Abstract
We used microarrays carrying most of the genes that are developmentally regulated in Dictyostelium to discover those that are preferentially expressed in prestalk cells. Prestalk cells are localized at the front of slugs and play crucial roles in morphogenesis and slug migration. Using whole-mount in situ hybridization, we were able to verify 104 prestalk genes. Three of these were found to be expressed only in cells at the very front of slugs, the PstA cell type. Another 10 genes were found to be expressed in the small number of cells that form a central core at the anterior, the PstAB cell type. The rest of the prestalk-specific genes are expressed in PstO cells, which are found immediately posterior to PstA cells but anterior to 80% of the slug that consists of prespore cells. Half of these are also expressed in PstA cells. At later stages of development, the patterns of expression of a considerable number of these prestalk genes changes significantly, allowing us to further subdivide them. Some are expressed at much higher levels during culmination, while others are repressed. These results demonstrate the extremely dynamic nature of cell-type-specific expression in Dictyostelium and further define the changing physiology of the cell types. One of the signals that affect gene expression in PstO cells is the hexaphenone DIF-1. We found that expression of about half of the PstO-specific genes were affected in a mutant that is unable to synthesize DIF-1, while the rest appeared to be DIF independent. These results indicate that differentiation of some aspects of PstO cells can occur in the absence of DIF-1.
The morphological structures seen during multicellular development of Dictyostelium are deceptively simple. About 10 h after the initiation of development, a tip forms at the top of each aggregate of 10,000 to 100,000 cells and leads the slug as it migrates. During culmination, cells at the anterior make a stalk tube, enter it, and vacuolize to form a stalk on which the prespore cells climb to get off the substratum before they encapsulate. Prestalk cells are found localized to the anterior 20% of slugs over a wide range of sizes. However, the prestalk zone is not homogeneous. By using reporter genes controlled by the regulatory regions of the prestalk genes, ecmA and ecmB, Jermyn et al. (13) were able to establish a new anatomy by revealing the different patterns of gene expression in cells at the very front of the slug (PstA cells), cells in a central core at the anterior (PstAB cells), and cells immediately posterior to PstA cells but anterior to prespore cells (PstO cells). Some of these demonstrations required subdivision of the regulatory regions to recognize the specific cell types. In order to determine how many of these patterns are normally found during development, we carried out a genome-wide survey by using microarrays to collect a large number of putative prestalk genes and then used in situ hybridization to recognize the prestalk cell types which expressed these genes.
Sequencing of the 34-Mb Dictyostelium genome is well along, and chromosome 2 has been assembled and partially annotated (9). Extrapolating from the number of genes recognized on chromosome 2 has led to an estimate of about 12,000 genes in the complete genome. By sequencing a very large number of cDNAs, mostly generated from mRNA prepared during development, we have identified about 6,000 independent expressed sequence tags (18). These clones have been microarrayed and used to characterize both developmentally regulated genes and cell-type-specific genes (references 10 and 27 and this work).
Whole-mount in situ hybridization can be used to recognize individual cell types on the basis of the abundance of established cell-type-specific genes in one cell type or another (8). We have previously used this technique to characterize expression patterns of cytoskeletal and cyclic AMP signaling genes in prestalk cell subtypes (17, 26). Using a combination of genome-wide discovery and in situ verification, we have now identified 3 genes enriched in PstA cells, 30 genes enriched in PstO cells, 10 genes enriched in PstAB cells, and 38 genes enriched in both PstA and -O cells. We also identified 23 genes which are evenly expressed in whole slugs but become highly enriched in prestalk cells during culmination. Expression of many of these prestalk genes is dynamically regulated at the Mexican hat, culmination, and fruiting-body stages, demonstrating that transcriptional control of cell-type-specific genes is far from simple.
MATERIALS AND METHODS
Microarray analysis.
The microarray analyses were performed by using DNA chips with 6,450 targets prepared at the BioGEM facility as previously described (10). Most of these targets were expressed sequence tags obtained from the Dictyostelium cDNA project (18). RNA was prepared from slugs of strain Ax4 after separation of dissociated cells on Percoll density gradients (3, 10). Equal amounts of RNAs prepared from prestalk cells or prespore cells were used to generate Cy3- or Cy5-labeled probes. Three independent determinations were carried out, and the ratios of prestalk to prespore mRNAs were averaged.
Whole-mount in situ hybridization.
Whole-mount in situ hybridization was performed on D. discoideum Ax2 cells in the slug, Mexican hat, and culmination stages. The patterns of gene expression seen on microarrays are not significantly different for strains Ax2 and Ax4 (27). Our previous in situ studies were carried out with strain Ax2 (17, 26), and we have continued to use this strain. After fixation, samples from each stage were mixed and hybridized in a single tube as described previously (8, 17, 26). Digoxigenin-labeled riboprobes were generated for the entire length of each cDNA clone for hybridization to either sense or antisense RNA (17, 26). The sense RNA probes gave only weak and diffuse signals (data not shown). Sequences of all cDNAs used in this study can be drawn from the Dicty-cDNA database (http://www.csm.biol.tsukuba.ac.jp/cDNAproject.html).
RESULTS
Microarray analyses.
Genome-wide surveys for genes that are preferentially expressed in either prespore or prestalk cells have recently become possible as the result of the international effort to sequence the Dictyostelium genome (9, 15, 16) and the Japanese Dictyostelium cDNA Project (18). We used 5,655 cloned inserts from the Japanese cDNA project together with 690 targets previously amplified from developmental genes to generate microarrays (10). Since the majority of the cDNAs were isolated from developing cells, this collection is likely to represent most developmentally regulated genes. After hybridization with Cy3- and Cy5-labeled probes, the ratios of prestalk to prespore mRNAs were determined. The results for each target can be found at http://www.biology.ucsd.edu/loomis-cgi/microarray/index.html.
All but one of the 25 genes found by microarray analyses to be enriched fivefold or more in prestalk cells at the slug stage were confirmed by in situ hybridization (Table 1). The microarray analyses indicated that another 80 genes were enriched at least 2.5-fold in prestalk cells. We were able to confirm 68 of these by in situ hybridization (Table 1). These results establish the reliability of microarray analyses.
TABLE 1.
Prestalk genes
| Accession no. | cDNA | Producta | Fold enrichment (Pst/Psp) |
|---|---|---|---|
| PstA genes | |||
| AU062194 | SLI271 | BCS1-like (3e-14) | 4.32 |
| AU061665 | SLF308 | Extracellular matrix protein (4e-05) | 3.02 |
| C94343 | SSK861 | Dictyostelium O-methyltransferase (4e-43) | 2.79 |
| PstAB genes | |||
| SLA128 pattern | |||
| AU060076 | SLA128 | Arabinofuranohydrolase (0.15) | 7 |
| AU060722 | SLB233 | Homologous to arabidopsis hypothetical protein (3e-09) | 5.99 |
| AU038850 | SSL558 | Dictyostelium cellulase (6e-09) | 4.11 |
| SLC388 pattern | |||
| AU060958 | SLC388 | Dictyostelium steroid isomerase | 4.29 |
| AU061846 | SLG322 | Carboxypeptidase (3e-16) | 3.26 |
| SSK348 pattern | |||
| AU062236 | SLI604 | Aconitate hydratase (4e-78) | 2.83 |
| AU074317 | SSK348 | Propionate-coenzyme A ligase (8e-30) | 2.22 |
| SSB312 pattern | |||
| AU060501 | SLK182 | ECMB homolog | 9.2 |
| AU071364 | SSB312 | Dictyostelium hypothetical protein AAL99319.1 (6e-11) (=SSC596) | 4.95 |
| C91223 | SSJ839 | Unknown | 4.35 |
| PstO genes | |||
| SSM184 pattern | |||
| AU038878 | SSL587 | Calcium binding protein 4A | 6.29 |
| AU072229 | SSD764 | Splicing factor 3A (0.007) | 3.58 |
| AU071425 | SSB476 | Unknown | 3.52 |
| AU037107 | SSB457 | Dictyostelium thioredoxin 1 | 3.26 |
| AU038948 | SSM184 | Unknown | 2.75 |
| AU073765 | SSI311 | Glycosylphosphatidylinositol phospholipase D1 | 2.77 |
| SLG775 pattern | |||
| C89959 | SSG694 | Dictyostelium thioredoxin 3 | 7.42 |
| AU074665 | SSL481 | Dictyostelium hypothetical protein AAL92 | 5.9 |
| AU060648 | SLK756 | Dictyostelium prestalk D11 protein | 4.86 |
| C91531 | SSK395 | Adducin 1 (alpha) (1e-35) | 4.29 |
| AU073534 | SSH476 | Dictyostelium hypothetical protein AAL92191.1 | 2.84 |
| AU061959 | SLG775 | Dictyostelium cystein-proteinase CPRB | 2.82 |
| SLH659 pattern | |||
| AU060263 | SLA769 | Dictyostelium hypothetical protein AAL92643.1 (GP64 homolog) | 10.9 |
| AU061762 | SLF774 | Dictyostelium β-glucosidase | 7.29 |
| AU037168 | SSB559 | Unknown | 7.22 |
| AU073811 | SSI424 | Unknown | 6.56 |
| AU038606 | SSL238 | Unknown | 6.28 |
| AU073596 | SSH630 | Dictyostelium hypothetical protein AAL93052.1 | 5.54 |
| AU062115 | SLH659 | δ-1-Pyrroline-5-carboxylate dehydrogenase (2e-66) | 5.36 |
| C93985 | SSC889 | Dictyostelium thioredoxin 2 | 3.92 |
| AU073683 | SSH823 | Unknown | 3.8 |
| AU074824 | SSM175 | Dictyostelium myosin light-chain kinase (MLCKA) (=SSJ249) | 3.32 |
| C91391 | SSK159 | Dictyostelium hypothetical protein AAL92274.1 (1e-15) (=SSB159) | 3.24 |
| AU072378 | SSE356 | Dictyostelium lysozyme | 2.96 |
| AU074364 | SSK495 | Potassium efflux system KEFA (8.7) | 2.78 |
| AU074982 | SSM642 | Retinal pigment epithelial protein (0.005) | 2.63 |
| AU071713 | SSC340 | Unknown | 2.39 |
| AU075086 | SSA210 | Unknown | 2.23 |
| AU073353 | SSG754 | Dictyostelium calcium/binding protein 1 | 2.17 |
| C90552 | SSI714 | Dictyostelium phosphatidylinositol transfer protein 2 | 2.01 |
| PstAO genes | |||
| SSH475 pattern | |||
| C92972 | SSF295 | Unknown | 9.6 |
| C84222 | SSB337 | Unknown | 9.2 |
| AU062233 | SLI566 | Homologous to yeast ACR1/protein homolog (7e-19) | 7.4/PICK> |
| C90923 | SSJ314 | Unknown | 5.8 |
| C91268 | SSJ728 | Unknown (=SSF823) | 5.49 |
| AU038875 | SSL584 | Unknown | 5.04 |
| AU074602 | SSL349 | Unknown | 4.76 |
| AU073356 | SSG767 | Unknown | 4.46 |
| C90189 | SSI228 | Extracellular endoglucanase (0.001) | 4.26 |
| AU074795 | SSL886 | Unknown | 4.1 |
| C92856 | SSF509 | Unknown | 4.01 |
| AU074211 | SSJ814 | Unknown (=SSM194) | 3.99 |
| AU073533 | SSH475 | Unknown | 3.23 |
| C92333 | SSD873 | Acetoin dehydrogenase (8e-14) | 3.2 |
| AU038842 | SSL550 | Unknown | 3.05 |
| AU062252 | SLI716 | Unknown | 2.89 |
| AU073109 | SSG159 | Unknown | 2.75 |
| AU060133 | SLA387 | β-1,4-endoglucanase (7e-19) (=SFE338) | 2.66 |
| C89985 | SSG805 | Unknown | 2.57 |
| AU061483 | SLE410 | Dictyostelium RASD | 2.49 |
| AU074481 | SSK868 | Dictyostelium extracellular matrix protein homolog (2e-20) | 2.37 |
| C91092 | SSJ538 | Dictyostelium P17 cyclic AMP-regulated protein | 2.3 |
| AU061713 | SLF535 | Dictyostelium kielin | 2.25 |
| AU071694 | SSC290 | Dictyostelium ECMB homolog (5e-26) | 2.22 |
| AU038846 | SSL554 | Cerebroside sulfate activator (4e-15) (=SSL531) | 2.2 |
| AU073407 | SSH123 | Dictyostelium hypothetical protein AAL92997.1 | 2.17 |
| SLE474 pattern | |||
| AU037288 | SSB721 | Dictyostelium hypothetical protein AAL92651.1 | 4.14 |
| AU035004 | SLE474 | Dictyostelium prestalk ECMA protein | 3.79 |
| AU060824 | SLB609 | Acid phosphatase (9e-17) (=VFB675) | 3.17 |
| C94063 | SSG357 | Dictyostelium CAR1; cyclic AMP Receptor 1 | 3.16 |
| AU062087 | SLH511 | Dictyostelium DOCA | 3.04 |
| AU071989 | SSD123 | Dictyostelium OSBPa | 2.7 |
| SSA854 pattern | |||
| AU072779 | SSA854 | Unknown | 5.85 |
| AU039124 | SSM416 | Unknown (=SSL853) | 5.08 |
| AU074011 | SSJ246 | Unknown | 3.88 |
| AU071496 | SSB661 | Unknown | 2.64 |
| AU060119 | SLA364 | Dictyostelium ACTIN5 | 2.41 |
| C90964 | SSJ359 | Unknown | 2.22 |
| Late prestalk genes | |||
| SSE634 pattern | |||
| C84694 | SSE634 | Phosphatidylglycerol/phosphatidylinositol transferase (1e-13) | 5 |
| AU074273 | SSK196 | Homolog of Neurospora glutamine dehydrogenase (2e-81) | 4.84 |
| AU060554 | SLK384 | Unknown | 3.8 |
| AU062051 | SLH367 | Glutamine synthase (4e-33) | 3.63 |
| AU071922 | SSC746 | Dictyostelium extracellular matrix protein | 3.6 |
| C93190 | SSM756 | Unknown | 2.92 |
| AU072597 | SSA348 | Dictyostelium hypothetical protein AAB06761.1 (2e-08) | 2.52 |
| AU060917 | SLC233 | Dictyostelium phosphoenolpyruvate carboxykinase | 2.46 |
| C89827 | SSG207 | Caffeic acid O-methyltransferase (7e-14) | 2.26 |
| C89911 | SSG614 | Dictyostelium δ5 fatty acid desaturase | 2.17 |
| AU034900 | SLE660 | Dictyostelium isocitrate lyase | 2 |
| AU071371 | SSB338 | Unknown | 4.84 |
| AU062192 | SLI252 | Dictyostelium DP87 precursor (4e-04) | 4.03 |
| AU052407 | SLD267 | Putative allantoinase precursor (5e-29) | 2.7 |
| SSG721 pattern | |||
| AU061693 | SLF442 | Dictyostelium malate synthase | 3.75 |
| C92120 | SSD115 | Homologous to Plasmodium falciparum hypothetical protein PFC0486 homolog (2e-10) | 3.13 |
| AU073340 | SSG721 | Oxalate/formate antiporter (3e-14) | 3.13 |
| AU053631 | SLJ273 | Dictyostelium cytosolic aldolase | 3.13 |
| AU037927 | SSE462 | Haloacid dehalogenase-like hydrolase (7e-14) | 2.64 |
| AU062141 | SLH752 | Dictyostelium hypothetical protein (4e-19) | 2.54 |
| AU062057 | SLH405 | Putative polyketide synthase module protein (1e-49) | 2.51 |
| AU072355 | SSE273 | Acyl coenzyme A oxidase (4e-08) | 2.47 |
| AU062008 | SLH173 | Dictyostelium Na/K-ATPase alpha4 (IonA) | 2.32 |
cDNA sequences were assigned to open reading frames found in the Dictyostelium genomic databases. Their protein products were compared to GenBank entries, and the most significant match is given, with the associated E value in parentheses. No E value is given for identity to established Dictyostelium proteins.
We also found 23 genes which were only marginally prestalk enriched at the slug stage and that gave clear indication of prestalk specificity by in situ hybridization of culminants (Table 1). Using a combination of genome-wide expression analyses and in situ hybridization, we have identified a total of 104 prestalk genes, most of which had not previously been recognized to be expressed in a cell-type-specific manner. Since in situ hybridization can distinguish the prestalk subtypes that accumulate specific mRNAs, the prestalk genes could be put into separate categories.
Expression patterns in migrating slugs.
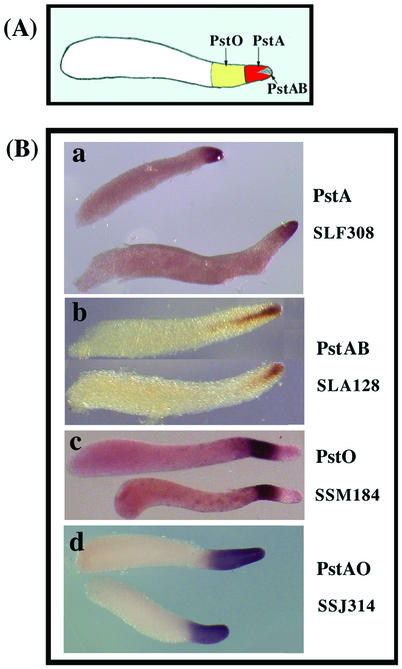
Classical fate mapping experiments have shown that cells at the anterior of slugs differentiate into stalk cells, while those at the posterior give rise to the spores (21). The first cells to enter the stalk tube at culmination are found in a central cone at the anterior of slugs and are marked by the expression of the prestalk gene ecmB (13). These few hundred cells are referred to as PstAB. Another marker of prestalk cells, ecmA, is expressed in all prestalk cells as the result of two independent regulatory modules, one of which is responsible for transcription in the most anterior cells, the PstA cells, and the other of which is responsible for transcription in the cells that lie immediately behind, the PstO cells (Fig. 1A). At culmination PstA cells enter the stalk directly, while PstO cells form upper and lower cups around the rising mass of prespore cells before some of them enter the stalk. Genes that are expressed in both PstA and PstO cells at the slug stage are referred to as PstAO.
FIG. 1.
Prestalk cell types. (A) Anatomy of a slug. PstA cells are at the very front, and PstO cells are immediately behind them. PstAB cells form a central core at the anterior. Prespore cells are found in the posterior. (B) Representative in situ hybridization patterns: mRNA recognized by cDNA clone SLF308 is found only in PstA cells, mRNA recognized by clone SLA128 is found only in PstAB cells, mRNA recognized by cDNA SSM184 is found only in PstO cells, and mRNA recognized by clone SSJ314 is found in both PstA and PstO cells.
Genes that gave at least a twofold-higher prestalk signal on the microarrays were analyzed by in situ hybridization. By examining the staining patterns of slugs, we identified 3 genes expressed only in PstA cells, 10 genes expressed exclusively in PstAB cells, 30 genes preferentially expressed in PstO cells, and another 38 genes expressed in all prestalk cells (Fig. 1B; Table1). At later stages of development, the patterns of expression of some of these genes showed dynamic changes that allowed us to further subdivide them into classes.
Expression of PstA genes during late development.
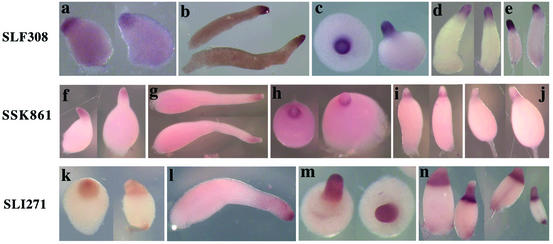
Three genes were found to be expressed only in the most anterior cells of slugs (Fig. 2). This pattern of cell type specificity is identical to the artificially generated pattern seen when reporter genes are put under the control of the PstA regulatory module of ecmA and confirms the unique properties of PstA cells (7). SLF308 was expressed at high levels in the most anterior cells throughout development (Fig. 2a to e). The level of SLK861 mRNA was low but detectable at the top of the tipped aggregates (Fig. 2f to j). It decreased in PstA cells as culmination proceeded but was still detectable at the late culmination stage. SLI271 mRNA was expressed in tipped aggregates and the most anterior slug cells but became abundant in all prestalk cells during culmination (Fig. 2k to n).
FIG. 2.
Developmental expression patterns of PstA genes. Spatial expression patterns of the three PstA genes that we identified in this study were analyzed by in situ hybridization. Expression patterns at the slug (b, g, and l), Mexican hat (c, h, and m), early culmination (d, i, and n), and late culmination (e, j, and n) stages are shown. SLF308 was expressed at high levels in the most anterior cells throughout development (a to e); expression of SSK861 decreased in PstA cells as culmination proceeded but transiently appeared in cells as they entered the stalk tube during early culmination (f to j). SLI271 recognized mRNA that became abundant in all prestalk cells during culmination (k to n).
It is of interest that SLK861 encodes a protein that is 75% identical to DIF-methyltransferase encoded by dmtA. This enzyme is responsible for the essential last step in biosynthesis of DIF-1, the hexaphenone intercellular signal that induces certain genes in PstO cells (25). dmtA is expressed exclusively in prespore cells, while SLK861 is a PstA gene. This raises the possibility that DIF-like signals affecting PstO genes emanate from multiple cell types.
PstAB genes.
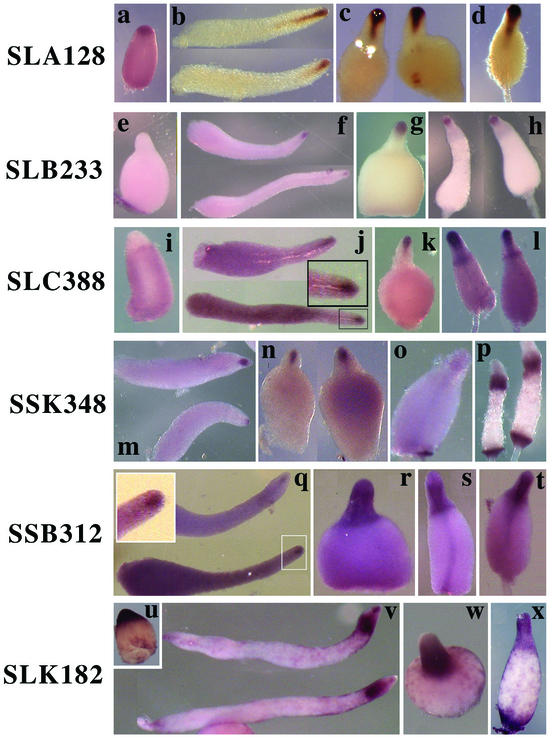
Three PstAB genes (SLA128, SLB233, and SSL558) showed essentially the same pattern of expression after the slug stage, as exemplified by SLA128 (Fig. 3a to d; Table 1) and SLB233 (Fig. 3e to h; Table 1). All of them are expressed in the central funnel of cells of migrating slugs (Fig. 3b and f) and at the top of rising culminants (Fig. 3d and h). However, there was a remarkable difference in expression pattern in the tipped aggregates. Both SLA128 (Fig. 3a) and SSL558 (data not shown) were expressed in the upper region, while SLB233 was not expressed until the slug stage (Fig. 3e and f). SLC388 (Fig. 3i to l) and SLG322 (data not shown) were both expressed at low levels in prespore cells at the tipped aggregate stage, expressed in the anterior funnel cells in slugs, and then became enriched in upper cup cells during culmination (Fig. 3l), the same pattern seen with ecmB (8). SSK348 (Fig. 3m to p) and SLI604 (data not shown) were expressed at the tipped aggregate stage, expressed in the anterior funnel cells in slugs, and then became abundant in both upper and lower cup cells during late culmination. The pattern of expression of three genes, exemplified by SSB312, spread after the slug stage from PstAB cells to all prestalk cells in culminants (Fig. 3q to t). SLK182 (Fig. 3u to x) showed this pattern but was also expressed in PstO cells in slugs. It was also expressed in cells scattered throughout the prespore region that might be anterior-like cells.
FIG. 3.
Expression of representative PstAB genes. In situ hybridization analysis of 10 PstAB genes identified in this study revealed that these genes are regulated in four distinct patterns, exemplified by SLA128 (a to d), SLB233 (e to h), SLC388 (i to l), SSK348 (m to p), SSB312 (q to t), or SLK182 (u to x). (a, e, i, and u) Tipped aggregate stage; (b, f, j, m, q, and v) slug stage; (c, g, k, n, r, and w) Mexican hat stage; (o, s, and x) early culmination stage; (d, h, l, p, and t) late culmination stage. The insets in panels j and q show a higher magnification of the anterior portion of a slug in a rectangle in each panel.
Genes homologous to ecmB and to the genes for steroid isomerase, propionate coenzyme A ligase, and aconitate hydratase are the members of the PstAB subtype (Table 1). One of the SLA128 class of genes, SSL558, encodes a product that is related to the Dictyostelium cellulase (2) and may be involved in deposition of the cellulose stalk tube.
PstO genes.
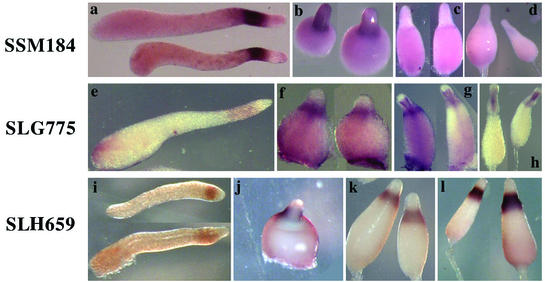
We found 30 genes that are expressed in PstO cells but not in PstA cells (Table 1). Most of these PstO genes were dominantly expressed in the tip region of tipped aggregates, while SSE634, SSC340, SLH659, SSA348, SSE356, and SSH476 did not show cell-type-specific expression (data not shown). A group of six genes, exemplified by SSM184, continue to be expressed in PstO cells during culmination but at reduced levels (Fig. 4a to d). A group of six genes, exemplified by SLG775, become more strongly expressed in PstO cells during culmination and are also expressed in cells at the top of the stalk tube (Fig. 4e to h). The remainder of the PstO genes continues to be strongly expressed in PstO cells throughout development, as exemplified by SLH659 (Fig. 4i to l).
FIG. 4.
Expression of representative PstO genes. We found 30 genes that are expressed in PstO cells but not in PstA cells. These genes can be grouped into three classes according to their expression patterns during culmination. SSM184 is a representative of six genes that are continuously expressed in PstO cells but at progressively reduced levels (a to d). SLG775 is a representative of six genes that become more strongly expressed in PstO cells during culmination and are also expressed in cells at the top of the stalk tube (e to h). SLH659 is a representative of the remainder of the PstO genes that continue to be strongly expressed in PstO cells throughout development (i to l). (a, e, and i) Slug stage; (b, f, and j) Mexican hat stage; (c, g, and k) early culmination stage; (d, h, and l) late culmination stage.
Previously characterized Dictyostelium genes that are expressed exclusively in PstO cells include cprB (cysteine proteinase) (20), cbpA (5) and cbpD (6) (calcium binding proteins 1 and 4), mlckA (myosin light-chain kinase 1) (17, 24), ampA (D11) (1), gluA (β-glucosidase) (4), and genes encoding phosphatidylinositol transfer protein 2 (23) and thioredoxins 1, 2, and 3 (28).
Genes expressed in all prestalk cells.
Most of the genes that are expressed in both PstA and PstO cells in the slug stage become preferentially restricted to PstO cells during culmination, as exemplified by SSH475 (Fig. 5a to f). There were 26 such genes in our collection (Table 1). Most of these genes encode novel proteins with no significant homology to known proteins, but this group also includes genes coding for RasD (12) and kielin (9), a dorsalizing factor in Xenopus.
FIG. 5.
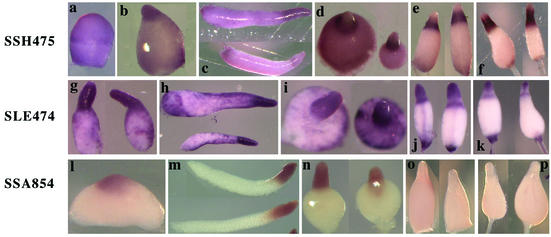
Genes expressed in PstA and PstO cells. We identified 38 PstAO genes that are expressed in both PstA and PstO cells in the slug stage. There were 26 genes that became preferentially restricted to PstO cells during culmination, as exemplified by SSH475 (a to f). SLE474 is a representative of six PstAO genes whose expression was maintained in all prestalk cells by early culmination (g to k). Their mRNAs could be seen in the vacuolized cells within the stalk tube (j). SSA854 is a representative of six PstAO genes that were expressed in both PstA and PstO cells up through early culmination and were subsequently repressed (l to p). (a) Mound stage; (b, g, and l) tipped aggregate stage; (c, h, and m) slug stage; (d, i, and n) Mexican hat stage; (e, j, and o) early culmination stage; (f, k, and p) late culmination stage.
A group of six genes that were expressed in all prestalk cells at the slug stage maintained this pattern through early culmination, as exemplified by SLE474 (Fig. 5g to k). Their mRNA could still be seen in the vacuolized cells within the stalk tube. This group includes the previously characterized Dictyostelium genes ecmA (8, 11, 13), docA (GenBank accession number AF020409), and carA (14, 26) (Table 1). On the other hand, six genes expressed in both PstA and PstO cells up through early culmination subsequently became repressed, as exemplified by SSA854 (Fig. 5l to p). All of these genes were also expressed in the tip region of a tipped aggregate, as seen in Fig. 5b, g, and l.
Genes expressed in prestalk cells only during culmination.
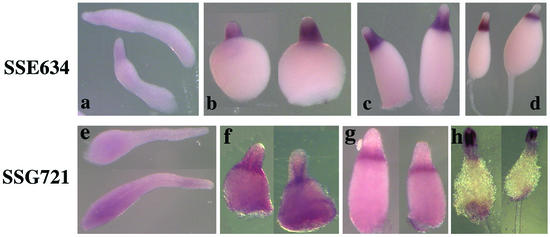
A considerable number of genes recognized on the microarrays to be slightly enriched in prestalk cells gave very little in situ signal at the slug stage. However, 14 of these genes were subsequently expressed at high levels in PstO cells, as exemplified by SSE634 (Fig. 6a to d). Another nine genes that failed to show cell type specificity at the slug stage subsequently accumulated to high levels in prestalk cells and could be seen at or around the top of the stalk tube, as exemplified by SSG721 (Fig. 6e to h).
FIG. 6.
Genes expressed in prestalk cells only during culmination. We found 23 genes which gave very little in situ signal at the slug stage. SSE634 is a representative of 14 of these genes that were subsequently expressed at high levels in PstO cells (a to d). Another nine genes, exemplified by SSG721, failed to show cell type specificity at the slug stage but were expressed in cells within the top of the stalk tube during culmination. Their mRNAs could be seen in vacuolized cells near the top of the stalk tube (e to h). (a and e) Slug stage; (b and f) Mexican hat stage; (c and g) early culmination stage; (d and h) late culmination stage.
Dictyostelium genes of interest that are expressed in prestalk cells late in development include those encoding isocitrate lyase (9) and malate synthase (9), which are involved in the malate shunt for gluconeogenesis (Table 1). Genes encoding aldolase (9), glutamate dehydrogenase (9), and phosphoenolpyruvate carboxykinase (9) are also members of this category. It appears that metabolic pathways may be significantly modified in prestalk cells late in development.
Temporal patterns of prestalk gene expression.
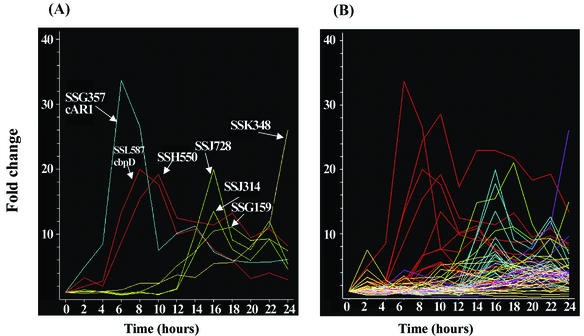
RNAs prepared from samples taken at 2-h intervals throughout filter development of strain Ax4 was compared to time-averaged RNAs following hybridization to the microarrays as previously described (10). The temporal patterns of accumulation of representative prestalk specific mRNAs are given in Fig. 7. Some of the prestalk mRNAs start to accumulate within the first 4 h of development, peak around 8 h of development, and decrease thereafter. Some of these genes may be expressed in all cells during aggregation and then become preferentially repressed in prespore cells, as has been previously found to be the case for ampA (D11) (1) and cprB and rasD (12). Other prestalk genes are expressed only after 10 h of development, and their mRNAs are found only in prestalk cells. Time course values for the each of the prestalk genes are available at http://www.biology.ucsd.edu/loomis-cgi/microarray/paperHL.html.
FIG. 7.
Expression patterns of representative prestalk genes. RNAs prepared at 2-h intervals throughout development were analyzed on microarrays. The fold increase relative to time-averaged RNAs prepared by pooling samples from different stages was normalized at the time of initiation of development. (A) Representative prestalk genes are indicated by their cDNA number (Table 1). (B) Developmental expression of 65 prestalk genes, including those marked in panel A. Five clusters were generated by K-means in the Genespring program and color coded. Values are the averages from four independent determinations.
DIF-dependent and DIF-independent PstO genes.
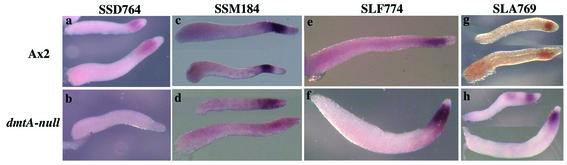
It has previously been shown that expression of reporter genes fused to the portion of the ecmA regulatory region that results in expression in PstO cells is dependent on DIF production (25). Cells carrying a null mutation in dmtA are unable to catalyze the last step in biosynthesis of DIF-1 and fail to express the PstO reporter unless DIF-1 is added exogenously. When we analyzed dmtA− cells throughout development by in situ hybridization with PstO genes, we found that 18 out of 30 PstO genes were expressed at a significantly lower level in the strain lacking DIF, as exemplified by SSD764 (Fig. 8a and b; Table 2). With some of these genes residual staining was seen to localize to PstO cells, as exemplified by SSM184 (Fig. 8c and d). On the other hand, expression of 12 PstO genes was not affected by the lack of DIF-1 in the dmtA− cells, as exemplified by SLF774 (gluA) and SLA769 (gp64 homolog) (Fig. 8e to h). These mutant cells lacking DIF clearly generate a PstO region where DIF-independent genes are expressed.
FIG. 8.
Expression of PstO genes in Ax2 and the dmtA− mutant lacking DIF. Thirty PstO genes were analyzed by in situ hybridization in both Ax2 and the dmtA− mutant lacking DIF at the slug stage. Among those, 18 genes were down-regulated in the mutant, as exemplified by SSD764 (a and b) and SSM184 (c and d). Although residual expression was detectable in SSM184, SSD764 was totally extinguished. In contrast to these genes, 12 genes were not affected, as exemplified by SLF774 (e and f) and SLA769 (g and h). (a, c, e, and g) Ax2; (b, d, f, and h) dmtA− mutant.
TABLE 2.
DIF-dependent and DIF-independent PstO genes
| Gene | Locus and/or product |
|---|---|
| Genes with significantly reduced expression in dmtA− null cells | |
| SLK756 | ampA (D11) |
| SSG754 | cbpA (calcium binding protein) |
| SLG775 | cprB (cystein proteinase) |
| SSM175 | mlckA (myosin light-chain kinase) |
| SSB457 | Thioredoxin 1 |
| SSC889 | Thioredoxin 2 |
| SSG694 | Thioredoxin 3 |
| SSI311 | Glycosylphosphatidylinositol phospholipase D1 |
| SLH659 | δ-1-Pyrroline-5-carboxylate dehydrogenase |
| SSD764 | Unknown |
| SSB476 | Unknown |
| SSM184 | Unknown |
| SSH476 | Unknown |
| SSI424 | Unknown |
| SSL238 | Unknown |
| SSH823 | Unknown |
| SSK495 | Unknown |
| SSA210 | Unknown |
| Genes expressed normally in dmtA− null cells | |
| SSL587 | cbpD (calcium binding protein) |
| SLF774 | gluA (β-glucosidase) |
| SSK395 | α-Adducin |
| SLA769 | GP64 homolog |
| SSE356 | Lysozyme |
| SSI714 | Phosphatidylinositol transferase 2 |
| SSL481 | Unknown |
| SSB559 | Unknown |
| SSH630 | Unknown |
| SSK159 | Unknown |
| SSM642 | Unknown |
| SSC340 | Unknown |
DISCUSSION
Genome-wide determination of expression patterns in isolated cell types by using microarrays can recognize most prestalk- and prespore-specific genes that are expressed at high enough levels to give robust signals. Since PstA and PstAB cells make up fewer than 10% of the total cells in a slug, some genes expressed exclusively in these subtypes may not have been recognized if they generate only low-abundance mRNAs at the slug stage. Nevertheless, from 160 genes chosen on the basis that they gave at least twice as strong a signal with probes generated from pooled prestalk cells than with probes generated from prespore cells, 3 were found by in situ hybridization to be expressed exclusively in PstA cells and 10 were found to be exclusively expressed in PstAB cells at the slug stage (Fig. 2 and 3). These results demonstrate that our microarray screen is effective in discovering genes expressed in only a minority of cells. However, microarray data alone are not sufficient to determine the subtypes of cells in which prestalk genes are expressed, nor can they recognize the changing patterns that occur during later development. In situ hybridization at multiple stages of development with the candidate prestalk genes has shown just how dynamic these patterns can be.
Although dozens of developmentally regulated Dictyostelium genes have been analyzed over the last 20 years, only a small number have been found to be expressed exclusively in one or another prestalk cell type. ecmB is an excellent marker of PstAB cells at the slug stage (11). It encodes a large extracellular matrix protein previously shown to be synthesized exclusively by prestalk cells (19). However, no other PstAB-specific gene has been encountered until now. Our combinatorial approach using two different techniques, genome-wide microarray analyses and in situ hybridization, has uncovered 10 PstAB genes that can be used as molecular markers, as well as indicating what unique physiological processes these cells may carry out.
The best marker for PstA cells has been an artificial construct in which one of the elements regulating ecmA is used to drive a reporter gene such as β-galactosidase (7). Our combinatorial approach found three genes that are naturally expressed only in PstA cells at the slug stage. Some of these are subsequently expressed in PstO cells.
The original marker for PstO cells was also an artificial construct in which another ecmA regulatory element is used to drive β-galactosidase (7). Other developmentally regulated genes, such as tipA (22), were preferentially expressed in PstO cells at the slug stage, but the list is short. We found 30 genes that are expressed only in PstO cells at the slug stage, and most retained this pattern throughout culmination.
ecmA itself is expressed in all prestalk cells as the result of the combined function of the regulatory elements (13). We found 38 genes expressed in all prestalk cells at the slug stage. Expression of most of these genes was repressed in PstA cells during culmination but continued at high levels in PstO cells. However, expression of six PstAO genes continued in both cell types throughout culmination. A few others were repressed in both prestalk cell types during culmination.
We discovered 23 genes that are strongly expressed in PstO cells but only following the initiation of culmination. Nine of these were also expressed in PstA cells during culmination. Many of these genes encode enzymes involved in gluconeogenesis from the products of protein catabolism, consistent with the demand for subunits from which to construct cell wall cellulose.
It is striking that 68 of the newly discovered prestalk genes are expressed in PstO cells in the slug stage. Two-thirds of these are also expressed in other prestalk cell types at later stages of development. Although their roles in slugs and culminants are unknown, it is noteworthy that such dynamic spatiotemporal changes in cells expressing these genes occur during development. We previously found that many genes related to myosin function dynamically changed their expression pattern from the PstAO cells at the slug stage to the upper cup cells during culmination (17).
These combined genome-wide expression studies coupled to in situ analyses have provided an expanded set of genes that can be used to characterize cell types. Comparison of the expression patterns of PstO genes in wild-type and dmtA− mutant cells further subdivided them into DIF-dependent and DIF-independent sets. Moreover, recognizing that dmtA− slugs have a PstO region even in the absence of DIF-1 helps explain how they are able to culminate and form fruiting bodies.
Acknowledgments
We thank Chris Thompson and Rob Kay for providing the dmtA− mutant strain and for constructive discussions. We also thank M. Tomisako, K. Nishio, and M. Yokoyama for their technical assistance with in situ hybridization.
Annotation of the microarrayed cDNAs benefited from the whole genome sequences generated by the Dictyostelium Sequencing Consortium: The Baylor Sequencing Center, Houston, Tex. (A. Kuspa and R. Gibbs), where sequencing is supported by the NIH, and the Institute of Biochemistry, Cologne, Germany, together with the Institute of Molecular Biotechnology, Jena, Germany (G. Glöckner, A. Rosenthal, L. Eichinger, and A. Noegel), where sequencing is supported by the Deutsche Forschungsgemeinschaft (no. 113/10-1 and 10-2). Sequencing in the United Kingdom was supported by the EUDICT consortium and by an MRC program grant to J. G. Williams, B. Barrel, R. R. Kay, and P. H. Dear. This work was supported by grants to W.F.L. from the NIH (GM60447 and GM62350) and the NSF Biocomplexity Program. It was also supported by grants from the Future of the Japan Society for the Promotion of Science to Y. Tanaka (JSPS-RFTF96L00105) and S. Kuhara, Kyushu University (JSPS-RFTF00L01412), and by Grants-in-Aid for Scientific Research on Priority Areas from the Ministry of Education Science, Sports, and Culture of Japan to M. Maeda (08283105) and Y. Tanaka (12206001).
REFERENCES
- 1.Barklis, E., B. Pontius, and H. F. Lodish. 1985. Structure of the Dictyostelium discoideum prestalk D11 gene and protein Mol. Cell. Biol. 5:1473-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blume, J. E., and H. L. Ennis. 1991. A Dictyostelium discoideum cellulase is a member of a spore germination-specific gene family. J. Biol. Chem. 266:15432-15437. [PubMed] [Google Scholar]
- 3.Borth, W., and D. Ratner. 1983. Different synthetic profiles and developmental fates of prespore versus prestalk proteins of Dictyostelium. Differentiation 24:213-219. [DOI] [PubMed] [Google Scholar]
- 4.Bush, J., J. Richardson, and J. Cardelli. 1994. Molecular cloning and characterization of the full-length cDNA encoding the developmentally regulated lysosomal enzyme beta-glucosidase in Dictyostelium discoideum. J. Biol. Chem. 269:1468-1476. [PubMed] [Google Scholar]
- 5.Coukell, B., J. Moniakis, and A. Grinberg. 1995. Cloning and expression in Escherichia coli of a cDNA encoding a developmentally regulated Ca(2+)-binding protein from Dictyostelium discoideum. FEBS Lett. 362:342-346. [DOI] [PubMed] [Google Scholar]
- 6.Dorywalska, M., B. Coukell, and A. Dharamsi. 2000. Characterization and heterologous expression of cDNAs encoding two novel closely related Ca(2+)-binding proteins in Dictyostelium discoideum. Biochim. Biophys. Acta 1496:356-361. [DOI] [PubMed] [Google Scholar]
- 7.Early, A. E., M. J. Gaskell, D. Traynor, and J. G. Williams. 1993. Two distinct populations of prestalk cells within the tip of the migratory Dictyostelium slug with differing fates at culmination. Development 118:353-362. [DOI] [PubMed] [Google Scholar]
- 8.Escalante, R., and W. F. Loomis. 1995. Whole-mount in situ hybridization of cell-type specific mRNAs in Dictyostelium. Dev. Biol. 171:262-266. [DOI] [PubMed] [Google Scholar]
- 9.Gloeckner, G., L. Eichinger, K. Szafranski, J. A. Pachebat, A. T. Bankier, P. H. Dear, R. Lehmann, C. Baumgart, G. Parra, J. F. Abril, R. Guigo, K. Kumpf, B. Tunggal, E. Cox, M. A. Quail, M. Platzer, A. Rosenthal, and A. A. Noegel. 2002. Sequence and analysis of chromosome 2 of Dictyostelium discoideum. Nature 418:79-85. [DOI] [PubMed] [Google Scholar]
- 10.Iranfar, N., D. Fuller, R. Sasik, T. Hwa, M. Laub, and W. F. Loomis. 2001. Expression patterns of cell-type-specific genes in Dictyostelium. Mol. Biol. Cell 12:2590-2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jermyn, K. A., and J. G. Williams. 1991. An analysis of culmination in Dictyostelium using prestalk and stalk-specific cell autonomous markers. Development. 111:779-787. [DOI] [PubMed] [Google Scholar]
- 12.Jermyn, K., and J. Williams. 1995. Comparison of the Dictyostelium rasD and ecmA genes reveals two distinct mechanisms whereby an mRNA may become enriched in prestalk cells. Differentiation 58:261-267. [DOI] [PubMed] [Google Scholar]
- 13.Jermyn, K. A., K. Duffy, and J. G. Williams. 1989. A new anatomy of the prestalk zone in Dictyostelium. Nature 340:144-146. [DOI] [PubMed] [Google Scholar]
- 14.Klein, P. S., T. J. Sun, C. L. Saxe III, A. R. Kimmel, R. L. Johnson, and P. N. Devreotes. 1988. A chemoattractant receptor controls development in Dictyostelium discoideum. Science 241:1467-1472. [DOI] [PubMed] [Google Scholar]
- 15.Kuspa, A., R. Sucgang, and G. Shaulsky. 2001. The promise of a protist: the Dictyostelium genome project. Funct. Integr. Genomics 1:279-293. [DOI] [PubMed] [Google Scholar]
- 16.Loomis, W. F., and A. Kuspa. 1997. The genome of Dictyostelium discoideum, p. 15-30. In Y. Maeda, K. Inouye, and I. Takeuchi (ed.), Dictyostelium—a model system for cell and developmental biology. Universal Academy Press, Tokyo, Japan.
- 17.Maeda, M., H. Kuwayama, M. Yokoyama, K. Nishio, T. Morio, H. Urushihara, M. Katoh, Y. Tanaka, T. Saito, H. Ochiai, K. Takemoto, H. Yasukawa, and I. Takeuchi. 2000. Developmental changes in spatial expression of genes involved in myosin function in Dictyostelium. Dev. Biol. 223:114-119. [DOI] [PubMed] [Google Scholar]
- 18.Morio, T., H. Urushihara, T. Saito, Y. Ugawa, H. Mizuno, M. Yoshida, R. Yoshino, B. N. Mitra, M. Pi, T. Sato, K. Takemoto, H. Yasukawa, J. Williams, M. Maeda, I. Takeuchi, H. Ochiai, and Y. Tanaka. 1998. The Dictyostelium developmental cDNA project: generation and analysis of expressed sequence tags from the first-finger stage of development. DNA Res. 5:335-340. [DOI] [PubMed] [Google Scholar]
- 19.Morrissey, J. H., K. M. Devine, and W. F. Loomis. 1984. The timing of cell-type-specific differentiation in Dictyostelium discoideum. Dev. Biol. 103:414-424. [DOI] [PubMed] [Google Scholar]
- 20.Pears,C. J., and J. G. Williams. 1987. Identification of a DNA sequence element required for efficient expression of a developmentally regulated and cAMP-inducible gene of Dictyostelium discoideum. EMBO J. 6:195-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raper, K. B. 1940. Pseudoplasmodium formation and organization in Dictyostelium discoideum. J. Elisha Mitchell Sci. Soc. 56:241-282. [Google Scholar]
- 22.Stege, J. T., G. Shaulsky, and W. F. Loomis. 1997. Sorting of the initial cell types in Dictyostelium is dependent on the tipA gene. Dev. Biol. 185:34-41. [DOI] [PubMed] [Google Scholar]
- 23.Swigart, P., R. Insall, A. Wilkins, and S. Cockcroft. 2000. Purification and cloning of phosphatidylinositol transfer proteins from Dictyostelium discoideum: homologues of both mammalian PITPs and Saccharomyces cerevisiae sec14p are found in the same cell. Biochem. J. 347:837-843. [PMC free article] [PubMed] [Google Scholar]
- 24.Tan, J. L., and J. A. Spudich. 1991. Characterization and bacterial expression of the Dictyostelium myosin light chain kinase cDNA. Identification of an autoinhibitory domain. J. Biol. Chem. 266:16044-16049. [PubMed] [Google Scholar]
- 25.Thompson, C. R., and R. R. Kay. 2000. The role of DIF-1 signaling in Dictyostelium development. Mol. Cell 6:1509-1514. [DOI] [PubMed] [Google Scholar]
- 26.Tsujioka, M., M. Yokoyama, K. Nishio, H. Kuwayama, T. Morio, M. Katoh, H. Urushihara, T. Saito, H. Ochiai, Y. Tanaka, I. Takeuchi, and M. Maeda. 2001. Spatial expression patterns of genes involved in cyclic AMP responses in Dictyostelium discoideum development. Dev. Growth Diff. 43:275-283. [DOI] [PubMed] [Google Scholar]
- 27.Van Driessche, N., C. Show, M. Katoh, T. Morio, R. Sucgang, M. Ibarra, H. Kuwayama, T. Saito, H. Urushihara, M. Maeda, I. Takeuchi, H. Ochiai, W. Eaton, J. Tollett, J. Halter, A. Kuspa, Y. Tanaka, and G. Shaulsky. 2002. Transcriptional profile of Dictyostelium development. Development 129:1543-1552. [DOI] [PubMed] [Google Scholar]
- 28.Wetterauer, B., J. P. Jacquot, and M. Veron. 1992. Thioredoxins from Dictyostelium discoideum are a developmentally regulated multigene family. J. Biol. Chem. 267:9895-9904. [PubMed] [Google Scholar]