Abstract
We describe here the isolation and characterization of a novel RNA-binding protein, RBP38, from Leishmania tarentolae mitochondria. This protein does not contain any known RNA-binding motifs and is highly conserved among the trypanosomatids, but no homologues were found in other organisms. Recombinant LtRBP38 binds single and double-stranded (ds) RNA substrates with dissociation constants in the 100 nM range, as determined by fluorescence polarization analysis. Downregulation of expression of the homologous gene, TbRBP38, in procyclic Trypanosoma brucei by using conditional dsRNA interference resulted in 80% reduction of steady-state levels of RNAs transcribed from both maxicircle and minicircle DNA. In organello pulse-chase labeling experiments were used to determine the stability of RNAs in mitochondria that were depleted of TbRBP38. The half-life of metabolically labeled RNA decreased from ∼160 to ∼60 min after depletion. In contrast, there was no change in transcriptional activity. These observations suggest a role of RBP38 in stabilizing mitochondrial RNA.
RNA-binding proteins are involved in the synthesis, processing, transport, translation, degradation, and stabilization of RNA (7, 24). The mitochondrial genome of trypanosomatids is composed of thousands of circular DNA molecules, which are catenated and form a dense body of DNA, called the kinetoplast DNA network. This consists of two types of molecules: maxicircles and minicircles. The maxicircle encodes two rRNAs, 18 mRNAs, and a few gRNAs (2). The minicircle encodes the majority of the gRNAs (28). The maxicircle protein coding regions are very compact, and many of the genes overlap extensively, leaving little room for regulatory sequences that might control transcription and RNA processing. It was shown previously that maxicircle transcription produces polycistronic RNAs that are processed to mature RNAs (16, 27). The presence of such precursors indicates that the mature RNAs are generated by 3′- and 5′-end processing, which, for several of the overlapping genes, eliminates a portion of the coding regions of the adjacent transcripts, which then must be degraded. Mitochondrial biogenesis in Trypanosoma brucei is developmentally regulated (23, 26, 29). In long slender bloodstream forms mitochondrial function is strongly repressed. During differentiation, the mitochondrion is partially activated in the short stumpy bloodstream form, but only in the insect midgut procyclic form is the mitochondrion fully active. This change in physiology is reflected in a stage-specific regulation of mitochondrial gene expression (15, 18). The levels of some mitochondrial transcripts in short stumpy cells are intermediate between the long slender and procyclic forms, whereas other transcripts are already at the procyclic levels. The transcription rates and processing of the rRNA genes are the same in the bloodstream and the procyclic forms, suggesting that the difference in abundance of the mature rRNAs is due to differential stability in the two forms of the life cycle (19). These findings suggest a regulation of mitochondrial gene expression at the level of RNA stability, and it is likely that RNA-binding proteins play a crucial role in this process.
We have isolated and characterized a novel 38-kDa RNA-binding protein from trypanosomatid mitochondria, and we present evidence for a role of this protein in RNA stability.
MATERIALS AND METHODS
Leishmania culture.
Leishmania tarentolae UC strain cells were grown in brain heart infusion (Difco) supplemented with hemin (10 μg/ml; Sigma). Cells were grown with rotation in a cell production roller apparatus (Bellco) at 27°C or in 15-liter quantities in a BioFlo IV fermentor (New Brunswick). The cells were harvested with a Pellicon tangential filter apparatus (Millipore). For the isolation of mitochondrial fractions, cells were harvested at late log phase (1.5 × 108 to 1.8 × 108 cells/ml) and were ruptured in 10 mM Tris-HCl (pH 7.9)-1 mM EDTA. Mitochondrial fractions were isolated by isopycnic flotation in Renografin gradients or in Percoll gradients as previously described (3). We have found (and as also noted in the published in organello procedure) (20) that Percoll gradients yield mitochondria that are more active in nucleotide uptake and biosynthesis in vitro, whereas Renografin gradients yield mitochondria of greater purity and higher yields and were used to obtain mitochondria for protein isolation.
LtRBP38 purification.
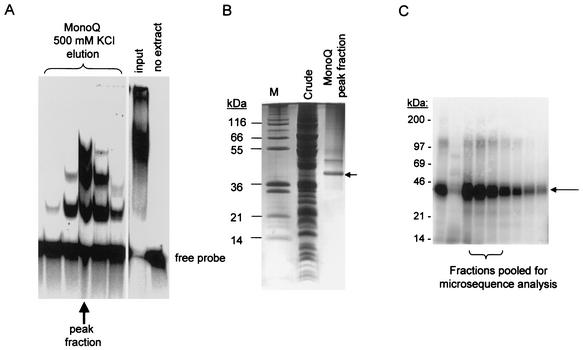
Two methods were developed independently which resulted in purification of the protein. In method 1, RNA-binding fractions were assayed by a gel shift assay by using a 5′-end-labeled RNA probe consisting of two annealed T7 transcribed partially complementary RNAs (substrate 3 in Fig. 4C). Mitochondria were solubilized in 1 mM CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}-0.5 μg of leupeptin/ml-1 μg of pepstatin/ml-0.01 μg of chymostatin/ml-0.1 mM benzamidine-0.1 mM phenylmethylsulfonyl fluoride and then clarified at 100,000 × g for 1 h. The supernate was loaded on a Blue-Sepharose column, and the flowthrough was collected. This was applied to a heparin-agarose column, which was developed with a 0 to 1 M KCl gradient. The peak of RNA gel shift activity eluting at 1 M KCl was collected and dialyzed against 50 mM KCl. This material was loaded on a MonoQ column that was developed with a 0 to 1 M KCl gradient (Fig. 1A). The peak of RNA gel shift activity eluted at 0.5 M KCl, as indicated by an arrow. A single major band migrating at 38 kDa and a minor band at ca. 50 kDa can be seen in a stained sodium dodecyl sulfate (SDS) gel of this fraction (Fig. 1B). The major 38-kDa band was eluted from the gel and subjected to tryptic digestion, and the tryptic peptides were microsequenced for protein identification.
FIG. 4.
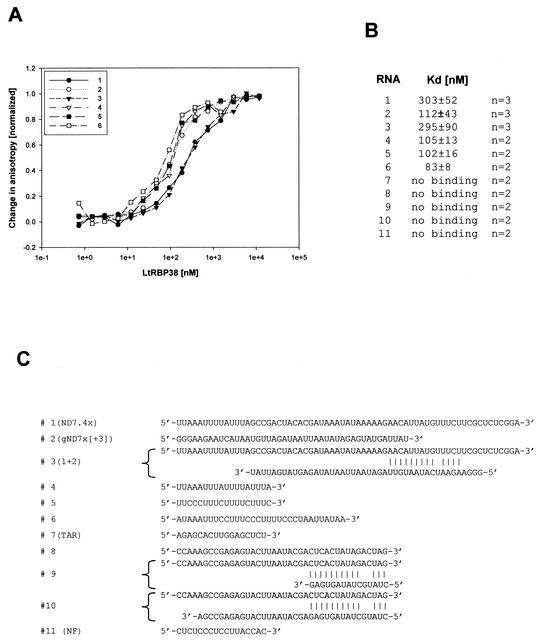
Binding of recombinant LtRBP38 to various RNA substrates. (A) The RNAs were 5′ labeled with fluorescein, and the binding was monitored in a Beacon 2000 fluorescence polarization system. The binding data were normalized, and the curves for an average of three measurements are shown. (B) The Kd values were calculated from the fitted curves. (C) Substrate RNAs: 1, ND7.4x (5); 2, gND7x[+3] (5); 3, RNA1+RNA2 annealed; 4, ND7-AU; 5, ND7-CU; 6, Cyb-CU (14). The following RNAs did not show binding: 7, TAR (13); 8, NR; 9, NR+comp1 annealed; 10, NR+comp2 annealed; 11, NF (25).
FIG. 1.
Isolation of LtRBP38 from mitochondrial extracts of L. tarentolae by using two different purification protocols. (A and B) Method 1 is a gel shift assay with a partially paired duplex RNA (substrate 3, panel C). Mitochondrial extracts were chromatographically enriched for RNA-binding activity by using Blue-Sepharose, heparin, and MonoQ columns. The final MonoQ column was developed with a 0 to 1 M KCl gradient. The collected fractions were used in the gel shift assay shown. (B) The 500 mM KCl peak fraction (indicated in panel A by an arrow) was analyzed on a SDS-polyacrylamide gel electrophoresis, and the gel was stained with Coomassie blue. The fraction contained a major 38-kDa band (arrow) that was isolated and subjected to microsequencing. (C) Method 2 is a gel shift assay for RNA binding to single-stranded RNA using uniformly labeled T7-transcribed gRPS12-III gRNA as the probe. The mitochondrial extract was chromatographically enriched with Q-Sepharose, S-Sepharose, and MonoQ columns. The final MonoQ column was developed by using a gradient of 50 to 500 mM KCl, and the peak activity was subjected to UV cross-linking, RNase digestion, and SDS-gel electrophoresis. The major radioactively labeled cross-link migrated at 38 kDa as a single band (Fig. 1C). This band was eluted from the gel (pooled from lanes 3 to 5) and subjected to N-terminal sequencing. The sequence was identical to the predicted amino acid sequence of the protein isolated by method 1: 27 amino acids from the N terminus.
In method 2, mitochondria were solubilized as described above, and the S-100 supernatant was loaded on a Q-Sepharose column. A gel shift assay for RNA-binding activity used uniformly labeled T7-transcribed gRPS12-III gRNA as the probe. The peak gel shift fractions eluting at 0.4 M KCl were then subjected to S-Sepharose chromatography by using a 50 to 500 mM KCl gradient. The peak fractions from S-Sepharose were subjected to chromatography on a MonoQ column by using a linear gradient of 50 to 500 mM KCl. Each fraction was subjected to UV cross-linking, RNase-digestion, SDS-gel electrophoresis, and autoradiography. The major radioactively labeled cross-link migrated at 38 kDa (Fig. 1C) and could be visualized as a single Coomassie blue-stained band (not shown). This protein was eluted from the gel and the N-terminal sequence of this protein was determined. This sequence was identical to the predicted N-terminal sequence of LtRBP38 isolated by method 1, 27 amino acids from the initial methionine (Fig. 2).
FIG. 2.
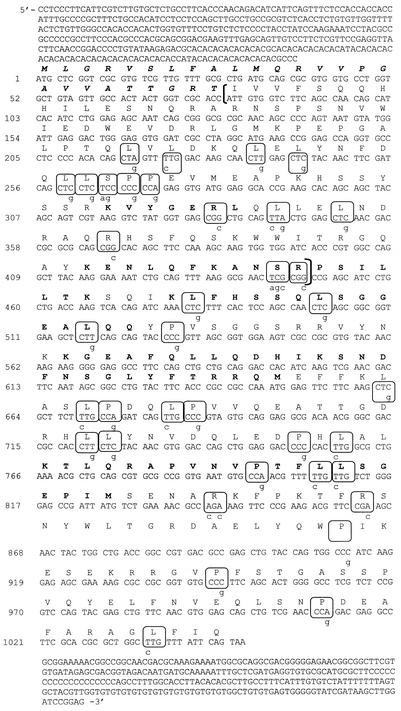
Nucleotide and amino acid sequence of LtRBP38. Nucleotide sequence and the predicted amino acid sequence are shown. The putative mitochondrial targeting signal consisting of the first 26 amino acids is shown in boldface italics. The six tryptic peptides used for cloning are highlighted in boldface. The 93-amino-acid fragment expressed for antiserum production is indicated in brackets. For the expression of the full-length gene, the boxed 37 codons, representing rare E. coli codons, were mutated as indicated by lowercase letters.
In vitro transcription and labeling of synthetic RNAs for gel shift assays.
RNA1 and RNA2 (Fig. 4C) were transcribed by using T7 RNA polymerase from plasmids containing the templates for the various transcripts under the control of a T7 promoter. The RNAs were dephosphorylated with calf intestinal alkaline phosphatase (Gibco-BRL), and RNA 1 was 5′ end labeled by incubation with [α-32P]ATP and T4 polynucleotide kinase for 1 h at 37°C (Gibco-BRL). The RNAs were gel purified by electrophoresis in 7 M urea-8% acrylamide and recovered by elution for 10 h at 4°C in 400 μl of 300 mM sodium acetate-1 mM EDTA buffer. The labeled RNA was precipitated with 1.2 ml of ethanol at −20°C for 15 min. Prior to the gel shift assay, the labeled RNA was annealed to its cognate gRNA by heating it at 70°C for 5 min, followed by slow cooling to room temperature. Uniformly 32P-labeled gRPS12-III gRNA was prepared by T7 transcription by using [α-32P]ATP and gel purified.
Gel shift assays were performed by incubation of the radioactively labeled RNA probes with extracts and fractions at 27°C for 30 min in 50 mM HEPES (pH 7.5)-200 mM KCl-0.2 mM EDTA-0.1 mM dithiothreitol (DTT). After incubation of the probe with protein fractions, binding was analyzed by electrophoretic separation on a 5% acrylamide native gel. Gels were transferred to 3MM filters (Whatman), dried under vacuum at 85°C for 45 min, and then exposed to a PhosphorImager screen (Molecular Dynamics).
Cloning and sequencing of LtRBP38 and TbRBP38.
Final purification of the 38-kDa protein isolated by method 1 from L. tarentolae was performed by SDS-polyacrylamide gel electrophoresis, followed by staining with Coomassie blue. After cutting out the protein band, an in-gel digestion with Endoproteinase Lys-C (Roche Diagnostics, Indianapolis, Ind.) was carried out as described previously (11). Gel-extracted peptides were then fractionated by using a Vydac Microbore C8 column (The Separations Group, Hesperia, Calif.), and individual peptides subjected to Edman degradation with a Procise protein sequencer (model 492; Applied Biosystems, Foster City, Calif.). These sequences were used to generate degenerate oligonucleotide primers for PCR amplification of the gene. All possible primer pairs were tested, and the largest amplified product was found to contain the sequences of all of the peptides obtained by microsequencing, indicating that the peptides were derived from a single polypeptide. Southern blot analysis showed that the polypeptide was encoded by a single-copy nuclear gene (not shown). A 5-kbp ClaI-XbaI DNA genomic fragment containing the entire LtRBP38 coding region was isolated. The peptides used for generation of degenerate oligonucleotides for cloning the gene are in boldface (Fig. 2). The full-length TbRBP38 gene was isolated from a T. brucei HindIII-EcoRI genomic library by hybridization screening with the L. tarentolae probe. A 3.3-kbp genomic fragment was sequenced and found to contain an open reading frame that encodes a protein of 338 amino acids of 38.5-kDa expected molecular mass and a calculated pI of 9.08. It shares 84% similarity and 70% identity with LtRBP38.
Expression of LtRBP38 in Escherichia coli.
The coding sequence was cloned into an E. coli expression vector but attempts to obtain high-level expression of this protein failed, most likely due to the large number of rare codons in the coding sequence. However, a 93-amino-acid fragment of the gene (indicated in brackets in Fig. 2) did express well in E. coli, and this mini-LtRBP38′ protein was used to generate polyclonal antibodies in rabbits (Covance). This antiserum could detect the RBP38 protein in mitochondrial extracts of L. tarentolae and T. brucei.
To express the full-length LtRBP38 for RNA-binding studies, the coding sequence was altered by recombinant PCR to replace all 37 rare codons with more-abundant E. coli codons. The altered codons are indicated in Fig. 2 by circles. The coding region omitting the mitochondrial targeting signal was cloned into pCRT7-TOPO and transformed into E. coli BL21, and the protein was isolated by standard metal affinity chromatography.
Fluorescence polarization analysis of RNA-binding.
Synthetic RNAs (Fig. 4) were purchased from Dharmacon and 5′ labeled with fluorescein as follows. RNAs were 5′ end activated withΥ-S-ATP by using T4 kinase. After heat inactivation of the enzyme for 30 min at 65°C, the RNA was incubated with 1 mM 5-iodoacetamidofluorescein for 30 min at 37°C. The nonincorporated ATP was removed by gel filtration through G-25. To determine dissociation constants, recombinant full-length LtRBP38 was incubated with 1 nM RNA in 50 mM HEPES (pH 7.5)-220 mM KCl-0.2 mM EDTA-0.1 mM DTT at 27°C for 30 min to reach equilibrium. The binding was then monitored in a Beacon 2000 fluorescence polarization system (Panvera), and the binding data were fitted to a one-site binding curve by using Sigmaplot.
Trypanosome culture and RNAi.
Procyclic T. brucei 427 and its derivatives were cultured in SDM-79 supplemented with 10% heat-inactivated serum (4). Bloodstream forms of the same strain were cultured in HMI-11 medium (12). To construct the pTbRBP38i vector for inducible RNA interference (RNAi) a 592-bp PCR fragment of the TbRBP38 gene was inserted into the HindIII/XhoI site of pJZM (30). The vector was linearized with NotI and transfected into procyclic T. brucei 29-13 as described earlier. RNAi was induced with 1 mg of tetracycline/liter, and the cells were maintained between 1 × 106 and 5 × 106 cells/ml.
RNA analysis.
Total RNA was isolated by the acid guanidium isothiocyanate method (6, 7, 24) and treated with RNase-free DNase I (BRL). For Northern blot analysis, 25 μg of total RNA were electrophoresed through a 1.2% formaldehyde-agarose gel and blotted onto a Zeta-Probe membrane. The membrane was hybridized and washed according to the manufacturer's instructions (standard protocol for random-primed DNA probes). Poisoned primer extension analysis with chain terminator nucleotides was performed with 30 μg of total RNA as described previously (7, 8, 24). To determine the relative steady-state RNA level, calmodulin mRNA was used as a standard. For normalization, primer 3813 (detecting the calmodulin mRNA) and a primer detecting a specific mitochondrial transcript were extended in the same reaction with ddGTP to terminate the elongation. To determine the editing pattern, primers were extended as described previously (1).
The following primers were used during the analysis: 3887 (A6), TATTTGATCTTATTCTATAACCTCC; 5002 (12S), GTAAAGGAGAGTAGGACTTG; 5003 (9S), TTTATAATCCTTATCATGGTATC; 5090 (TbRBP38), GTTTGCCAGCTGTTCTACG; and 5135 (gCyb-II), TCCCTAGAGAGTAGTTATCCTCCCCATTACTCAG. The primers 3807 (Murf2), 3808 (Co1), 4080 (Co3), 3812 (Cyb), 3813 (calmodulin), and 4282 (ND7) are described elsewhere (1).
To visualize gRNAs, 1 μg of total RNA was labeled by using guanylyltransferase. A 20-μl reaction contained 50 mM Tris-HCl (pH 8.0), 1 mM MgCl2, 5 mM DTT, 10 μCi of [α-32P]GTP, and 2 μl of guanylyltransferase. The reaction was stopped with 0.5% SDS and 10 μl of 7.5 M ammonium acetate. The reaction products were ethanol precipitated and analyzed on a denaturing 8% acrylamide-8 M urea gel.
Recombinant guanylyltransferase was expressed in E. coli BL21(DE3) containing the plasmid pET-His-D1/D2 (obtained from Stuart Shuman) (17). Bacteria were lysed by sonication in 50 mM Tris-HCl (pH 7.5)-0.15 M NaCl-0.1% Triton X-100-10% sucrose. The lysate was clarified by centrifugation at 30,000 × g for 30 min and mixed with 1 ml of nickel-nitrilotriacetic acid (NTA)-agarose for 1 h. The slurry was transferred into a column and washed with 20 mM Tris-HCl (pH 7.9)-0.5 M NaCl-1 mM phenylmethylsulfonyl fluoride-10% glycerol-20 mM imidazole. The proteins were eluted with 250 mM imidazole. The pooled fractions were dialyzed against 25 mM Tris-HCl (pH 8.0)-0.5 mM EDTA-1 mM DTT-0.05% Triton X-100-5% glycerol and applied to a phosphocellulose column. The proteins were eluted stepwise with 50 mM Tris-HCl (pH 8.0)-1 mM EDTA-2 mM DTT-0.1% Triton X-100-10% glycerol containing 0.05, 0.1, 0.5, and 1 M NaCl. The coexpressed subunits were recovered as a heterodimer in the 1 M NaCl elution step.
Protein analysis.
Total cell protein was extracted in phosphate-buffered saline containing 0.4% Triton X-100. Then, 20 μg of soluble protein was analyzed by Western blotting with rabbit antisera to LtRBP38 and mitochondrial Hsp70 (22). Blots were developed by using the SuperSignal West Pico chemiluminescent system (Pierce).
In organello RNA labeling and degradation.
For metabolic labeling, cells were cultured for ∼72 h with or without tetracycline, reaching a cell density of ca. 5 × 106 cells/ml. Cells were ruptured under hypotonic conditions, and mitochondria were isolated on Percoll gradients (10). Mitochondrial pellets were resuspended at a density of 100 mg/ml (wet weight) in storage buffer (0.25 M sucrose, 20 mM Tris-HCl [pH 7.9], 2 mM EDTA, 50% glycerol) and frozen at −80°C. Metabolic labeling of mitochondria was adapted from Militello et al. (20). Mitochondrial vesicles were washed in transcription buffer (0.6 M sorbitol, 5 mM HEPES [pH 7.6], 3 mM potassium phosphate [pH 7.6], 6 mM KCl, 10 mM MgCl2, 1 mM EDTA, 2 mM 2-mercaptoethanol) and collected by centrifugation at 3,800 × g. Mitochondria were resuspended at 25 mg (wet weight)/ml (∼1 mg of protein/ml) in transcription buffer containing 0.1 mM ATP, 0.1 mM CTP, 0.1 mM UTP, and 100 μCi of [α-32P]GTP/ml. For pulse-chase experiments, mitochondria were labeled for 10 min at 27°C and chased with 2 mM unlabeled GTP. At each time point, 20-μl aliquots were taken and lysed with 15 μl of stop buffer (0.5 M sodium phosphate; 0.5% SDS), spotted onto DE81 nitrocellulose filters, washed extensively with 0.5 M sodium pyrophosphate, and counted in a Beckman scintillation counter. The kinetic data were imported into the Sigmaplot program and fitted to an exponential decay curve: y = a · e−rt + b, where a = span, b = plateau, r = rate of degradation, and t = time. To determine the degradation rates, the data were subjected to a linear transformation by using the following calculation: −rt = ln[(y − b)/a].
GenBank accession numbers.
The sequences reported in this study were submitted to GenBank under accession numbers AY18728 and AY187286.
RESULTS
Isolation and cloning of RBP38.
During our search for RNA-binding proteins involved in mitochondrial RNA metabolism, we isolated a novel 38-kDa protein from the mitochondria of L. tarentolae by using gel shift assays. Two assays were developed to isolate proteins that bind either to a T7-transcribed gRNA or to a partially paired duplex representing a substrate similar to an mRNA-gRNA partial duplex formed during editing. With both assays we isolated the same 38-kDa RNA-binding protein, which we called LtRBP38. The LtRBP38 gene was PCR amplified from genomic DNA by using back-translated degenerate primers obtained from six internal tryptic peptide sequences of the isolated protein, and this product was used as a probe to isolate a genomic fragment containing the entire gene with flanking sequences. The sequence of the gene and the 5′- and 3′-flanking regions are shown in Fig. 2. The N-terminal sequence (IVVFSQQHHILESNQRAR) of the protein isolated from purified mitochondria by cross-linking to a radioactively labeled substrate RNA (method 2 in Materials and Methods) was identical to the predicted N-terminal amino acid sequence of the LtRBP38 gene 27 amino acids downstream of the initiating methionine, suggesting the presence of a mitochondrial targeting sequence consisting of the first 26 amino acids (boldface and italics in Fig. 2). In addition, several computer programs (SignalP, Predotar, Mitoprot, and TargetP) indicated the presence of a mitochondrial targeting signal in the N-terminal region of the predicted protein, and the predicted cleavage site is close that that determined experimentally. This is strong evidence for RBP38 being a mitochondrial protein. Besides the mitochondrial targeting signal, no known motifs, including any known RNA-binding motifs, could be identified, and no homologues outside the trypanosomatids were found.
The T. brucei homologue, TbRBP38, was cloned as described in Materials and Methods. Database searching showed the presence of homologues in Leishmania major and Trypanosoma cruzi. An alignment of the amino acid sequences of the full-length RBP38 genes from L. tarentolae, L. major, and T. brucei is shown in Fig. 3, demonstrating the high conservation of this sequence within the trypanosomatids. Northern analysis in T. brucei bloodstream and procyclic cells showed that a transcript of ∼2.3 kb is present in both stages of the life cycle, and Western analysis showed that the protein is present in both stages (data not shown).
FIG. 3.
Sequence alignment of RBP38 from T. brucei (Tb, accession no. AY187285), L. tarentolae (Lt, accession no. AY187286), and L. major (Lm, accession no. AL138558) showing the high conservation within the trypanosomatids. Identical amino acids are boxed in black, conservative changes are boxed in gray.
RNA-binding properties of LtRBP38.
LtRBP38 had been isolated by using RNA gel shift assays. The recombinant protein also showed RNA-binding activity, and this was quantitated by using a fluorescence polarization assay. Since the protein was first isolated by using a model gRNA-mRNA partial hybrid RNA substrate, we systematically compared gRNA/mRNA model substrates, gRNA or mRNA alone, short polymers, short nonstructured and structured RNAs, and AU- or GC-rich RNAs. Eleven different synthetic RNA substrates (Fig. 4C) were 5′ end labeled with fluorescein, and the changes in fluorescence polarization anisotropy after binding to the protein were monitored. It became apparent that there was no specificity for the gRNA/mRNA or for RNAs involved in editing. As shown in Fig. 4A and B, RNAs 1 to 6 showed binding to LtRBP38 at the 100 to 300 nM Kd range, whereas RNAs 7 to 11 did not show any detectable binding. There was little difference between the binding of the single-stranded RNA1 and RNA2 and the annealed RNA1 plus RNA2. RNA1 and RNA2 could each potentially form secondary structures alone, but this was not examined experimentally. However, RNA substrates 9 and 10, which were annealed to produce partially duplexed structures, showed no binding, nor did the small hairpin RNA7 or RNA11, which could not form secondary structures. There appears to be some specificity of binding, but the specific structural or sequence signals responsible cannot be deduced from these data. Further work is required to ascertain the nature of this specificity.
RBP38 is essential in procyclic T. brucei.
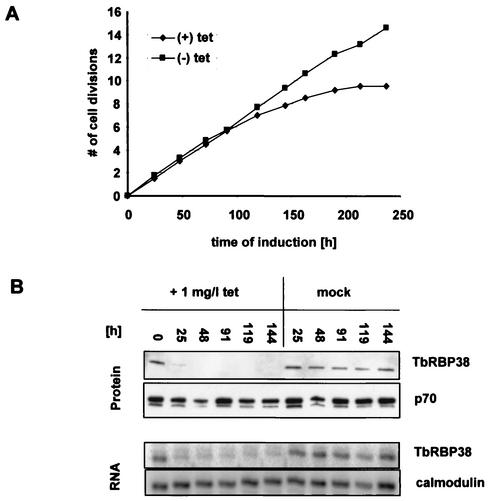
To assess the function of RBP38 in vivo we attempted to disrupt both alleles in procyclic T. brucei. Several attempts to make null mutants failed (data not shown), suggesting that the gene may be essential in this stage of the life cycle. In an alternative approach with conditional RNAi, we were able to downregulate TbRBP38 expression. A 592-bp fragment of the gene was cloned into the pJZM vector, and the resulting vector was transformed into T. brucei strain 29-13, a cell line expressing T7 RNA polymerase and tetracycline repressor (30). After induction of RNAi with 1 mg of tetracycline/liter, the cells showed severe growth inhibition within 5 to 6 days, confirming that the gene is essential for viability (Fig. 5A). The downregulation of TbRBP38 expression is shown in Fig. 5B. Protein and RNA samples were harvested at the indicated time points after induction with tetracycline or mock treatment. For the Western blot, 20 μg of total cell protein was loaded per lane and analyzed with anti-LtRBP38 antiserum. The protein was greatly reduced after 25 to 48 h. There was also a significant decrease in TbRBP38 mRNA after 26 h, as shown by primer extension (Fig. 5B). Calmodulin mRNA, a cytosolic RNA unaffected by RNAi, was used as a normalization signal.
FIG. 5.
Downregulation of TbRBP38 expression in procyclic T. brucei by inducible RNAi. RNAi was induced by the addition of 1 mg of tetracycline/liter. (A) Cumulative growth curve with or without tetracycline. Cell growth is expressed as the number of cell divisions. (B) Analysis of TbRBP38 downregulation in induced and mock-treated cells. A total of 20 μg of total protein was loaded per lane and then analyzed with anti-LtRBP38 antisera. p70 was used as a loading control. Next, 35 μg of RNA per time point was analyzed by primer extension as described in Materials and Methods. Calmodulin was used as a loading control.
Downregulation of TbRBP38 affects the steady-state level of total mitochondrial RNA in vivo.
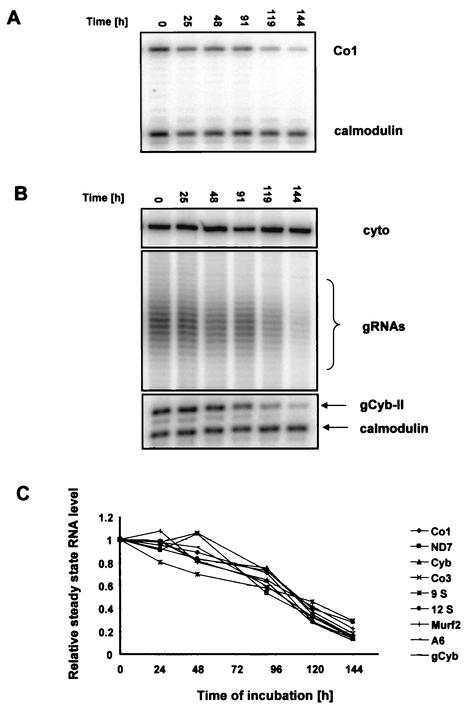
To quantitate changes in mitochondrial steady-state RNA levels during RNAi, we performed poisoned primer extension analysis of 8 of the 18 maxicircle-encoded RNAs. We had previously shown that the addition of tetracycline to the parental 29-13 cell line had no effect on mitochondrial RNA abundance (1). In all reactions, a primer complementary to the cytosolic calmodulin mRNA was included to obtain a normalization signal. During the induction of RNAi, the mitochondrial RNA levels dropped continuously, reaching 20% of the initial values within 6 days (Fig. 6A and C).
FIG. 6.
Downregulation of TbRBP38 affects the steady-state level of total mitochondrial RNA in vivo. Total RNA was isolated at the indicated time points after induction of RNAi. Relative abundance of specific RNAs was determined by poisoned primer extension with 35 μg of RNA per reaction. The cytoplasmic calmodulin mRNA was used as the normalization signal. (A) Representative autoradiogram detecting the maxicircle-encoded Co1 transcript. (B) Minicircle-encoded gRNAs (total population) were visualized by labeling 1 μg of total RNA with guanylyltransferase and [γ-32P]GTP. The cytoplasm-specific 5S rRNA was also labeled and was used as a loading control. In the lower panel is shown the abundance of a specific gRNA (gCyb-II), monitored by primer extension as described above. (C) Quantitation of relative steady-state levels of nine mitochondrial transcripts during induction of RNAi.
We then investigated if only maxicircle-encoded RNAs were affected or if the abundance of the minicircle-encoded gRNAs was also changed. The isolated RNA was 5′ end labeled with [α-32P]GTP by using guanylyltransferase, which specifically labels gRNAs. As shown Fig. 6B, middle panel, a dramatic decrease of gRNA abundance during RNAi was observed. The cytoplasmic 5S rRNA is also 5′ end labeled with guanylyltransferase and [α-32P]GTP, and this served as a loading control (Fig. 6B, upper panel). To further confirm these results, we monitored one specific gRNA (gCyb-II) during the RNAi time course by primer extension, and this gRNA showed a kinetics of loss similar to that of the total gRNA population (Fig. 6B, lower panel). The results for all of the RNAs analyzed are presented graphically in Fig. 6C.
Downregulation of TbRBP38 reduced RNA stability in isolated mitochondria.
The observed changes in steady-state RNA levels could be caused either by reduced rates of transcription or by an effect on RNA stability. To test these hypotheses, we performed pulse-chase labeling experiments with mitochondria isolated under hypotonic rupture conditions from cells treated with or without tetracycline for ∼72 h.
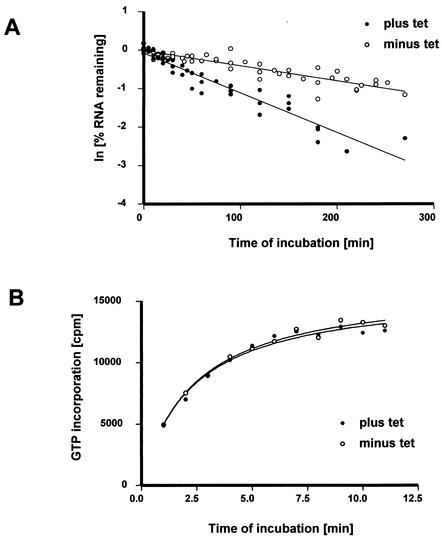
In organello metabolic labeling experiments to study RNA stability in isolated mitochondria from T. brucei have been described previously (10, 20, 21). The isolated mitochondria were incubated with [α-32P]GTP for 10 min, followed by a chase with an excess of cold GTP. The mitochondria were lysed, and the lysates spotted onto DE81 filters for detection of incorporated radioactivity. Exponential decay curves were fitted to the data points with good fidelity (r2 > 0.98), and decay rates were calculated from the pooled data obtained from six independent mitochondrial isolations (Fig. 7A). In the mock-treated mitochondria, ca. 50% of the pulse-labeled RNA degraded with a half-life of ca. 160 min. These results are consistent with published data obtained with wild-type T. brucei mitochondria (21). Depletion of TbRBP38 by RNAi significantly (Student t test, P < 0.001) reduced the half-life of total mitochondrial RNA from ca. 160 to ca. 58 min (Fig. 7A), but no change in the amount of RNA degraded could be detected. These data suggest a possible role of TbRBP38 in RNA stability. In contrast, there was no significant (Student t test, P < 0.88) effect of RNAi on the incorporation of [α-32P]GTP into RNA (Fig. 7B). This suggests that transcription was not affected by depletion of TbRBP38.
FIG. 7.
Downregulation of TbRBP38 affects RNA stability in organello. Mitochondria were isolated ∼72 h after induction of RNAi (plus tet) or mock treatment (minus tet). Mitochondrial RNA was metabolically labeled with [α-32P]GTP, harvested at the indicated time points, and quantitated by using a filter-binding assay. (A) Degradation kinetics of mitochondrial RNA in organello. The kinetic data collected from six independent mitochondrial isolations were linearized, and the decay rates were calculated from the pooled data. (B) Kinetics of [α-32P]GTP incorporation into mitochondrial RNA as a marker for transcription.
Downregulation of TbRBP38 does not affect RNA editing.
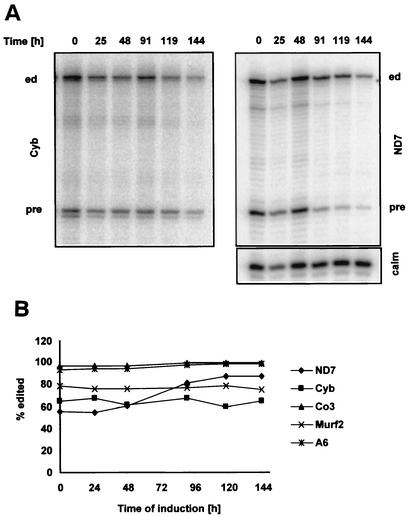
Twelve of the 18 mitochondrial mRNAs in T. brucei undergo U-insertion/deletion editing to produce translatable mRNAs (for a review, see reference 9). The effect of TbRBP38 downregulation on RNA editing in vivo was analyzed for 5 of the 12 edited mRNAs by primer extension, with a downstream primer that allowed the detection of both the edited and the preedited sequences in the same reaction (Fig. 8A). In four of the five mRNAs analyzed, no significant change in the abundance of edited transcripts editing could be detected. The only exception was the ND7 transcript, for which the percentage of editing increased. As the calmodulin primer extension loading control shows, the increase in editing was rather due to a more rapid degradation of the pre-mRNA than to an increase of the edited mRNA (Fig. 8A, right panel). These data suggest that RBP38 is not involved in RNA editing.
FIG. 8.
Downregulation of TbRBP38 does not affect RNA editing in vivo, as shown by primer extension analysis of mitochondrial mRNAs (35 μg of RNA per reaction) during RNAi. (A) Time course analysis of Cyb (left panel) and ND7 (right panel) mRNAs detecting edited (ed) and preedited (pre) RNA. The cytoplasmic calmodulin mRNA (calm) was analyzed in the same reaction as an internal control for loading and assay performance. (B) Quantitative analysis of five edited mRNAs. The percentage of edited mRNA was calculated.
DISCUSSION
The trypanosomatid mitochondrial RNA-binding protein, RBP38, was serendipitously discovered as a protein that produced an electrophoretic gel shift of a labeled partially duplexed RNA probe and could also be UV cross-linked to a single-stranded RNA probe. The Kd of binding of recombinant LtRBP38 to several synthetic RNAs (Fig. 4C, RNA1 to RNA6) is in the 100 nM range. There is some specificity in that several other RNAs (Fig. 4C, RNA7 to RNA11) are not bound. The binding specificity does not involve AU-rich regions, since the same RNA with A's changed to C's worked just as well (Fig. 4C, RNA4 and -5). TAR RNA (Fig. 4C, RNA7), which forms a small duplex hairpin, does not bind, nor does a completely unstructured small RNA (Fig. 4C, RNA11). The nature of this specificity is not yet determined.
In the present study we demonstrate that RBP38 is essential for the growth of T. brucei procyclics. Induction of conditional RNAi resulted in the downregulation of TbRBP38 expression on the RNA and protein levels and resulted in a severe growth inhibition. The mitochondrial genome encodes three types of RNAs: rRNA, mRNA, and gRNA. We monitored the change in steady-state levels of all three types of RNAs during a 6-day time course after induction of RNAi. Analysis of the two maxicircle-encoded rRNAs and 6 of the 18 mRNAs revealed a reduction in RNA abundance to ca. 20% of the original levels. We examined representative mRNAs that are never edited (COI), 5′ end edited (Cyb, Murf2), pan-edited (A6, CO3), and developmentally regulated (ND7). Since all mRNA transcripts analyzed changed in the same way, we believe that it is very likely that all mitochondrial mRNA transcripts are affected by the downregulation of TbRBP38.
The majority of gRNAs are encoded on minicircles and contain a triphosphate at the 5′ end, enabling us to specifically visualize all gRNAs by the labeling total cell RNA by using guanylyltransferase and [α-32P]GTP (2, 28). We found that the gRNA abundance changed during the time course in a fashion similar to that of the maxicircle-encoded transcripts. Moreover, the analysis of a specific gRNA transcript (gCyb-II) by primer extension confirmed the initial observation from the labeling experiments with guanylyltransferase capping. This eliminated the possibility that we might be monitoring the loss of the phosphates rather than the loss of the entire population of gRNAs. We conclude that all mitochondrial transcripts are lost after induction of RNAi of TbRBP38.
The reduction in RNA abundance after depletion of TbRBP38 from the mitochondrion can be caused either by the reduction of transcription or by an increase in RNA turnover due to a reduction of stability or an increase in the rate of degradation. In organello pulse-chase labeling experiments have been successfully used in previous studies analyzing transcription and RNA turnover in trypanosome mitochondria (10, 20, 21). We adapted these procedures to compare mitochondrial RNA metabolism in mitochondria isolated from cells depleted of TbRBP38 with mitochondria from mock-treated cells. Mitochondrial vesicles were harvested 72 h postinduction, a time point when TbRBP38 was depleted from the mitochondria, but the overall cell morphology was yet not altered. [α-32P]GTP was used to metabolically label RNA in organello, since it is the only nucleotide that is not incorporated into RNA by any known posttranscriptional processing event. This ensured that we were only monitoring transcriptional activity or RNA turnover and that the results from those experiments were not superimposed with results from any other RNA-processing event such as U-insertion/deletion editing or CCA turnover in tRNAs. Since downregulation of TbRBP38 affected RNA abundance of all mitochondrial transcripts in a similar fashion in vivo, total mitochondrial RNA stability was monitored in organello rather than the stability of specific transcripts. In mock-treated cells only ca. 50% of the labeled RNA is subjected to degradation with an average half-life of ca. 160 min. These results are consistent with the previous data for in organello RNA stability in wild-type mitochondria of T. brucei. When these results were compared to the data obtained with mitochondria depleted of TbRBP38, we observed a significant decrease in the half-life of the labeled RNA to ca. 60 min. In contrast, when we monitored the incorporation of GTP into RNA as a marker for transcriptional activity, we were not able to detect a difference between mitochondria isolated from TbRBP38-depleted cells versus mock-treated cells.
These data are consistent with an involvement of RBP38 in the stabilization of total mitochondrial RNA. RBP38 could either act as an inhibitor of degradation by interfering with the mitochondrial degradation enzymes or by stabilizing the RNA. Since RBP38 is an RNA-binding protein, it seems more likely that it binds to the RNA, thus preventing its degradation. There is a possible caveat in that the mitochondria were isolated from cells subjected to several days of RNAi downregulation of RBP38, which is eventually lethal, and there may be secondary changes in mitochondrial proteins. However, the observed increase in decay cannot be directly attributed to such secondary changes since there was no effect on mitochondrial transcription or editing at this stage of the RNAi.
The editing pattern of mitochondrial RNAs in cells depleted of TbRBP38 was also examined. We monitored 5 of the 12 edited mRNAs but could only find minor changes in editing. ND7 was the only transcript in which the percentage of edited RNA changed significantly, but this change is probably caused by different stabilities of edited and preedited RNAs and not by an involvement of RBP38 in editing. In addition, Western analysis of the purified l-complex in L. tarentolae showed that LtRBP38 is not an integral part of this core editing complex (R. Aphasizhev and L. Simpson, unpublished data). However, we cannot, of course, exclude the possibility that RBP38 is a multifunctional protein that also acts as an editing accessory factor for specific transcripts.
Acknowledgments
This research was supported in part by a grant from the National Institutes of Health (AI09102) to L.S.
We thank all members of the Simpson laboratory for advice and discussion. We also thank Jose Luis Vazquez-Ibar for advice on the fluorescence polarization assays, Gabriela Lopez for help with the statistical analysis, and Stuart Shuman for the pET-His-Da/D2 plasmid.
REFERENCES
- 1.Aphasizhev, R., S. Sbicego, M. Peris, S. H. Jang, I. Aphasizheva, A. M. Simpson, A. Rivlin, and L. Simpson. 2002. Trypanosome mitochondrial 3′-terminal uridylyl transferase (TUTase): the key enzyme in U-insertion/deletion RNA editing. Cell 108:637-648. [DOI] [PubMed] [Google Scholar]
- 2.Blum, B., N. Bakalara, and L. Simpson. 1990. A model for RNA editing in kinetoplastid mitochondria: “guide” RNA molecules transcribed from maxicircle DNA provide the edited information. Cell 60:189-198. [DOI] [PubMed] [Google Scholar]
- 3.Braly, P., L. Simpson, and F. Kretzer. 1974. Isolation of kinetoplast-mitochondrial complexes from Leishmania tarentolae. J. Protozool. 21:782-790. [DOI] [PubMed] [Google Scholar]
- 4.Brun, R., and Schonenberger. 1979. Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 36:289-292. [PubMed] [Google Scholar]
- 5.Byrne, E. M., G. J. Connell, and L. Simpson. 1996. Guide RNA-directed uridine insertion RNA editing in vitro. EMBO J. 15:6758-6765. [PMC free article] [PubMed] [Google Scholar]
- 6.Chomczynski, P., and N. Sacchi. 1987. Single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162:156-159. [DOI] [PubMed] [Google Scholar]
- 7.Dodson, M. L., A. J. Kurtz, and R. S. Lloyd. 2002. T4 endonuclease V: use of NMR and borohydride trapping to provide evidence for covalent enzyme-substrate imine intermediate. Methods Enzymol. 354:202-207. [DOI] [PubMed] [Google Scholar]
- 8.Estévez, A. M., F. Kierszenbaum, E. Wirtz, F. Bringaud, J. Grunstein, and L. Simpson. 1999. Knockout of the glutamate dehydrogenase gene in bloodstream Trypanosoma brucei in culture has no effect on editing of mitochondrial mRNAs. Mol. Biochem. Parasitol. 100:5-17. [DOI] [PubMed] [Google Scholar]
- 9.Estévez, A. M., and L. Simpson. 1999. Uridine insertion/deletion RNA editing in trypanosome mitochondria: a review. Gene 240:247-260. [DOI] [PubMed] [Google Scholar]
- 10.Harris, M. E., D. R. Moore, and S. L. Hajduk. 1990. Addition of uridines to edited RNAs in trypanosome mitochondria occurs independently of transcription. J. Biol. Chem. 265:11368-11376. [PubMed] [Google Scholar]
- 11.Hellman, U., C. Wernstedt, J. Gonez, and C. H. Heldin. 1995. Improvement of an “in-gel” digestion procedure for the micropreparation of internal protein fragments for amino acid sequencing. Anal. Biochem. 224:451-455. [DOI] [PubMed] [Google Scholar]
- 12.Hirumi, H., and K. Hirumi. 1989. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 75:985-989. [PubMed] [Google Scholar]
- 13.Ippolito, J. A., and T. A. Steitz. 1998. A 1.3-Å resolution crystal structure of the HIV-1 trans-activation response region RNA stem reveals a metal ion-dependent bulge conformation. Proc. Natl. Acad. Sci. USA 95:9819-9824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kabb, A. L., L. M. Oppegard, B. A. McKenzie, and G. J. Connell. 2001. A mRNA determinant of gRNA-directed kinetoplastid editing. Nucleic Acids Res. 29:2575-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koslowsky, D. J., G. R. Riley, J. E. Feagin, and K. Stuart. 1992. Guide RNAs for transcripts with developmentally regulated RNA editing are present in both life cycle stages of Trypanosoma brucei. Mol. Cell. Biol. 12:2043-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koslowsky, D. J., and G. Yahampath. 1997. Mitochondrial mRNA 3′ cleavage polyadenylation and RNA editing in Trypanosoma brucei are independent events. Mol. Biochem. Parasitol. 90:81-94. [DOI] [PubMed] [Google Scholar]
- 17.Luo, Y., X. Mao, L. Deng, P. Cong, and S. Shuman. 1995. The D1 and D12 subunits are both essential for the transcription termination factor activity of vaccinia virus capping enzyme. J. Virol. 69:3852-3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michelotti, E., and S. Hajduk. 1987. Developmental regulation of trypanosome mitochondrial DNA gene expression. J. Biol. Chem. 262:927-932. [PubMed] [Google Scholar]
- 19.Michelotti, E. F., M. E. Harris, B. Adler, A. F. Torri, and S. L. Hajduk. 1992. Trypanosoma brucei mitochondrial ribosomal RNA synthesis, processing and developmentally regulated expression. Mol. Biochem. Parasitol. 54:31-42. [DOI] [PubMed] [Google Scholar]
- 20.Militello, K. T., M. L. Hayman, and L. K. Read. 2000. Transcriptional and posttranscriptional in organello labeling of Trypanosoma brucei mitochondrial RNA. Int. J. Parasitol. 30:643-647. [DOI] [PubMed] [Google Scholar]
- 21.Militello, K. T., and L. K. Read. 2000. UTP-dependent and -independent pathways of mRNA turnover in Trypanosoma brucei mitochondria. Mol. Cell. Biol. 20:2308-2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peris, M., A. M. Simpson, J. Grunstein, J. E. Liliental, G. C. Frech, and L. Simpson. 1997. Native gel analysis of ribonucleoprotein complexes from a Leishmania tarentolae mitochondrial extract. Mol. Biochem. Parasitol. 85:9-24. [DOI] [PubMed] [Google Scholar]
- 23.Priest, J. W., and S. L. Hajduk. 1994. Developmental regulation of Trypanosoma brucei cytochrome c reductase during bloodstream to procyclic differentiation. Mol. Biochem. Parasitol. 65:291-304. [DOI] [PubMed] [Google Scholar]
- 24.Ross, J. 1995. mRNA stability in mammalian cells. Microbiol. Rev. 59:423-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rubio, M. A., X. Liu, H. Yuzawa, J. D. Alfonzo, and L. Simpson. 2000. Selective importation of RNA into isolated mitochondria from Leishmania tarentolae. RNA 6:988-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schneider, A. R., H. M. Stapleton, J. Cornwell, and J. E. Baker. 2001. Recent declines in PAH, PCB, and toxaphene levels in the northern Great Lakes as determined from high resolution sediment cores. Environ. Sci. Technol. 35:3809-3815. [DOI] [PubMed] [Google Scholar]
- 27.Simpson, A., N. Neckelmann, V. de la Cruz, M. Muhich, and L. Simpson. 1985. Mapping and 5′ end determination of kinetoplast maxicircle gene transcripts from Leishmania tarentolae. Nucleic Acids Res. 13:5977-5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sturm, N. R., and L. Simpson. 1990. Kinetoplast DNA minicircles encode guide RNAs for editing of cytochrome oxidase subunit III mRNA. Cell 61:879-884. [DOI] [PubMed] [Google Scholar]
- 29.Vickerman, K. 1985. Developmental cycles and biology of pathogenic trypanosomes. Br. Med. Bull. 41:105-114. [DOI] [PubMed] [Google Scholar]
- 30.Wang, Z. F., J. C. Morris, M. E. Drew, and P. T. Englund. 2000. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 275:40174-40179. [DOI] [PubMed] [Google Scholar]