYT (abcd) |
| % TCA cycle (considering scrambling around FUM) |
| PYRMIT (abc) → ACCOAMIT (bc) + CO (a) |
| OAAMIT (abcd) + ACCOAMIT (ef) → ICIT (dcbafe) |
| ICIT (abcdef) → AKG (abcef) + CO (d) |
| AKG (abcde) → FUM (bcde) + CO (a) |
| FUM (abcd) + FUM (efgh) → OAAMIT (abcd) + OAAMIT (hgfe) |
| OAAMIT (abcd) → FUM (abcd) |
| % Transports |
| OAAMIT (abcd) → OAACYT (abcd) |
| OAACYT (abcd) → OAAMIT (abcd) |
| ACCOACYT (ab) → ACCOAMIT (ab) |
| PYRCYT (abc) → PYRMIT (abc) |
| % Threonine, serine, and glycine metabolism (all enzymes are assumed to be cytoplasmic) |
| G3P (abc) → SER (abc) |
| SER (abc) → GLY (ab) + C1 (c) |
| SER (abc) + GLY (de) + C1 (f) → SER (def) + GLY (ab) + C1 (c) |
| OAACYT (abcd) → THR (abcd) |
| THR (abcd) → GLY (ab) + ACA (cd) |
| GLY (ab) + ACA (cd) + THR (efgh) → THR (abcd) + GLY (ef) + ACA (gh) |
| % Malic enzyme (malate decarboxylation, mitochondrial) |
| OAAMIT (abcd) → PYRMIT (abc) + CO (d) |
| % Drain of intermediates to macromolecules |
| G6P (abcdef) → G6POUT (abcdef) |
| P5P (abcde) → P5POUT (abcde) |
| E4P (abcd) → E4POUT (abcd) |
| G3P (abc) → G3POUT (abc) |
| PEP (abc) → PEPOUT (abc) |
| PYRCYT (abc) → PYRCYTOUT (abc) |
| PYRMIT (abc) → PYRMITOUT (abc) |
| OAACYT (abcd) → OAACYTOUT (abcd) |
| AKG (abcde) → AKGOUT (abcde) |
| ACCOACYT (ab) → ACCOACYTOUT (ab) |
| ACCOAMIT (ab) ACCOAMITOUT (ab) |
| SER (abc) → SEROUT (abc) |
| GLY (ab) → GLYOUT (ab) |
| C1 (a) → C1OUT (a) |
| GLYC (abc) → GLYCOUT (abc) |
| THR (abcd) → THROUT (abcd) |
| % Excreted products |
| ETH (ab) → ETHOUT (ab) |
| ACE (ab) → ACEOUT (ab) |
| % CO2 evolution |
| CO (a) → COOUT (a) |
Standard three-letter abbreviations are used for the amino acids. Other abbreviations are as follows: ACA, acetaldehyde; ACE, acetate; ACEOUT, acetate excreted to the medium; ACCOACYT, acetyl-CoA in the cytosol; ACCOACYTOUT, cytosolic acetyl-CoA used for biomass formation; ACCOAMIT, acetyl-CoA in the mitochondria; ACCOAMITOUT, mitochondrial acetyl-CoA used for biomass formation; AKG, α-ketoglutarate; AKGOUT, α-ketoglutarate used for biomass formation; CO, carbon dioxide; E4P, erythrose-4-phosphate; E4POUT, erythrose-4-phosphate used for biomass formation; ETH, ethanol; ETHOUT, ethanol excreted to the medium; F6P, fructose-6-phosphate; FUM, fumarate; GLC, glucose; GLYC, glycerol; GLYOUT, glycine used for biomass formation; G3P, glyceraldehyde-3-phosphate; G3POUT, glyceraldehyde-3-phosphate used for biomass formation; G6P, glucose-6-phosphate; G6POUT, glucose-6-phosphate used for biomass formation; ICIT, isocitrate; MAL, malate; OAA, oxaloacetate; OAACYT, oxaloacetate in the cytosol; OAACYTOUT, cytosolic oxaloacetate used for biomass formation; OAAMIT, oxaloacetate in the mitochondria; PEP, phosphoenolpyruvate; PEPOUT, phosphoenolpyruvate used for biomass formation; P5P, pentose 5-phosphate; P5POUT, pentose 5-phosphate used for biomass formation; PP pathway, pentose phosphate pathway; PYR, pyruvate; PYROUT, pyruvate used for biomass formation; SEROUT, serine used for biomass formation; S7P, sedoheptulose-7-phosphate; THROUT, threonine used for biomass formation.
REFERENCES
- 1.Beltzer, J. P., S. R. Morris, and G. B. Kohlhaw. 1988. Yeast LEU4 encodes mitochondrial and nonmitochondrial forms of alpha-isopropylmalate synthase. J. Biol. Chem. 263:368-374. [PubMed] [Google Scholar]
- 2.Boles, E., P. de Jong-Gubbels, and J. T. Pronk. 1998. Identification and characterization of MAE1, the Saccharomyces cerevisiae structural gene encoding mitochondrial malic enzyme. J. Bacteriol. 180:2875-2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen, S., J. S. Brockenbrough, J. E. Dove, and J. P. Aris. 1997. Homocitrate synthase is located in the nucleus in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 16:10839-10846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christensen, B., and J. Nielsen. 1999. Metabolic network analysis. A powerful tool in metabolic engineering. Adv. Biochem. Eng. Biotechnol. 66:209-231. [PubMed] [Google Scholar]
- 5.Christensen, B., and J. Nielsen. 2000. Metabolic network analysis of Penicillium chrysogenum using 13C-labeled glucose. Biotechnol. Bioeng. 68:652-659. [PubMed] [Google Scholar]
- 6.Christensen, B., and J. Nielsen. 1999. Isotopomer analysis using GC-MS. Metab. Eng. 1:282-290. [DOI] [PubMed] [Google Scholar]
- 7.de Graaf, A. A., K. Striegel, R. M. Wittig, B. Laufer, G. Schmitz, W. Wiechert, G. A. Sprenger, and H. Sahm. 1999. Metabolic state of Zymomonas mobilis in glucose-, fructose-, and xylose-fed continuous cultures as analysed by 13C- and 31P-NMR spectroscopy. Arch. Microbiol. 171:371-385. [DOI] [PubMed] [Google Scholar]
- 8.de Jong-Gubbels, P., J. Bauer, P. Niederberger, I. Stückrath, P. Kötter, J. P. van Dijken, and J. T. Pronk. 1998. Physiological characterisation of a pyruvate-carboxylase-negative Saccharomyces cerevisiae mutant in batch and chemostat cultures. Antonie Leeuwenhoek 74:253-263. [DOI] [PubMed] [Google Scholar]
- 9.de Jong-Gubbels, P., P. Vanrolleghem, S. Heijnen, J. P. van Dijken, and J. T. Pronk. 1995. Regulation of carbon metabolism in chemostat cultures of Saccharomyces cerevisiae grown on mixtures of glucose and ethanol. Yeast 11:407-418. [DOI] [PubMed] [Google Scholar]
- 10.Dickinson, J. R., I. W. Dawes, A. S. Boyd, and R. L. Baxter. 1983. 13C NMR studies of acetate metabolism during sporulation of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 19:5847-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duboc, P. 1997. Transient growth of Saccharomyces cerevisiae, a quantitative approach. Ph.D. Thesis. Swiss Federal Institute of Technology, Lausanne, Switzerland.
- 12.Falco, S. C., K. S. Dumas, and K. J. Livak. 1985. Nucleotide sequence of the yeast ILV2 gene which encodes acetolactate synthase. Nucleic Acids Res. 13:4011-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandez, E., M. Fernandez, F. Moreno, and R. Rodicio. 1993. Transcriptional regulation of the isocitrate lyase encoding gene in Saccharomyces cerevisiae. FEBS Lett. 333:238-242. [DOI] [PubMed] [Google Scholar]
- 14.Flikweert, M. T., M. de Swaaf, J. P. van Dijken, and J. T. Pronk. 1999. Growth requirements of pyruvate-decarboxylase-negative Saccharomyces cerevisiae. FEMS Microbiol Lett. 174:73-79. [DOI] [PubMed] [Google Scholar]
- 15.Gombert, A. K., M. M. dos Santos, B. Christensen, and J. Nielsen. 2001. Network identification and flux quantification in the central metabolism of Saccharomyces cerevisiae under different conditions of glucose repression. J. Bacteriol. 183:1441-1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones. E. W., and G. R. Fink. 1982. Regulation of amino acid and nucleotide biosynthesis in yeast, p 181-300. In J. N. Strathern, E. W. Jones, and J. R. Broach (ed.), The molecular biology of the yeast Saccharomyces: metabolism and gene expression, Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 17.Lee, W.-N. P., L. O. Byerley, E. A. Bergner, and J. Edmond. 1991. Mass isotopomer analysis: theoretical and practical considerations Biol. Mass Spectrom. 20:451-458. [DOI] [PubMed] [Google Scholar]
- 18.Lee, W.-N. P., E. A. Bergner, and Z. K. Guo. 1992. Mass isotopomer pattern and precursor-product relationship. Biol. Mass Spectrom. 21:114-122. [DOI] [PubMed] [Google Scholar]
- 19.Maaheimo, H., J. Fiaux, Z. P. Cakar, J. E. Bailey, U. Sauer, and T. Szyperski. 2001. Central carbon metabolism of Saccharomyces cerevisiae explored by biosynthetic fractional 13C labeling of common amino acids. Eur. J. Biochem. 268:2464-2479. [DOI] [PubMed] [Google Scholar]
- 20.Marx, A., A. A. de Graaf, W. Wiechert, L. Eggeling, and H. Sahm. 1996. Determination of the fluxes in the central metabolism of Corynebacterium glutamicum by nuclear magnetic resonance spectroscopy combined with metabolite balancing. Biotechnol. Bioeng. 49:111-129. [DOI] [PubMed] [Google Scholar]
- 21.Monschau, N., K. P. Stahmann, H. Sahm, J. B. McNeil, and A. L. Bognar. 1997. Identification of Saccharomyces cerevisiae GLY1 as a threonine aldolase: a key enzyme in glycine biosynthesis. FEMS Microbiol. Lett. 150:55-60. [DOI] [PubMed] [Google Scholar]
- 22.Oura, E. 1983. Biomass from carbohydrates. Bio/Technology 3:3-42. [Google Scholar]
- 23.Oura, E. 1972. The effect of aeration on the growth energetics and biochemical composition of baker's yeast, with an appendix: reactions leading to the formation of yeast cell material from glucose and ethanol. Ph.D. thesis. Helsinki University, Helsinki, Finland.
- 24.Petersen, S., A. A. de Graaf, L. Eggeling, M. Mollney, W. Wiechert, and H. Sahm. 2000. In vivo quantification of parallel and bidirectional fluxes in the anaplerosis of Corynebacterium glutamicum. J. Biol. Chem. 275:35932-35941. [DOI] [PubMed] [Google Scholar]
- 25.Pronk, J. T., H. Y. Steensma, and J. P. van Dijken. 1999. Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 16:1607-1633. [DOI] [PubMed] [Google Scholar]
- 26.Ratledge, C., and J. Wynn. 2000. Understanding microbial obesity. ASM News 50:181-185. [Google Scholar]
- 27.Sauer, U., V. Hatzimanikatis, J. E. Bailey, M. Hochuli, T. Szyperski, and K. Wuthrich. 1997. Metabolic fluxes in riboflavin-producing Bacillus subtilis. Nat. Biotechnol. 15:448-452. [DOI] [PubMed] [Google Scholar]
- 28.Szyperski, T. 1995. Biosynthetically directed fractional 13C-labeling of proteinogenic amino acids. An efficient analytical tool to investigate intermediary metabolism. Eur. J. Biochem. 232:433-448. [DOI] [PubMed] [Google Scholar]
- 29.Takada, Y., and T. Noguchi. 1985. Characteristics of alanine:glyoxylate aminotransferase from Saccharomyces cerevisiae, a regulatory enzyme in the glyoxylate pathway of glycine and serine biosynthesis from tricarboxylic acid-cycle intermediates. Biochem. J. 231:157-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van den Berg, M., and H. Y Steensma. 1995. ACS2, a Saccharomyces cerevisiae gene encoding acetyl-coenzyme A synthetase, essential for growth on glucose. Eur. J. Biochem. 231:704-713. [DOI] [PubMed] [Google Scholar]
- 31.van den Berg, M., P. de Jong-Gubbels, and H. Y. Steensma. 1998. Transient mRNA responses in chemostat cultures as a method of defining putative regulatory elements: application to genes involved in Saccharomyces cerevisiae acetyl-coenzyme A metabolism. Yeast 14:1089-1104. [DOI] [PubMed] [Google Scholar]
- 32.Verduyn, C., E. Postma, W. A. Scheffers, and J. P. van Dijken. 1992. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast 8:501-517. [DOI] [PubMed] [Google Scholar]
- 33.Verduyn, C., A. H. Stouthamer, W. A. Scheffers, and J. P. van Dijken. 1991. A theoretical evaluation of growth yields of yeasts. Antonie Leeuwenhoek 59:49-63 [DOI] [PubMed] [Google Scholar]
- 34.Verduyn, C. 1991. Physiology of yeasts in relation to biomass yields. Antonie Leeuwenhoek 60:325-353. [DOI] [PubMed] [Google Scholar]
- 35.Wiechert, W., and A. A. de Graaf. 1997. Bidirectional reaction steps in metabolic networks. 1. Modeling and simulation of carbon isotope labeling experiments. Biotechnol. Bioeng. 55:101-117. [DOI] [PubMed] [Google Scholar]
- 36.Xavier, K. B., M. S. da Costa, and H. Santos. 2000. Demonstration of a novel glycolytic pathway in the hyperthermophilic archaeon Thermococcus zilligii by 13C-labeling experiments and nuclear magnetic resonance analysis. J. Bacteriol. 182:4632-4636. [DOI] [PMC free article] [PubMed] [Google Scholar]