Abstract
To isolate genes encoding coenzyme B12-dependent glycerol and diol dehydratases, metagenomic libraries from three different environmental samples were constructed after allowing growth of the dehydratase-containing microorganisms present for 48 h with glycerol under anaerobic conditions. The libraries were searched for the targeted genes by an activity screen, which was based on complementation of a constructed dehydratase-negative Escherichia coli strain. In this way, two positive E. coli clones out of 560,000 tested clones were obtained. In addition, screening was performed by colony hybridization with dehydratase-specific DNA fragments as probes. The screening of 158,000 E. coli clones by this method yielded five positive clones. Two of the plasmids (pAK6 and pAK8) recovered from the seven positive clones contained genes identical to those encoding the glycerol dehydratase of Citrobacter freundii and were not studied further. The remaining five plasmids (pAK2 to -5 and pAK7) contained two complete and three incomplete dehydratase-encoding gene regions, which were similar to the corresponding regions of enteric bacteria. Three (pAK2, -3, and -7) coded for glycerol dehydratases and two (pAK4 and -5) coded for diol dehydratases. We were able to perform high-level production and purification of three of these dehydratases. The glycerol dehydratases purified from E. coli Bl21/pAK2.1 and E. coli Bl21/pAK7.1 and the complemented hybrid diol dehydratase purified from E. coli Bl21/pAK5.1 were subject to suicide inactivation by glycerol and were cross-reactivated by the reactivation factor (DhaFG) for the glycerol dehydratase of C. freundii. The activities of the three environmentally derived dehydratases and that of glycerol dehydratase of C. freundii with glycerol or 1,2-propanediol as the substrate were inhibited in the presence of the glycerol fermentation product 1,3-propanediol. Taking the catalytic efficiency, stability against inactivation by glycerol, and inhibition by 1,3-propanediol into account, the hybrid diol dehydratase produced by E. coli Bl21/pAK5.1 exhibited the best properties of all tested enzymes for application in the biotechnological production of 1,3-propanediol.
1,3-Propanediol is a monomer employed in the industrial production of polyester fibers, polyurethanes, and cyclic compounds. The microbial formation of 1,3-propanediol from glycerol has been known for a number of years (20). Glycerol is fermented by a dismutation process involving two pathways. Through one pathway, glycerol is dehydrogenated by the NAD+-linked glycerol dehydrogenase to dihydroxyacetone, which is then phosphorylated and funneled to glycolysis by dihydroxyacetone kinase (15). Through the reductive branch of the pathway, glycerol is dehydrated by the coenzyme B12-dependent glycerol dehydratase to form 3-hydroxypropionaldehyde, which is reduced to the major fermentation product 1,3-propanediol by the NADH-linked 1,3-propanediol dehydrogenase, thereby regenerating NAD+ (16, 17). The four key enzymes of this pathway are encoded by the dha regulon, the expression of which is induced when dihydroxyacetone or glycerol is present (17). The usefulness of natural producers such as Citrobacter freundii, Klebsiella pneumoniae, Clostridium butyricum, and Clostridium pasteurianum for industrial-scale production of 1,3-propanediol has been well studied (2, 7, 13, 24, 33). Drawbacks for industrial processes employing these organisms are the requirement for the expensive starting material glycerol as well as the strong inhibition of 1,3-propanediol production and formation of by-products during fermentation in the presence of inexpensive cosubstrates such as glucose (11). Recently, Genencor International and DuPont circumvented these problems by construction of a recombinant Escherichia coli strain for the large-scale production of 1,3-propanediol from glucose. They combined two natural pathways: glucose to glycerol and glycerol to 1,3-propanediol (11). Glycerol is formed by the recombinant E. coli strain via the glycolytic intermediate dihydroxyacetone 3-phopshate by using dihydroxyacetone 3-phosphate dehydrogenase and glycerol 3-phosphate phosphatase from Saccharomyces cerevisiae. Subsequently, glycerol is converted to 1,3-propanediol by glycerol dehydratase and 1,3-propanediol dehydrogenase (see above). The limiting step of this process is the activity of the glycerol dehydratase (1, 2, 7). Glycerol dehydratases (EC 4.2.1.30) and the related diol dehydratases (EC 4.2.1.28) can catalyze the conversion of glycerol, 1,2-propanediol, and 1,2-ethanediol to the corresponding aldehydes (17). These enzymatic reactions are known to proceed by a radical mechanism involving coenzyme B12 as an essential cofactor. Glycerol dehydratases as well as diol dehydratases are multisubunit (α2β2γ2) enzymes and undergo irreversible inactivation by glycerol during catalysis (5, 17, 40, 48, 50). Inactivation by glycerol involves irreversible cleavage of the Co-C bond of coenzyme B12, forming 5′-deoxyadenosine and an alkylcobalamin-like species. Irreversible inactivation is then brought about by tight binding of the modified coenzyme (17, 48). Such suicide inactivation seems enigmatic, since glycerol is the growth substrate and the dehydratase is essential for the conversion of glycerol to 1,3-propanediol. It has been shown for the dehydratases of C. freundii, Klebsiella oxytoca, and K. pneumoniae that the glycerol-inactivated enzymes are reactivated by a protein complex consisting of two different subunits (26, 32, 40, 47).
The goal of this study was to isolate coenzyme B12-dependent glycerol and diol dehydratases by construction and screening of complex libraries and then to identify the dehydratase among the recovered dehydratases that possesses the best suitability for the biotechnological production of 1,3-propanediol with respect to inactivation by glycerol and inhibition by 1,3-propanediol. The DNA used for the preparation of the so-called metagenomic libraries was directly isolated from different environmental samples after a short enrichment of glycerol-fermenting microorganisms. The screening of environmental libraries for genes encoding dehydratases was based either on enzyme activity of recombinant E. coli strains or on nucleotide sequence. The genes encoding the targeted dehydratases were recovered from the resulting positive E. coli strains and sequenced. Subsequently, the corresponding gene products were purified and the sensitivity of the proteins to inactivation by glycerol and inhibition by 1,3-propanediol was analyzed.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The plasmids used in this study are shown in Table 1. The E. coli strains DH5α (4), ECL707 (43), and BL21 (Invitrogen, Karlsruhe, Germany) were used as hosts for the cloning experiments, the activity-based screening procedures, and production of the dehydratases, respectively. Salmonella enterica serovar Typhimurium TA100 was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (Braunschweig, Germany).
TABLE 1.
Plasmids
| Plasmid | Relevant characteristicsa | Reference(s) or source |
|---|---|---|
| pSK+ | Apr, lac promoter, pMB1 replicon | Stratagene |
| pSTV28 | Cmlr, lac promoter, p15A replicon | Takara |
| pET100/D | Apr, T7 promoter, pMB1 replicon | Invitrogen |
| pET101/D | Apr, T7 promoter, pMB1 replicon | Invitrogen |
| pRD1 | pWE15::32,000-bp Sau3AI fragment containing the dha regulon (dhaBCDEFGHIKRT genes) of C. freundii | 15, 16, 41 |
| pFL1 | pWE15::13,500-bp Sau3AI fragment containing dhaBCEFGHIT genes of Clostridium pasteurianum | 28 |
| pMS2 | pSK(+)::5,500-bp HindIII-BstEII fragment containing the dhaBCEF genes of C. freundii | 41 |
| pCS111 | pKK223-3::2,212-bp EcoI-HindIII fragment containing the His6-tagged dhaFG genes of C. freundii under control of the tac promoter | 40 |
| pCS120 | pSK(+)::2,731-bp EcoRI-HindIII fragment containing the His6-tagged dhaBCE genes of C. freundii under control of the lac promoter | 40 |
| pAK1 | pSTV28::8,536-bp BpiI-Bsp119I fragment containing the dhaDGHIKRT genes of C. freundii | This study |
| pAK2 | pSK(+)::7,115-bp Bsp143I fragment containing dhaBCEFIT∗ genes derived from enrichment cultures | This study |
| pAK3 | pSK(+)::2,157-bp Bsp143I fragment containing dhaB∗CEF∗ genes derived from enrichment cultures | This study |
| pAK4 | pSK(+)::1,750-bp Bsp143I fragment containing pduC∗DE∗ genes derived from enrichment cultures | This study |
| pAK5 | pSK(+)::4,236-bp Bsp143I fragment containing the pduC∗DEGHJ genes derived from enrichment cultures | This study |
| pAK6 | pSK(+)::4,332-bp Bsp143I fragment containing parts of the C. freundii dha regulon (not further characterized) | This study |
| pAK7 | pSK(+)::6,295-bp Bsp143I fragment the containing dhaBCEF∗GH∗IT genes derived from enrichment cultures | This study |
| pAK8 | pSK(+)::6,805-bp Bsp143I fragment containing parts of the C. freundii dha regulon (not further characterized) | This study |
| pAK2.1 | pET100/D-TOPO::2,699-bp PCR product containing dhaBCE genes from pAK2 under control of the T7 promoter | This study |
| pAK3.1 | pET100/D-TOPO::2,699-bp PCR product containing a hybrid dhaB gene derived from C. freundii and pAK3 and the dhaCE genes of pAK3 under control of the T7 promoter | This study |
| pAK5.1 | pET101/D-TOPO::2,884-bp PCR product containing a hybrid pduC gene derived from S. enterica serovar Typhimurium and pAK5 and the pduDE genes of pAK5 under control of the T7 promoter | This study |
| pAK7.1 | pET101/D-TOPO::2,696-bp PCR product containing the dhaBCE genes of pAK7 under control of the T7 promoter | This study |
*, partial gene.
Media and growth conditions.
E. coli was routinely grown in Luria-Bertani (LB) medium at 30°C (4). For activity-based screening of environmental libraries, recombinant E. coli strains were grown under anaerobic conditions in M9 medium (31), which was supplemented with glycerol (100 mM) and coenzyme B12 (1 mg/liter) and solidified with agar (15 g/liter). The enrichment of glycerol-fermenting microorganisms was performed under anaerobic conditions for 24 h in a medium (pH 7.5) containing, per liter, the following: K2HPO4, 14.0 g; KH2PO4, 6.0 g; (NH4)2SO4, 3.0 g; MgSO4 · 7H2O, 0.2 g; CoCl2 · 6H2O, 0.0119 g; yeast extract, 0.2 g; cysteine-HCl, 0.2 g; and trace element solution SL4 (34), 1 ml. The medium was supplemented with 100 mM glycerol. The enrichment was initiated by adding 10 g (wet weight) of environmental sample to anaerobic flasks (1 liter) containing enrichment medium (500 ml). For production of the His6-tagged dehydratases, recombinant E. coli strains were grown in LB medium at 37°C. All growth media for E. coli strains harboring plasmids contained 100 μg of ampicillin per ml and/or 25 μg of chloramphenicol per ml to maintain the presence of the plasmids.
Preparation of cell extracts.
Cells of the stationary growth phase from 500-ml cultures were harvested by centrifugation at 6,000 × g for 20 min, washed once with 100 mM potassium phosphate buffer (pH 8.0), and resuspended in 2 to 3 ml of the same buffer. The cells were disrupted by French pressing (1.38 × 108 Pa), and the extract was cleared by centrifugation at 32,000 × g and 4°C for 30 min.
Purification of His6-tagged proteins.
His6-tagged proteins were purified from the cell extracts by nickel affinity chromatography on Ni-nitrilotriacetic acid agarose (Qiagen, Hilden, Germany) according to the manufacturer's instructions. After the column (1 by 5 cm; 3-ml bed volume) was loaded with 5 ml of cell extract (approximately 30 mg/ml), it was washed with 15 ml of buffer (300 mM KCl in 50 mM potassium phosphate [pH 8.0]) containing 10 mM imidazole followed by 15 ml of the same buffer containing 20 mM imidazole. Proteins bound to the resin were eluted with buffer containing 250 mM imidazole. Fractions harboring homogeneous His6-tagged proteins were pooled and used for the experiments. The His6-tagged glycerol dehydratase and the His6-tagged DhaFG complex of C. freundii were purified from cell extracts of recombinant E. coli strains harboring pCS120 and pCS111, respectively, as described previously (40).
General protein techniques.
Electrophoresis under nondenaturing conditions was performed on polyacrylamide gradient slab gels (4 to 28%) at 4°C in Tris-glycine buffer (pH 8.3) by the method of Andersson et al. (3). For calculation of the native molecular mass, a commercial high-molecular-mass calibration kit of standard proteins was used (Amersham Pharmacia, Freiburg, Germany). Activity staining of glycerol dehydratase was performed as described by Tobimatsu et al. (45) with 1,2-propanediol as the substrate.
Enzyme assays.
The determination of glycerol or diol dehydratase activity was based on the ability of the aldehydes formed during the dehydratase reaction to react with 3-methyl-2-benzothiazolinone hydrazone (MBTH). The resulting azine derivatives were detected spectrophotometrically in the presence of FeCl3. The assay mixture contained 60 μl of apoenzyme solution, 100 μl of reaction buffer (80 mM HEPES [pH 8.2]), and 20 μl of substrate (1 M glycerol or 1 M 1,2-propanediol). The reaction was initiated by addition of 20 μl of coenzyme B12 solution (0.12 mM). After incubation at 37°C for 1 min, the enzyme reaction was terminated by addition of 100 μl of MBTH solution (28 mM MBTH in 375 mM glycine buffer [pH 2.7]) and incubation at 100°C for 3 min. Subsequently, the samples were cooled on ice and 1 ml of FeCl3 solution (12.2 mM FeCl3 in H2O) was added. After 15 min of incubation at room temperature, the amount of aldehyde formed was determined from the absorbance at 670 nm. The apparent molar extinction coefficient at 670 nm for the colored product from 3-hydroxypropionaldehyde is 5.23 × 103 M−1 cm−1. The Km values for glycerol were calculated from standard Lineweaver-Burk plots. The Ki values for 1,3-propanediol were determined from standard Lineweaver-Burk plots derived from experiments in which an increasing amount of 1,3-propanediol (0 to 2 M) was added to the assay mixture. The inactivation of B12-dependent dehydratases by glycerol and the reactivation of glycerol-inactivated holoenzymes by the DhaFG complex of C. freundii were performed and assayed as described previously (40). The activities of glycerol dehydrogenase, dihydroxyacetone kinase, and 1,3-propanediol dehydrogenase were determined as described previously (7, 15, 16). Protein concentrations were determined by the method of Bradford (8) with bovine serum albumin as a standard.
Molecular procedures.
Manipulations of DNA, PCR, and transformation of plasmids into E. coli were done according to routine procedures (4) unless otherwise specified. The Göttingen Genomics Laboratory (Göttingen, Germany) determined the DNA sequences. Sequence analysis was performed with the Genetics Computer Group program package (18).
(i) Construction of environmental DNA libraries.
The isolation of DNA from environmental samples was performed as described previously (14, 23). The purified DNA was partially digested with Bsp142I and, in order to avoid cloning of very small DNA fragments, was size fractionated by sucrose density centrifugation (10 to 40% [wt/vol]). Fractions containing DNA fragments of >2 kb were ligated into BamHI-digested pBluescript SK(+) [pSK(+)], and the products were then transformed into E. coli DH5α. This strain was employed for maintenance and amplification of the environmental DNA libraries.
(ii) Amplification of internal fragments of genes encoding dehydratases.
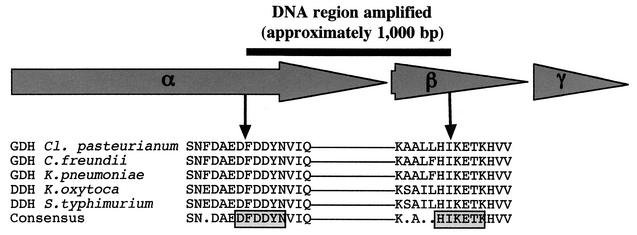
For the identification of environmental libraries harboring dehydratase genes by PCR, degenerate oligonucleotides (GAYTTYGAYGAYTAYAAY and YTTNGTYTCYTTDATRTG) were employed, which were derived from regions conserved in all known sequences for dehydratases (Fig. 1). In this way, approximately 1,000-bp internal fragments of the entire gene regions encoding dehydratases were amplified. To cover the different annealing temperatures of the degenerated primer pair, 10 PCRs per sample were performed in a gradient cycler. For calibration of the PCRs, the genes encoding the dehydratases of C. freundii (41) and Clostridium pasteurianum (28, 29) were used as positive controls. The reactions were performed by using the isolated recombinant plasmids pMS2 and pFL1, respectively, as sources for the dehydratase genes of these organisms. In addition, small amounts of the recombinant plasmids were mixed with large amounts of plasmids from environmental libraries or the vector used for the construction of the libraries and then subjected to PCR. The best results with respect to the nonappearance of unspecific by-products and reliability of the PCR were obtained by using the following reaction mixture: 100 μl of Mg-free buffer (MBI Fermentas, St. Leon-Rot, Germany), a 200 μM concentration of each of the four deoxynucleoside triphosphates, 2.5 mM MgCl2, a 2 μM concentration of each of the primers, 2 U of Taq polymerase (MBI Fermentas), and 100 to 200 ng of the recombinant plasmids harboring the environmental DNA as the template. The reactions were initiated at 94°C (1 min); followed by 30 cycles of 95°C (1 min), a temperature gradient ranging from 27 to 47°C (1 min), and 72°C (1 min); and ended with incubation at 72°C for at least 5 min. The dehydratase-specific 1,000-bp PCR products obtained during the amplification step were cloned by the TA method, which takes advantage of the terminal transferase activity of Taq polymerase. This enzyme added a single 3′-A overhang to each end of the PCR product. Subsequently, the PCR products were directly cloned into pCR2.1-TOPO by using the TOPO TA cloning kit (Invitrogen, Karlsruhe, Germany) according to the manufacturer's instructions. In order to confirm that the cloned PCR products were derived from dehydratase genes, the sequences were determined. Libraries revealing a specific PCR product were used as starting material for screening of the genes encoding dehydratases.
FIG. 1.
Conserved regions of coenzyme B12-dependent glycerol dehydratases (GDHs) and diol dehydratases (DDHs) selected for generation of degenerate oligonucleotides. Sequences from C. freundii (41); Clostridium pasteurianum (29); K. pneumoniae (44); K. oxytoca (45); and S. enterica serovar Typhimurium (6) are shown.
(iii) Sequence-based screening by colony hybridization.
The colonies of the recombinant E. coli strains of each environmental library were imprinted on Hybond N membranes (Amersham Pharmacia) as specified by the manufacturer. Membranes were prehybridized for 1 to 3 h at 60°C in the buffer described by Ausubel et al. (4). Hybridization with radiolabeled DNA fragments was carried out for 16 h at 60°C (4). The cloned dehydratase-specific 1,000-bp PCR products obtained during the PCR step were used as probes for capturing the corresponding clones in the correlating environmental library by colony hybridization. The probes were labeled with [α-32P]dATP by using the random-primer DNA labeling system (Gibco/BRL, Eggenstein, Germany) as recommended by the manufacturer.
(iv) Construction of plasmid pAK1.
In order to facilitate activity-based screening of the environmental libraries, the recombinant cosmid pRD1 containing the entire dha regulon of C. freundii (15, 16) was digested with BpiI and Bsp119I. Subsequently, the resulting 8.6-kb fragment, which harbored all of the genes necessary for anaerobic glycerol breakdown except the glycerol dehydratase gene, was treated with Klenow fragment and ligated into pSTV28 (Takara, Kyoto, Japan) previously linearized with SmaI. The resulting recombinant plasmid (pAK1) contained a replication origin of p15A that is compatible with the one of the vectors used for library construction.
(v) Complementation of the incomplete dehydratase-encoding DNA regions and high-level expression of the dehydratase genes.
The incomplete dehydratase-encoding gene regions of pAK3 to -5 were complemented by PCR-based approaches, using the missing part of the closely related genes encoding the α subunits of the dehydratases of C. freundii (pAK3) and S. enterica serovar Typhimurium (pAK4 and -5) for complementation. The first step was the separate amplification of the incomplete dehydratase-encoding regions of pAK3 to -5 by using the following sets of oligonucleotides: for pAK3, CGGTACCGAACTACGACAACATGTTTG (primer 3A) and TCACTGACTGCCTTTACGCAG (primer 3B); for pAK4, TCCGGTTATTCCGCGGTGCCGAACTAC (primer 4A) and TCAAGGATGCGATCGTCT (primer 4B); and for pAK5, TGCTGCGCCAGATAATTGAAGACGTAC (primer 5A) and ATCGTCACCTTTGACTTT (primer 5B). The missing parts of genes encoding the α subunit were amplified from pMS2 of C. freundii (41) or genomic DNA of S. enterica serovar Typhimurium, including overhangs (25 to 30 bases) complementary to the 5′ termini of the incomplete dehydratase-encoding gene regions. In order to achieve high-level expression and subsequent purification of the complemented glycerol and diol dehydratases by nickel affinity chromatography, synthetic sites were added (underlined in the sequences below), which allowed a directional cloning of the complemented dehydratase-encoding gene regions into pET100/D or pET101/D by employing the pET100 or pET101 directional TOPO expression kit (Invitrogen). For this purpose, the following sets of oligonucleotides were employed: for C. freundii (pAK3), CACCATGAGAAGATCAAAACGATTC (primer CFA) and CAAACATGTTGTCGTAGTTCGGTACCG (primer CFB); for S. enterica serovar Typhimurium (pAK4), CACCATGAGATCGAAAAGATTTGA (primer STA) and GTAGTTCGGCACGGCGGAATAACCGGA (primer STB); and for S. enterica serovar Typhimurium (pAK5), primer STA and GTACGTCTTCAATTATCTGGCGCAGCA (primer STC). The PCRs were performed as described for the amplification of the internal dehydratase-encoding fragments, but Herculase (Stratagene, San Diego, Calif.) was used instead of Taq as the DNA polymerase. The obtained PCR products were isolated, purified, and used as templates together with the corresponding PCR products derived from pAK3 to -5 for the subsequent assembly PCRs, which were performed in the presence of two primers that are complementary to the ends of the desired hybrid dehydratase-encoding gene regions. The primer pairs CFA-3B, STA-4B, and STA-5B were used for the complementation of the incomplete dehydratase-encoding regions of pAK3, pAK4, and pAK5, respectively. Finally, all obtained PCR products were cloned into pET100/D or pET101/D as recommended by the manufacturer (Invitrogen). In order to achieve high-level expression of the completely cloned dehydratase genes located on pAK2 and pAK7, the plasmids pAK2.1 and pAK7.1, respectively, were constructed by employing the same cloning kit. For this purpose, the coding regions were amplified from pAK2 and pAK7 by PCR using the following sets of primers: for pAK2.1, CACCATGAGAAGATCAAAACGATCC and TTACTGGCTGCCTTTACGCAG; and for pAK7.1, CACCATGAGAAGATCAAAACGATTTG and TTACTGGCTGCCTTTACGCAG.
Nucleotide sequence accession numbers.
The nucleotide sequences of the inserts of pAK2 to -5 and pAK7 have been deposited in the GenBank database under accession numbers AY205332 to AY205335 and AY205336, respectively.
RESULTS
Construction and selection of environmental DNA libraries.
The enrichments of glycerol-fermenting microorganisms were performed with samples derived from three environments. The first environment was sediment of the river Grone (Germany), the second environment was soil from a sugar beet field collected near Göttingen (Germany), and the third habitat consisted of sediment collected from the Solar Lake (Egypt). After inoculation of the enrichment cultures and growth for 1 day at 30°C, microorganisms were pelleted, used to inoculate fresh medium, and grown for another day. The enrichment of glycerol-fermenting microorganisms was confirmed in all cultures by detection of the characteristic product of glycerol fermentation, 1,3-propanediol, in all culture supernatants (data not shown). Subsequently, the cells and the remaining environmental matrix substances were harvested by centrifugation. The genomic DNA was isolated from the resulting pellets by direct lysis of the microorganisms present without prior removal of remaining environmental compounds and cloned into pSK(+) as described previously (23). The quality of the three different libraries produced was controlled by determination of the average insert size and the percentage of recombinant plasmids containing inserts. The three libraries revealed average insert sizes of 3.3 to 5.0 kb (Table 2). The percentage of plasmids containing inserts was approximately 70 to 80%. No significant differences between the different environmental samples were observed during the preparation of the libraries.
TABLE 2.
Screening of three environmental libraries for genes encoding coenzyme B12-dependent glycerol and diol dehydratases
| Library | Sample site | Average insert size (bp) | No. of E. coli clones tested by:
|
No. of dehydratase-positive E. coli clones by:
|
||
|---|---|---|---|---|---|---|
| Activity-based screening | Sequence-based screening | Activity-based screening | Sequence-based screening | |||
| I | River Grone | 3,300 | 80,000 | 32,000 | 1 (pAK8) | 2 (pAK6-7) |
| II | Sugar beet field | 5,000 | 240,000 | 65,000 | 1 (pAK2) | 2 (pAK4-5) |
| III | Solar Lake | 3,400 | 240,000 | 61,000 | 0 | 1 (pAK3) |
The presence of genes encoding glycerol and diol dehydratases in the libraries was verified by PCR. The PCRs were performed employing the isolated recombinant plasmids of the libraries as templates and degenerated primers, which were derived from regions conserved in all known sequences for dehydratases (Fig. 1). The expected 1,000-bp region was amplified in all libraries. In order to confirm that the PCR products were derived from dehydratase genes, the PCR products were cloned by the TA method and the sequences of several clones from each library were determined. Nucleotide sequence analyses of the cloned PCR products revealed that almost all harbored genes that are similar to genes encoding the structural subunits of coenzyme B12-dependent glycerol or diol dehydratases of enteric bacteria. The recorded identities ranged from 82 to 100% (data not shown). The similarity to dehydratases of enteric bacteria is advantageous for the subsequent analyses, since E. coli is the host for expression of the genes. Previous work has shown that high-level expression of genes encoding dehydratases from other enteric bacteria, such as C. freundii, S. enterica serovar Typhimurium, and K. oxytoca, is feasible in E. coli (6, 40, 41, 45, 46). In addition, E. coli recognizes the natural promoters of the dehydratase genes from other enteric bacteria, which is required for activity-based screening of the libraries.
Thus, these results revealed that the constructed libraries could serve as starting material for identification of the genes encoding the entire dehydratase. The screening strategies for obtaining these genes were based either on enzyme activity or on sequence.
Activity-based screening of environmental DNA libraries.
In order to perform an activity-based screening for genes encoding glycerol or diol dehydratases, an E. coli strain was constructed which contained and expressed all genes necessary for anaerobic glycerol breakdown of C. freundii except the genes coding for coenzyme B12-dependent glycerol dehydratase. The only way for such an E. coli strain to survive under anaerobic conditions in M9 medium with glycerol as the sole carbon and energy source is complementation by genes that confer a glycerol or diol dehydratase activity. To clone the region of the C. freundii dha regulon lacking the dehydratase genes, an 8.6-kb BpiI-Bsp119I fragment of the recombinant cosmid pRD1 (Table 1) was isolated and cloned into pSTV28 as described in Materials and Methods. Sequencing of the resulting recombinant plasmid (pAK1) confirmed that the genes encoding glycerol dehydratase are missing and the genes encoding the other three key enzymes of glycerol fermentation (glycerol dehydrogenase, dihydroxyacetone kinase, and 1,3-propanediol dehydrogenase) are present. Furthermore, the plasmid pAK1 possesses the replication origin of p15A and the chloramphenicol acetyltransferase gene. It is compatible with the vector [pSK(+)] used for construction of the environmental libraries, which harbors the replication origin of pMB1 and the β-lactamase gene. Thus, pAK1 and the recombinant plasmids of the libraries could be stably cotransformed to E. coli cells, and cotransformants were readily selected by resistance to both chloramphenicol and ampicillin. The E. coli strain ECL707 (43), which lacks the activities of the genes responsible for aerobic glycerol breakdown by E. coli, was used as the host for pAK1. As expected, E. coli ECL707/pAK1 showed no growth under anaerobic conditions on M9 agar plates with glycerol as the sole carbon and energy source. Growth was restored by supplementing the medium with 60 mM pyruvate. In order to test the production of the three remaining key enzymes of glycerol fermentation by E. coli ECL707/pAK1, the specific enzyme activities were determined in cell extracts of this strain grown in the presence of glycerol and pyruvate. The specific enzyme activities of glycerol dehydrogenase, dihydroxyacetone kinase, and 1,3-propanediol dehydrogenase were 4.57, 0.07, and 0.88 U/mg, respectively. These activities were in the same range as those in cell extracts of C. freundii and E. coli ECL707/pRD1, which harbors the entire dha regulon of C. freundii (28). Subsequently, the entire screening procedure was controlled by transferring pMS2 (41) or pFL1 (28) harboring the genes encoding glycerol dehydratase of C. freundii or Clostridium pasteurianum, respectively, into E. coli ECL707/pAK1. Both resulting E. coli strains, ECL707/pAK1/pMS2 and ECL707/pAK1/pFL1, showed growth under anaerobic conditions on M9 agar medium containing glycerol as the sole carbon and energy source, in contrast to the case for E. coli ECL707/pAK1. Thus, only recombinant E. coli strains harboring a gene conferring glycerol dehydratase activity could grow under the conditions employed. In addition, E. coli ECL707/pAK1/pMS2 and ECL707/pAK1/pFL1 exhibited glycerol dehydratase activity in crude extracts and produced 1,3-propanediol (data not shown). These results demonstrated that the above-described screening system is suitable for the recovery of genes conferring glycerol or diol dehydratase activity.
In order to identify novel glycerol and diol dehydratases, the three constructed environmental DNA libraries were used to transform E. coli ECL707/pAK1. Subsequently, the resulting recombinant E. coli strains were screened for dehydratase activity as described above. Two different E. coli clones out of approximately 560,000 tested clones were obtained during the initial screening procedure. In order to confirm that the glycerol fermentation-positive phenotype of both clones is plasmid-encoded, the recombinant plasmids were isolated, retransformed into E. coli, and the resulting clones were screened again on glycerol-containing M9 plates under anaerobic conditions. Both recombinant plasmids, designated pAK2 and pAK8, conferred a stable glycerol fermentation-positive phenotype to the resulting recombinant E. coli strains ECL707/pAK1/pAK2 and ECL707/pAK1/pAK8. Plasmid pAK2 was obtained from library II, and pAK8 was obtained from library I (Table 2). The insert sizes were 7,115 and 6,805 bp, respectively. Enzymatic analysis revealed glycerol dehydratase activity in crude extracts of E. coli ECL707/pAK1/pAK2 and ECL707/pAK1/pAK8. The recorded specific dehydratase activities were 0.8 and 0.9 U/mg, respectively (Table 3).
TABLE 3.
Characterization of pAK2 to pAK8 and the corresponding E. coli clones
| Plasmid or organism | Insert size (bp) | Size (bp) of gene encoding the following dehydratase subunit, molecular mass (kDa) of the corresponding gene product:
|
G+C content (%) of dehydratase-encoding DNA region | Dehydratase activity (U/mg) of the corresponding clone (U/mg)a | ||
|---|---|---|---|---|---|---|
| α | β | γ | ||||
| pAK2 | 7,115 | 1,668; 60.5 | 585; 21.3 | 429; 16.1 | 55.73 | 0.8 |
| pAK3 | 2,157 | 729b; NDc | 585; 21.3 | 429; 16.1 | 54.98 | 0 |
| pAK4 | 1,750 | 741b; ND | 678; 24.1 | 363b; ND | 53.66 | 0 |
| pAK5 | 4,236 | 45b; ND | 678; 24.2 | 519; 19.4 | 52.29 | 0 |
| pAK6 | 4,332 | ND | ND | ND | ND | 0.7 |
| pAK7 | 6,295 | 1,668; 60.4 | 585; 21.4 | 429; 16.1 | 56.33 | 1.2 |
| pAK8 | 6,805 | ND | ND | ND | ND | 0.9 |
| C. freundii | 1,668; 60.4 | 585; 21.5 | 429; 16.1 | 55.92 | ND | |
| S. enterica serovar Typhimurium | 1,665; 60.3 | 675; 24.2 | 522; 19.1 | 56.65 | ND | |
Specific coenzyme B12-dependent dehydratase activity with glycerol as the substrate in crude extracts.
Partial codon sequence.
ND, not determined.
Sequence-based screening of environmental DNA libraries.
For the isolation of dehydratase-encoding genes, the three constructed libraries were also screened by a sequence-based approach. Therefore, the recombinant plasmids of the three constructed libraries were transformed into E. coli DH5α and grown on LB-ampicillin agar plates at 37°C, and the resulting colonies were imprinted on filter disks. Subsequently, E. coli clones harboring genes encoding glycerol or diol dehydratases were identified by colony hybridization. The cloned dehydratase-specific 1,000-bp PCR products (see above) were used as probes for capturing the corresponding clones in the correlating environmental library. In this way, five positive clones (E. coli DH5α/pAK3 to E. coli DH5α/pAK7) were recovered by screening of approximately 158,000 E. coli colonies (Table 2). Two plasmids were obtained from library I, two were obtained from library II, and one was obtained from library III (Table 2). The insert sizes of pAK3 to pAK7 were in the range of 1,750 to 6,295 bp (Table 3).
The plasmids pAK3 to -7 were tested for complementation of E. coli ECL707/pAK1 (see above). The transformation of pAK6 and -7 into E. coli ECL707/pAK1 restored growth in M9 medium containing glycerol as the sole carbon and energy source under anaerobic conditions, and significant glycerol dehydratase activity was detected in crude extracts (Table 3). The other three recombinant plasmids failed to complement E. coli ECL707/pAK1. In addition, dehydratase activity was not detectable in crude extracts of the corresponding clones. Since the sequence-based screening procedure is not selective for full-length genes (14), the lack of activity of the latter E. coli clones might be due to an incompletely cloned dehydratase-encoding DNA region.
Molecular analyses.
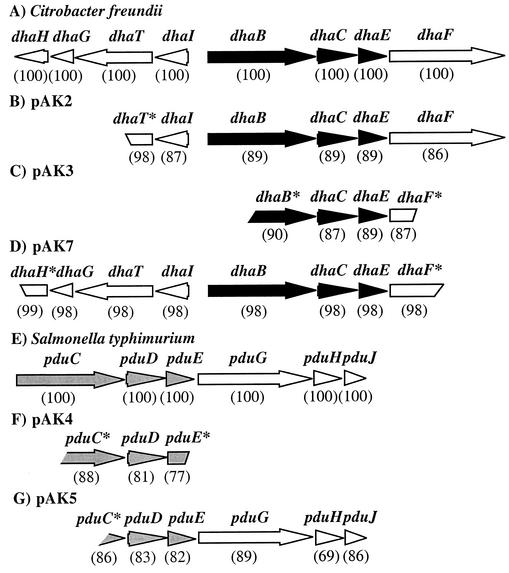
The inserts of pAK2 to pAK8 were sequenced and compared to the sequences in the National Center for Biotechnology Information databases. All known coenzyme B12-dependent glycerol and diol dehydratases are encoded by three structural genes (Fig. 1) (17). Partial sequencing of pAK6 and pAK8 revealed that the two inserts were identical to the glycerol dehydratase-encoding gene region of C. freundii. Therefore, these plasmids were not studied further. Sequencing of the remaining five plasmids showed that all contained open reading frames encoding subunits of coenzyme B12-dependent glycerol or diol dehydratases that are different from the known genes (Fig. 2). Two of the plasmids (pAK2 and pAK7) contained entire genes encoding the three structural subunits of dehydratases; the other three plasmids (pAK3 to -5) lacked different parts of the gene encoding the large subunit of dehydratase (Fig. 2; Table 3). The insert of pAK4 also lacked the region encoding the C-terminal part of the small subunit. These results are in accordance with those obtained during enzymatic analyses, since only pAK2 and pAK7 conferred B12-dependent dehydratase activity on the corresponding E. coli clones. Generally, the three structural genes encoding dehydratases located on pAK2 to -5 and pAK7 were arranged in the same order (large, medium, and small genes) as in the genomes of known dehydratase-containing microorganisms such as C. freundii, S. enterica serovar Typhimurium, Clostridium pasteurianum, Lactobacillus collinoides, and K. pneumoniae (6, 17, 29, 39, 41, 44). The nucleotide sequences of the presumptive dehydratase-encoding genes located on pAK2, -3, and -7 revealed striking similarities to genes encoding the three structural subunits of glycerol dehydratases from other organisms, such as C. freundii (41), Clostridium pasteurianum (29), and K. pneumoniae (44), whereas the dehydratase genes encoded by pAK4 and -5 were strongly related to the three genes encoding diol dehydratases of characterized microorganisms such as S. enterica serovar Typhimurium (6) and K. oxytoca (45) (data not shown). In addition, the lengths of the entirely cloned genes for the α, β, and γ subunits and the deduced molecular masses of the gene products were highly similar to those of the corresponding genes and gene products for glycerol or diol dehydratases of the above-mentioned microorganisms (Table 3). The three dehydratase-encoding genes (dhaBCE) of pAK2, -3, and -7 were most similar to the dhaBCE genes of C. freundii, which encode a glycerol dehydratase (41). The recorded identities were 89, 87 to 90, and 98%, respectively (Fig. 2). This resulted in 24, 21, and 7 amino acid substitutions, respectively, over the length of all partially or entirely cloned dehydratase-encoding subunits. In addition, the G+C content of the dehydratase-encoding gene regions of pAK2, -3, and -7 was similar to the one of C. freundii (Table 3). In comparison to the cloned presumptive glycerol dehydratase-encoding genes, the DNA sequences of the genes (pduCDE) encoding the three subunits of the putative diol dehydratases located on pAK4 and -5 were more distantly related to known genes coding for this type of enzyme. The dehydratase genes of pAK4 and -5 exhibited the highest similarities to the pduCDE genes of S. enterica serovar Typhimurium (77 to 88% and 82 to 86% identity, respectively), which encode the corresponding subunits of a diol dehydratase (6). The amino acid sequences deduced from the completely and partially cloned genes revealed 58 (pAK4) and 41 (pAK5) substitutions compared to the amino acid sequences of the diol dehydratase subunits from S. enterica serovar Typhimurium. In addition, the G+C contents of the cloned dehydratase-encoding DNA regions of pAK4 and -5 (53.66 and 52.29%, respectively) were significantly lower than that of S. enterica serovar Typhimurium (56.65%).
FIG. 2.
Genetic organization of the DNA region carrying genes participating in glycerol fermentation (A to D) and part of the region carrying the genes for 1,2-propanediol utilization (E to G). (A) C. freundii (15, 16, 41); (B) pAK2 (this study); (C) pAK3 (this study); (D) pAK7 (this study); (E) S. enterica serovar Typhimurium (6, 17); (F) pAK4 (this study); (G), pAK5 (this study). Arrows and arrowheads represent length, location, and orientation of potential genes. Black and gray arrows indicate open reading frames encoding homologs of the three structural subunits of glycerol dehydratase from C. freundii (dhaBCE) and of diol dehydratase from S. enterica serovar Typhimurium (pduCDE), respectively. Interrupted arrows and asterisks represent incomplete genes. The nucleotide sequence identities with respect to the corresponding genes of the dha regulon from C. freundii (A to D) and the pdu operon of S. enterica serovar Typhimurium (E to G) are shown in parentheses. dhaBCE and pduCDE encode the three structural subunits of glycerol dehydratase and diol dehydratase, respectively; dhaFG and pduGH encode reactivation factors of B12-dependent dehydratases; dhaHI encode a putative adenosyltransferase; dhaT encodes 1,3-propanediol dehydrogenase; and pduJ encodes a paralog of E. coli detox protein.
Several amino acid residues are essential for the binding of cobalamin by the diol dehydratase of K. oxytoca (42). The amino acid residues Thrα172, Gluα205, Thrα222, Aspα234, and Metα373 in the α subunit and Aspβ112, Asnβ150, and Glnβ156 in the β subunit form hydrogen bonds to the peripheral amide side chains of the corrin ring or participate in hydrogen bonding (42). Corresponding amino acid residues are also conserved in glycerol dehydratase of K. pneumoniae (51). Amino acid residues corresponding to these residues were found in all amino acid sequences deduced from the cloned parts of the genes which encode the α and β subunits of the dehydratases located on pAK2 to -5 and pAK7. In addition to the above-mentioned conserved residues, the hydroxyl group Serβ122 is hydrogen bonded to the amide oxygen of the g-acetamide side chain of the corrin ring in glycerol dehydratase of K. pneumoniae. In diol dehydratase of K. oxytoca, the corresponding residue is Proβ155, which cannot form the hydrogen bond (51). This difference between glycerol and diol dehydratases was also present in the sequences of the cloned genes encoding the β subunit of dehydratase. The β-subunits encoded by pAK2, -3, and -7 possessed a serine residue in the corresponding position, whereas the β subunits encoded by pAK4 and -5 harbored a proline residue in the corresponding position. Thus, this result confirmed that glycerol dehydratases were encoded by pAK2, -3, and -7 and that diol dehydratases were encoded by pAK4 and -5. The relationship of the cloned dehydratase-encoding DNA regions to either glycerol dehydratases or diol dehydratases was also revealed by inspection of the genes and the gene organization in the neighborhood of the dehydratase genes. Glycerol dehydratase is a key enzyme for the dihydroxyacetone pathway of glycerol fermentation and its genes are located in the dha regulon, whereas diol dehydratase is a key enzyme for the anaerobic degradation of 1,2-diols and its genes are part of the pdu operon (17). In the vicinity of the dehydratases encoded by pAK2, -3, and -7, genes which are typical for the dha regulon were located, such as dhaT (pAK2 and pAK7), dhaI (pAK2 and pAK7), dhaF (pAK2, -3, and -7), and dhaG (pAK7) (Fig. 2). The products of the latter two genes are involved in reactivation of glycerol dehydratase (40). On the other hand, downstream of the dehydratase-encoding genes of pAK5, homologs of the pduGHJ genes of S. enterica serovar Typhimurium were located, which are characteristic for the pdu operon (Fig. 2). In addition, these genes were arranged in the same order as in S. enterica serovar Typhimurium and other 1,2-propanediol-utilizing enteric bacteria (17). Such a comparison cannot be done for pAK4, since only parts of the dehydratase-encoding gene region were cloned (Fig. 2).
In summary, two complete and three incomplete dehydratase-encoding gene regions were located on the inserts of pAK2 to -5 and pAK7. The sequence analyses indicated that three of these regions (pAK2, -3, and -7) coded for glycerol dehydratases and that two (pAK4 and -5) coded for diol dehydratases.
Complementation of the incompletely cloned dehydratase-encoding gene regions.
In order to obtain enzyme activity for the subsequently performed biochemical characterization of the dehydratases, the incomplete dehydratase-encoding gene regions of pAK3 to -5 were complemented by PCR-based approaches by using the missing parts of the closely related genes encoding the large dehydratase subunits of C. freundii (pAK3) and S. enterica serovar Typhimurium (pAK4 and -5) for assembly (see Materials and Methods). This strategy was successful for pAK3 and pAK5. The targeted hybrid PCR products containing the complemented entire dehydratase-encoding gene regions were recovered (data not shown). All attempts to generate such a hybrid PCR product of the dehydratase-encoding gene region located on pAK4 failed.
To obtain high-level expression of the genes encoding the dehydratases, the hybrid PCR products derived from pAK3 and pAK5 were cloned into the expression vectors pET100/D and pET101/D, respectively, thereby placing the dehydratase genes under control of the isopropyl-1-thio-β-d-galactoside (IPTG)-inducible T7 promoter and adding a sequence encoding a His6 tag. The fidelity of the PCR products and the cloning steps was confirmed by sequencing of the resulting constructs, pAK3.1 and pAK5.1, respectively. Both plasmids were transformed into E. coli Bl21. Subsequently, the corresponding E. coli strains Bl21/pAK3.1 and Bl21/pAK5.1 were grown in LB medium, and the production of the dehydratases was induced by addition of IPTG. Analyses of the specific dehydratase activities in crude extracts of both recombinant E. coli strains revealed that the enzyme produced by E. coli Bl21/pAK5.1 was active (8.7 U/mg), whereas the enzyme formed by E. coli Bl21/pAK3.1 was inactive. Attempts to obtain enzyme activity of the latter dehydratase by variation of the growth and induction conditions or by changing the cloning vector failed (data not shown).
Purification, molecular masses, substrate specificities, and kinetic parameters of the environmentally derived dehydratases.
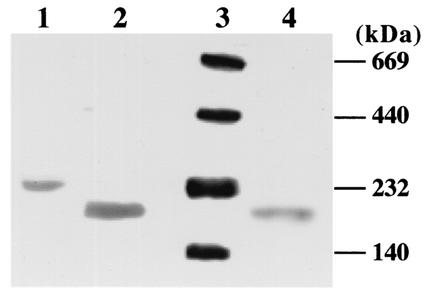
In order to facilitate purification of the other dehydratases encoded by pAK2 and pAK7, the dhaBCE genes of these plasmids were also placed under the control of the T7 promoter by cloning in the expression vectors pET100/D and pET101/D, respectively. In addition, sequences encoding His6 tags provided by these vectors were added to the 5′ end (pAK2) or the 3′ end (pAK7) of the coding regions during cloning. The resulting constructs, pAK2.1 and pAK7.1, respectively, were transformed into E. coli Bl21. The gene products were produced in E. coli, and the specific dehydratase activities were measured in crude extracts as described above. Both recombinant E. coli strains Bl21/pAK2.1 and Bl21/pAK7.1 exhibited significant coenzyme B12-dependent dehydratase activity with glycerol as the substrate (14.5 and 14.1 U/mg, respectively). Subsequently, the dehydratases produced by E. coli Bl21/pAK2.1, E. coli Bl21/pAK7.1, and E. coli Bl21/pAK5.1 (see above) were purified from cell extracts by affinity chromatography on Ni-nitrilotriacetic acid agarose. The specific dehydratase activities of the final enzyme preparations with glycerol as the substrate were 119.5, 111.8, and 96.4 U/mg, respectively. Analytical sodium dodecyl sulfate-polyacrylamide gel electrophoresis of the three purified enzymes revealed that all three typical dehydratase subunits (α, β, and γ) were present (data not shown). The molecular masses of the three subunits (approximately 60 kDa [α], 21 to 24 kDa [β], and 16 to 19 kDa [γ]) were in the same range as described for the corresponding subunits of glycerol and diol dehydratases from other organisms (6, 17, 29, 41, 44, 45, 46) and as expected from the sequences (Table 3). Separation of the purified dehydratases of E. coli Bl21/pAK2.1, E. coli Bl21/pAK5.1, and E. coli Bl21/pAK7.1 by gradient gel polyacrylamide electrophoresis under nondenaturing conditions and activity staining gave single bands corresponding to native molecular masses of 190,000, 235,000, and 190,000 Da, respectively, including the amino acid residues added during the subcloning procedures (Fig. 3). No activity staining was observed when coenzyme B12 was omitted from the reaction mixture (data not shown). These results indicated a subunit composition of α2β2γ2 for all three environmentally derived dehydratases, which is in agreement with the compositions reported for coenzyme B12-dependent glycerol and diol dehydratases derived from characterized microorganisms (17, 29, 41, 46).
FIG. 3.
Nondenaturing polyacrylamide gel electrophoresis and activity staining of the dehydratases produced by E. coli strains Bl21/pAK2.1, Bl21/pAK5.1, and Bl21/pAK7.1. The purified enzymes (2 μg) were subjected to electrophoresis under nondenaturing conditions on polyacrylamide gradient slab gels (4 to 28%). The protein bands were stained as described in Materials and Methods. Lanes: 1, purified dehydratase of E. coli Bl21/pAK5.1; 2, purified dehydratase of E. coli Bl21/pAK2.1; 3, molecular mass markers; 4, purified dehydratase of E. coli Bl21/pAK7.1.
The dehydratases produced in E. coli Bl21/pAK2.1, E. coli Bl21/pAK5.1, and E. coli Bl21/pAK7.1 were capable of catalyzing the coenzyme B12-dependent conversion of glycerol, 1,2-propanediol, and 1,2-ethanediol to 3-hydroxypropionaldehyde, propionaldehyde, and acetaldehyde, respectively, as is typical of all characterized coenzyme B12-dependent glycerol and diol dehydratases from other organisms (17). Like glycerol dehydratases of other organisms, the presumptive glycerol dehydratases of E. coli Bl21/pAK2.1 and E. coli Bl21/pAK7.1 were most active with glycerol, followed by 1,2-propanediol and 1,2-ethanediol. In comparison, the presumptive diol dehydratase of E. coli Bl21/pAK5.1 preferred 1,2-propanediol as a substrate, like diol dehydratases of other organisms (data not shown).
To characterize the catalytic properties of the dehydratases produced by E. coli Bl21/pAK2.1, E. coli Bl21/pAK5.1, E. coli Bl21/pAK7.1, and C. freundii (control), the kinetic parameters of the purified enzymes were determined (Table 4). The Km values of the glycerol dehydratases purified from E. coli Bl21/pAK2.1 and E. coli Bl21/pAK7.1 for glycerol were 1.3 and 1.7 mM, respectively. These were in reasonable agreement with the Km values (0.73 to 1.6 mM) that have been reported for coenzyme B12-dependent glycerol and diol dehydratases derived from enteric bacteria (5, 35, 46), whereas the Km values of the hybrid diol dehydratase of E. coli Bl21/pAK5.1 and the glycerol dehydratase of C. freundii (0.4 and 0.5 mM, respectively) were lower than the reported values. The kcat values (turnover numbers) of the three environmentally derived dehydratases for glycerol were in the same range (354 to 378 s−1). The greatest turnover number (425 s−1) was recorded for the glycerol dehydratase of C. freundii (Table 4). The analyses of the catalytic efficiencies, calculated as kcat/Km, revealed significant differences between the four dehydratases. The catalytic efficiencies of the hybrid diol dehydratase produced by E. coli Bl21/pAK5.1 and the glycerol dehydratase of C. freundii were 3.2 to 4.5 and 2.9 to 4.1 times, respectively, greater than those of the glycerol dehydratases derived from E. coli Bl21/pAK2.1 and E. coli Bl21/pAK7.1 (Table 4).
TABLE 4.
Apparent kinetic parameters of the dehydratases purified from E. coli Bl21/pAK2.1, E. coli Bl21/pAK5.1, E. coli Bl21/pAK7.1, and C. freundii for the substrate glycerola
| Enzyme | Sp act (U/mg) | Km (mM) | kcat (s−1) | kcat/Km (mM−1 · s−1) |
|---|---|---|---|---|
| E. coli Bl21/pAK2.1 | 119.5 | 1.3 | 378 | 291 |
| E. coli Bl21/pAK5.1 | 96.4 | 0.4 | 378 | 944 |
| E. coli Bl21/pAK7.1 | 111.8 | 1.7 | 354 | 208 |
| C. freundii | 134.1 | 0.5 | 425 | 850 |
The specific dehydratase activities and Km values of the purified enzymes for the substrate glycerol were determined as described in Materials and Methods. The kcat values were calculated based on the specific activities and the determined molecular weights of the native dehydratases (pAK2.1, 190,000 [this study]; pAK5.1, 235,000 [this study]; pAK7.1, 190,000 [this study]; C. freundii, 190,000 [29]).
Suicide inactivation and reactivation of the environmentally derived dehydratases.
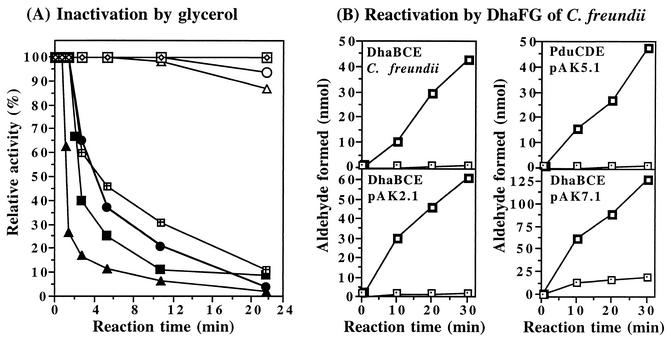
The dehydratases produced by E. coli Bl21/pAK2.1, E. coli Bl21/pAK5.1, E. coli Bl21/pAK7.1, and C. freundii (control) were subject to suicide inactivation by glycerol, which is characteristic of all known coenzyme B12-dependent glycerol and diol dehydratases (5, 17, 48, 50). As depicted in Fig. 4A, the activities of the enzymes decreased to 17, 65, 40, and 60%, respectively, within 3 min of incubation in the presence of glycerol. The remaining activities after 22 min were 2, 4, 9, and 11%, respectively. No significant inactivation occurred with 1,2-propanediol as the substrate. Thus, the presumptive diol dehydratase produced by E. coli Bl21/pAK5.1 and the glycerol dehydratase of C. freundii were best with respect to resistance against suicide inactivation by glycerol.
FIG. 4.
Inactivation by glycerol (A) and DhaFG-dependent reactivation (B) of the dehydratases encoded by pAK2.1, pAK5.1, and pAK7.1. The activities of the dehydratase with glycerol or 1,2-propanediol as the substrates (A) and the amount of propionaldehyde formed (B) were determined as described in Materials and Methods. (A) Dehydratase activity during glycerol dehydration (pAK2.1, ▴; pAK5.1, •; pAK7.1, ▪; C. freundii, ⊠) and corresponding dehydratase activity during 1,2-propanediol dehydration (pAK2.1, ▵; pAK5.1, ○; pAK7.1, □; C. freundii, ⋄). Activities are expressed relative to the initial activities obtained with glycerol (pAK2.1, 44.2 U/ml; pAK5.1, 15.7 U/ml; pAK7.1, 48.8 U/ml; C. freundii, 27.3 U/ml) or 1,2-propanediol (pAK2.1, 14.0 U/ml; pAK5.1, 16.4 U/ml; pAK7.1, 10.7 U/ml; C. freundii, 7.9 U/ml). (B) Glycerol-inactivated dehydratases were prepared as described previously (40). The glycerol-inactivated dehydratases (1 to 2 μg) were incubated with 1,2-propanediol as the substrate for the indicated time periods with 10 μg of purified DhaF-DhaG complex of C. freundii, 21 μM coenzyme B12, and 12 mM ATP/Mg2+ (□) or without DhaF-DhaG complex and with 21 μM coenzyme B12 and 12 mM ATP/Mg2+ (⊡).
For the glycerol dehydratase of C. freundii and other dehydratases, a complex of two proteins was identified as the reactivation factor of glycerol-inactivated dehydratases (26, 40, 47). The ability of the purified reactivation factor (DhaFG) of C. freundii to reactivate the glycerol-inactivated dehydratases produced by E. coli Bl21/pAK2.1, E. coli Bl21/pAK5.1, and E. coli Bl21/pAK7.1 was examined with 1,2-propanediol as the substrate after inactivation of the enzymes by glycerol (Fig. 4B). In the presence of ATP, Mg2+, and coenzyme B12, the purified DhaFG complex was able to reactivate the three environmentally derived dehydratases and the glycerol-inactivated glycerol dehydratase of C. freundii (control). The propionaldehyde formation increased linearly with time in the presence of the reactivation factor, whereas no significant increase in propionaldehyde formation was recorded in the absence of the DhaFG complex (Fig. 4B). No reactivation of the enzymes was observed also in the absence of ATP, Mg2+, or coenzyme B12 (data not shown). Subsequently, the extents of reactivation were determined by comparison of the amounts of propionaldehyde formed by the glycerol-inactivated dehydratases in the presence of the reactivation factor with those formed by the noninactivated dehydratases. The extents of reactivation were 81% (pAK2.1), 62% (pAK5.1), 85% (pAK7.1), and 89% (C. freundii) after 30 min of incubation (data not shown).
Inhibition of the dehydratase activity by 1,3-propanediol.
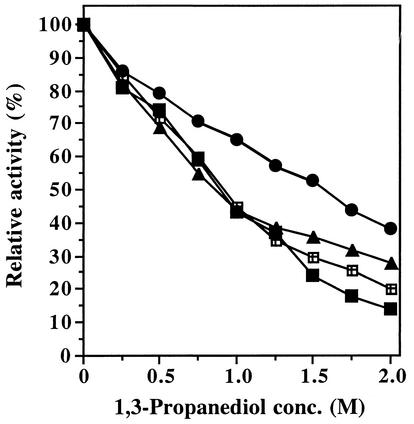
The industrial production of 1,3-propanediol by the recombinant E. coli strain developed by Genencor and DuPont is performed by employing a batch fermentation process. The final concentration of 1,3-propanediol is in the range of 2 M (11). It has been mentioned in one report that 1,3-propanediol is not a substrate for the reactions catalyzed by coenzyme B12-dependent dehydratases but manifests a competitive inhibition towards the substrates dehydrated by dehydratases (35). In order to investigate inhibition of the purified dehydratases, the enzyme activities were determined in the presence of increasing 1,3-propanediol concentrations with 100 mM 1,2-propanediol as the substrate. The purified dehydratases of E. coli Bl21/pAK2.1, E. coli Bl21/pAK5.1, E. coli Bl21/pAK7.1, and C. freundii were sensitive to inhibition by 1,3-propanediol. As depicted in Fig. 5, the activity of the enzymes decreased to 81 to 86% in the presence of 0.25 mM 1,3-propanediol and to approximately 44% (pAK2.1, pAK7.1, and C. freundii) and 66% (pAK5.1) in the presence of 1 M 1,3-propanediol. The glycerol dehydratase of C. freundii and the presumptive glycerol dehydratases encoded by pAK2.1 and pAK7.1 showed quite similar inhibition profiles. In comparison, the presumptive diol dehydratase encoded by pAK5.1 was less sensitive to inhibition by 1,3-propanediol at all tested concentrations; even in the presence of 2 M 1,3-propanediol, the remaining enzyme activity was approximately 40% of that recorded in the absence of 1,3-propanediol. Similar results were obtained when 100 or 2.5 mM glycerol instead of 1,2-propanediol was used as the substrate (data not shown). The apparent Ki values for 1,3-propanediol determined with 100 mM 1,2-propanediol as the substrate were 0.85 M (pAK2.1), 1.36 M (pAK5.1), 0.93 M (pAK7.1), and 0.93 M (C. freundii). Thus, the hybrid diol dehydratase purified from E. coli Bl21/pAK5.1 showed the greatest resistance against inhibition by 1,3-propanediol of all tested enzymes.
FIG. 5.
Inhibition of the dehydratases encoded by pAK2.1, pAK5.1, and pAK7.1 by 1,3-propanediol. The dehydratase activity was determined as described in Materials and Methods with 100 mM 1,2-propanediol as the substrate in the presence of increasing concentrations of 1,3-propanediol (pAK2.1, ▴; pAK5.1, •; pAK7.1, ▪; C. freundii, ⊠). Activities are expressed relative to the initial activities obtained in the absence of 1,3-propanediol (pAK2.1, 14.0 U/ml; pAK5.1, 28.3 U/ml; pAK7.1, 26.1 U/ml; C. freundii, 9.8 U/ml).
DISCUSSION
Classical enrichment of microorganisms with the desired traits and new molecular strategies relying on direct isolation and insertion of environmental (metagenomic) DNA into libraries and then screening for the targeted genes have both proven to be powerful tools for the identification of novel biocatalysts, natural products, and new molecular structures (9, 12, 19, 21, 22, 23, 25, 30, 37, 38). In this study, we have combined both approaches for the isolation of genes encoding coenzyme B12-dependent glycerol and diol dehydratases. These enzymes are of considerable industrial importance, since they catalyze the limiting step of biotechnological 1,3-propanediol production (1, 2, 7, 17).
We were able to isolate seven plasmids (pAK2 to -8) by activity- and sequence-based screening of metagenomic libraries, which were constructed from environmental samples after enriching dehydratase-containing microorganisms by incubation with glycerol as the sole carbon and energy source under anaerobic conditions. This enrichment strategy has been chosen since the activity of the targeted dehydratases is essential for growth under these conditions (2, 7, 13, 17, 24, 28, 49). DNA sequencing of the seven plasmids revealed striking similarities to the genes encoding coenzyme B12-dependent glycerol and diol dehydratases and to the gene organization of the dehydratase-encoding DNA regions of glycerol- and diol-fermenting microorganisms such as C. freundii and S. enterica serovar Typhimurium. Two of the dehydratase-encoding gene regions (pAK6 and pAK8) were identical to that of C. freundii and were not studied further. The sequences of the other five showed differences (2 to 23%) from known dehydratase genes (Fig. 2). The greatest differences from known dehydratase genes were recorded for the partially cloned presumptive diol dehydratase-encoding gene regions of pAK4 and -5, which were most similar to the corresponding region of S. enterica serovar Typhimurium. In both cases, the G+C content of this region was significantly lower than that in S. enterica serovar Typhimurium. This indicated a codon usage of the unknown microorganisms from which the DNA fragments of pAK4 and -5 were derived that is different from that in S. enterica serovar Typhimurium.
The high degree of identity to known genes encoding the targeted type of enzyme is different from results from screening programs of metagenomic libraries, which have been prepared without enrichment steps. Most of the genes and the corresponding gene products discovered by screening of unbiased environmental libraries are entirely novel, similar to genes with unknown functions, or weakly related or unrelated to known genes encoding the targeted functions (9, 22, 23, 37). There may be several reasons for the high identity of the isolated dehydratase genes to the corresponding genes from enteric bacteria. Generally, the use of enrichment steps is associated with the loss of microbial diversity present in environmental samples and favors the fast-growing and culturable part of a microbial population such as enteric bacteria (10). A similar preference for the isolation of genes related to enteric bacteria by combining enrichments with metagenomic techniques has been reported for genes involved in biotin synthesis (19) and formation of carbonyls from short-chain polyols (27). Nevertheless, the results of both studies showed that a diverse set of genes conferring the targeted reaction is still recovered by employing enrichments.
In the case of the plasmids derived from the activity screen (pAK2 and pAK8), the similarity to dehydratase genes from enteric bacteria could also be a result of the screening procedure, which was based on complementation of an E. coli mutant. This allowed the detection of cloned genes whose promoters were recognized by the E coli host strain. The use of other hosts might have increased the diversity and the number of the resulting clones. However, in almost all published reports E. coli has been used as the host for metagenomic libraries during activity-based screening programs (9, 12, 14, 22, 23, 30, 37), since a variety of molecular tools are available for this organism. In addition, a recombinant E. coli strain is employed for the industrial-scale production of 1,3-propanediol (11). A more important reason for the similarity to known genes may originate from the targeted enzyme class. All coenzyme B12-dependent glycerol and diol dehydratases from individual microorganisms characterized so far, such as those from C. freundii (41), Clostridium pasteurianum (29), K. oxytoca (45), K. pneumoniae (44), L. collinoides (39), and S. enterica serovar Typhimurium (6), are encoded by three genes that are transcribed in the following order: large gene (α subunit), medium gene (β subunit), and small gene (γ subunit). More important, each subunit has significant amino acid sequence similarity to the analogous subunit from the other organisms. The amino acid sequence similarities vary from approximately 54 to 99%. This indicated that the structure and the sequence of B12-dependent dehydratases were highly conserved during evolution and that all of these enzymes were derived from a common ancestor.
In three cases, we were able to obtain enzyme activity and subsequently to purify the environmentally derived dehydratases. The molecular masses of both presumptive glycerol dehydratases purified from E. coli Bl21/pAK2.1 and E. coli Bl21/pAK7.1 and the hybrid diol dehydratase purified from E. coli Bl21/pAK5.1 were 190 and 235 kDa, respectively. This is in accordance with the native molecular masses reported for other glycerol dehydratases, such as GldABC of K. pneumoniae (188 kDa) or DhaBCE of C. freundii (190 kDa), and for other diol dehydratases, such as PddABC of K. oxytoca (220 kDa). All three enzymes were sensitive to competitive inhibition by 1,3-propanediol, but the diol dehydratase located on pAK5.1 was less sensitive than the other tested enzymes. In addition, all enzymes showed substrate spectra and substrate preferences typical for coenzyme B12-dependent glycerol and diol dehydratases.
Glycerol and diol dehydratases belong to class II of coenzyme B12-containing enzymes, which bind the cobalamin in the base-on form (42). Binding of the coenzyme to the apoenzyme activates the Co-C bond of the coenzyme, and the radical reaction is initiated by substrate-induced homolytic cleavage of the Co-C bond. The radical intermediates formed during the catalytic cycle must maintain their high reactivity at the active site and must become extinct in the only way destined for the reaction. Once a radical intermediate is quenched by side reactions or escapes from the active site, regeneration of the coenzyme is not feasible. This leads not only to cessation of the catalytic cycle but also to inactivation of the enzyme, since the modified coenzyme remains tightly bound to the enzyme and is not exchangeable with free intact coenzyme B12. Such undesirable side reactions occur in the case of glycerol and diol dehydratases, and these enzymes are subject to mechanism-based suicide inactivation by the growth substrate glycerol and other substrates (5). The diol dehydratase encoded by pAK5.1 and the glycerol dehydratases encoded by pAK2.1 and pAK7.1 were rapidly inactivated by glycerol (Fig. 4A). The diol dehydratase was less sensitive to glycerol inactivation than both glycerol dehydratases. In addition, the inhibition profile of the diol dehydratase was more similar to that of the glycerol dehydratase of C. freundii. This result was surprising, since it has been established for the B12-dependent dehydratases of Klebsiella that diol dehydratase undergoes inactivation by glycerol at a higher rate than glycerol dehydratase (5, 36). It has been proposed that this difference is connected to the different affinities for the S and R isomers of the substrate. The glycerol dehydratase shows almost equal affinity toward the S and R isomers, whereas diol dehydratase prefers the S isomer (51). It has been reported that the dehydratase-(R)-glycerol complex is predominantly responsible for the product-forming reaction, while the dehydratase-(S)-glycerol complex results primarily in the inactivation reaction (5). Therefore, the less marked preference of the glycerol dehydratase for the S isomer explains why it is inactivated by glycerol at a lower rate than the diol dehydratase (5, 51). Thus, it can be concluded that the environmentally derived diol dehydratase purified from E. coli Bl21/pAK5.1 behaves more like a glycerol dehydratase with respect to isomer preference.
Reactivation of glycerol-inactivated dehydratases is required for anaerobic growth on glycerol, since glycerol dehydratases or the mutually related diol dehydratases are essential for glycerol fermentation of microorganisms such as C. freundii, K. pneumoniae, and Clostridium pasteurianum (7, 17, 28, 49). It has been shown that glycerol-inactivated glycerol and diol dehydratases undergo rapid reactivation by exchange of the modified coenzyme for intact coenzyme B12 in the presence of ATP, Mg2+, and a heterodimeric protein complex, i.e., DhaFG of C. freundii (40), DdrAB of K. oxytoca (32, 47), and GdrAB of K. pneumoniae (26). Genes encoding homologs of these reactivating factors are located in the neighborhood of all known genes encoding the three structural subunits of B12-dependent glycerol and diol dehydratases. Such genes (dhaFG or pduGH) were also partially or entirely present in the sequences derived from the inserts of pAK2, pAK3, pAK5, and pAK7 (Fig. 2). The requirement of the three characterized environmentally derived coenzyme B12-dependent dehydratases for a reactivating factor was confirmed by the ability of the purified DhaFG complex of C. freundii to cross-activate the glycerol-inactivated enzymes. The diol dehydratase was reactivated to a lesser extent than both glycerol dehydratases. This is in accordance with our previous results with the reactivation factor of C. freundii, since it prefers glycerol dehydratases of enteric bacteria as substrates (40).
In conclusion, the hybrid diol dehydratase produced by E. coli Bl21/pAK5.1 revealed the best properties of all tested enzymes, including glycerol dehydratase of C. freundii, for an application in the biotechnological production of 1,3-propanediol with recombinant E. coli strains with respect to catalytic efficiency, resistance against suicide inactivation by glycerol, and inhibition by 1,3-propanediol. One drawback is the reactivation of the glycerol-inactivated diol dehydratase by the reactivation factor DhaFG of C. freundii, since the extent of reactivation was the least of all tested enzymes. To overcome this problem, reactivation factors specific for diol dehydratases, such as DdrAB or PduGH, can be employed.
Acknowledgments
The work was supported by a grant from Genencor International and DuPont.
REFERENCES
- 1.Abbad-Andaloussi, S., E. Guedon, E. Spiesser, and H. Petitdemange. 1996. Glycerol dehydratase activity: the limiting step for 1,3-propanediol production by Clostridium butyricum DSM 5431. Lett. Appl. Microbiol. 22:311-314. [Google Scholar]
- 2.Ahrens, K., K. Menzel, A. Zeng, and W. Deckwer. 1998. Kinetic, dynamic, and pathway studies of glycerol metabolism by Klebsiella pneumoniae in anaerobic continuous culture. III. Enzymes and fluxes of glycerol dissimilation and 1,3-propanediol formation. Biotechnol. Bioeng. 59:544-552. [DOI] [PubMed] [Google Scholar]
- 3.Andersson, L. O., H. Borg, and M. Mikaelsson. 1972. Molecular weight estimation of proteins by electrophoresis in polyacrylamide gel of graded porosity. FEBS Lett. 20:199-202. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1987. Current protocols in molecular biology. John Wiley & Sons, New York, N.Y.
- 5.Bachovchin, W. W., R. G. Eagar, K. W. Moore, and J. H. Richards. 1977. Mechanism of action of adenosylcobalamin: glycerol and other substrate analogues as substrates and inactivators for propanediol dehydratase—kinetics, stereospecificity, and mechanism. Biochemistry 16:1082-1092. [DOI] [PubMed] [Google Scholar]
- 6.Bobik, T. A., Y. Xu, R. M. Jeter, K. E. Otto, and J. R. Roth. 1997. Propanediol utilization genes (pdu) of Salmonella typhimurium: three genes for the propanediol dehydratase. J. Bacteriol. 179:6633-6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boenigk, R., S. Bowien, and G. Gottschalk. 1993. Fermentation of glycerol to 1,3-propanediol in continuous cultures of Citrobacter freundii. Appl. Microbiol. Biotechnol. 38:453-457. [Google Scholar]
- 8.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Brady, S. F., C. J. Chao, J. Handelsman, and J. Clardy. 2001. Cloning and heterologous expression of a natural product biosynthetic gene cluster from eDNA. Org. Lett. 3:1981-1984. [DOI] [PubMed] [Google Scholar]
- 10.Caldwell, D. E., G. M. Wolfaardts, D. R. Kober, and J. R. Lawrence. 1997. Cultivation of microbial consortia and communities, p. 79-90. In J. H. Hurst, G. R. Knudsen, M. J. McInerey, L. D. Stetzenbach, and M. V. Walter (ed.), Manual of environmental microbiology. ASM Press, Washington, D.C.
- 11.Chotani, G., T. Dodge, A. Hsu, M. Kumar, R. LaDuca, D. Trimbur, W. Weyler, and K. Sanford. 2000. The commercial production of chemicals using pathway engineering. Biochim. Biophys. Acta 1543:434-455. [DOI] [PubMed] [Google Scholar]
- 12.Cottrell, M. T., J. A. Moore, and D. L. Kirchman. 1999. Chitinases from uncultured microorganisms. Appl. Environ. Microbiol. 65:2553-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dabrock, B., H. Bahl, and G. Gottschalk. 1992. Parameters effecting solvent production by Clostridium pasteurianum. Appl. Environ. Microbiol. 58:1233-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daniel, R. 2002. Construction of environmental libraries for functional screening of enzyme activity, p. 63-78. In S. Brakmann and K. Johnsson (ed.), Directed molecular evolution of proteins. Wiley-VCH Verlag GmbH, Weinheim, Germany.
- 15.Daniel, R., K. Stuertz, and G. Gottschalk. 1995. Biochemical and molecular characterization of the oxidative branch of glycerol utilization by Citrobacter freundii. J. Bacteriol. 177:4392-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daniel, R., R. Boenigk, and G. Gottschalk. 1995. Purification of the 1,3-propanediol dehydrogenase from Citrobacter freundii and cloning, sequencing and overexpression of the corresponding gene in Escherichia coli. J. Bacteriol. 177:2151-2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daniel, R., T. A. Bobik, and G. Gottschalk. 1998. Biochemistry of coenzyme B12-dependent glycerol and diol dehydratases and organization of the encoding genes. FEMS Microbiol. Rev. 22:553-566. [DOI] [PubMed] [Google Scholar]
- 18.Devereux, J., P. Haeberli, and O. Smithies. 1984. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 12:387-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Entcheva, P., W. Liebl, A. Johann, T. Hartsch, and W. Streit. 2001. Direct cloning from enrichment cultures, a reliable strategy for isolation of complete operons and genes from microbial consortia. Appl. Environ. Microbiol. 67:89-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freund, A. 1881. Über die Bildung und Darstellung von Trimethylenalkohol aus Glycerin. Monatsh. Chem. 2:636-641.
- 21.Healy, F. G., R. M. Ray, H. C. Aldrich, A. C. Wilkie, L. O. Ingram, and K. T. Shanmugam. 1995. Direct isolation of functional genes encoding cellulases from the microbial consortia in a thermophilic, anaerobic digester maintained on lignocellulose. Appl. Microbiol. Biotechnol. 43:667-674. [DOI] [PubMed] [Google Scholar]
- 22.Henne, A., R. A. Schmitz, M. Bömeke, G. Gottschalk, and R. Daniel. 2000. Screening of environmental DNA libraries for the presence of genes conferring lipolytic activity on Escherichia coli. Appl. Environ. Microbiol. 66:3113-3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henne, A., R. Daniel, R. A. Schmitz, and G. Gottschalk. 1999. Construction of environmental DNA libraries in Escherichia coli and screening for the presence of genes conferring utilization of 4-hydroxybutyrate. Appl. Environ. Microbiol. 65:3901-3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Homann, T., C. Tag, H. Biebl, W.-D. Deckwer, and B. Schink. 1990. Fermentation of glycerol to 1,3-propanediol by Klebsiella and Citrobacter strains. Appl. Microbiol. Biotechnol. 33:121-126. [Google Scholar]
- 25.Hoster, F., R. Daniel, and G. Gottschalk. 2001. Isolation of a new Thermoanaerobacterium thermosaccharolyticum strain (FH1) producing a thermostable dextranase. J. Gen. Appl. Microbiol. 47:187-192. [DOI] [PubMed] [Google Scholar]
- 26.Kajiura, H., K. Mori, T. Tobimatsu, and T. Toraya. 2001. Characterization and mechanism of action of a reactivating factor for adenosylcobalamin-dependent glycerol dehydratase. J. Biol. Chem. 276:36514-36519. [DOI] [PubMed] [Google Scholar]
- 27.Knietsch, A., T. Waschkowitz, S. Bowien, A. Henne, and R. Daniel. 2003. Construction and screening of metagenomic libraries derived from enrichment cultures: generation of a gene bank for genes conferring alcohol oxidoreductase activity on Escherichia coli. Appl. Environ. Microbiol. 69:1408-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luers, F., M. Seyfried, R. Daniel, and G. Gottschalk. 1997. Glycerol conversion to 1,3-propanediol by Clostridium pasteurianum: cloning and expression of the gene encoding 1,3-propanediol dehydrogenase. FEMS Microbiol. Lett. 154:337-345. [DOI] [PubMed] [Google Scholar]
- 29.Macis, L., R. Daniel, and G. Gottschalk. 1998. Properties and sequence of the coenzyme B12-dependent glycerol dehydratase of Clostridium pasteurianum. FEMS Microbiol. Lett. 164:21-28. [DOI] [PubMed] [Google Scholar]
- 30.Majernik, A., G. Gottschalk, and R. Daniel. 2001. Screening of environmental DNA libraries for the presence of genes conferring Na+(Li+)/H+ antiporter activity on Escherichia coli: characterization of the recovered genes and the corresponding gene products. J. Bacteriol. 183:6645-6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Mori, K., T. Tobimatsu, T. Hara, and T. Toraya. 2041. 1997. Characterization, sequencing, and expression of the genes encoding a reactivating factor for glycerol-inactivated adenosylcobalamin-dependent diol dehydratase. J. Biol. Chem. 272:32034-32041. [DOI] [PubMed] [Google Scholar]
- 33.Petitdemange, H., C. Dürr, S. Abbad-Andaloussi, and G. Raval. 1995. Fermentation of raw glycerol to 1,3-propanediol by new strains of Clostridium butyricum. J. Ind. Microbiol 15:498-502. [Google Scholar]
- 34.Pfennig, N., and K. D. Lippert. 1966. Über das Vitamin B12-Bedürfnis phototropher Schwefelbakterien. Arch. Microbiol. 55:245-256. [Google Scholar]
- 35.Poznanskaya, A. A., and T. L. Korsova. 1979. Adenosylcobalamin-dependent glycerol dehydratase interaction with substrates and their analogs, p. 431-436. In B. Zagalak and W. Friedrich (ed.), Vitamin B12. Walter de Gruyter & Co., Berlin, Germany.
- 36.Poznanskaya, A. A., M. I. Yakushea, and V. A. Yakovlev. 1977. Study of the mechanism of action of adenosylcobalamin-dependent glycerol dehydratase from Aerobacter aerogenes. II. The inactivation kinetics of glycerol dehydratase complexes with adenosylcobalamin and its analogs. Biochim. Biophys. Acta 484:236-243. [DOI] [PubMed] [Google Scholar]
- 37.Rondon, M. R., P. R. August, A. D. Bettermann, S. F. Brady, T. H. Grossman, M. R. Liles, K. A. Loiacono, B. A. Lynch, L. A. MacNeil, C. Minor, C. L. Tiong, M. Gilman, M. S. Osburne, J. Clardy, J. Handelsman, and R. M. Goodman. 2000. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 66:2541-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saddler, J. N., and A. W. Khan. 1979. Cellulose degradation by a new isolate from sewage sludge, a member of the Bacteroidaceae family. Can J. Microbiol. 25:1427-1432. [DOI] [PubMed] [Google Scholar]
- 39.Sauvageot, N., C. Muller, A. Hartke, Y. Auffray, and J.-M. Laplace. 2002. Characterisation of the diol dehydratase pdu operon of Lactobacillus collinoides. FEMS Microbiol. Lett. 209:69-74. [DOI] [PubMed] [Google Scholar]
- 40.Seifert, C., S. Bowien, G. Gottschalk, and R. Daniel. 2001. Identification and expression of the genes and purification and characterization of the gene products involved in reactivation of coenzyme B12-dependent glycerol dehydratase of Citrobacter freundii. Eur. J. Biochem. 268:2369-2378. [DOI] [PubMed] [Google Scholar]
- 41.Seyfried, M., R. Daniel, and G. Gottschalk. 1996. Cloning, sequencing, and overexpression of the genes encoding coenzyme B12-dependent glycerol dehydratase of Citrobacter freundii. J. Bacteriol. 178:5793-5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shibata, N., J. Masuda, T. Tobimatsu, T. Toraya, K. Suto, Y. Morimoto, and N. Yasuoka. 1999. A new mode of B12 binding and the direct participation of a potassium ion in enzyme catalysis: X-ray structure of diol dehydratase. Struct. Fold. Des. 7:997-1008. [DOI] [PubMed] [Google Scholar]
- 43.Sprenger, G. A., B. A. Hammer, E. A. Johnson, and E. C. C. Lin. 1989. Anaerobic growth of Escherichia coli on glycerol by importing genes of the dha regulon from Klebsiella pneumoniae. J. Gen. Microbiol. 135:1255-1262. [DOI] [PubMed] [Google Scholar]
- 44.Tobimatsu, T., M. Azuma, H. Matsubara, H. Takatori, T. Niida, K. Nishimoto, H. Satoh, R. Hayashi, and T. Toraya. 1996. Cloning, sequencing, and high level expression of the genes encoding adenosylcobalamin-dependent glycerol dehydrase of Klebsiella pneumoniae. J. Biol. Chem. 271:22352-22357. [DOI] [PubMed] [Google Scholar]
- 45.Tobimatsu, T., T. Hara, M. Sakaguchi, Y. Kishimoto, Y. Wada, M. Isoda, T. Sakai, and T. Toraya. 1995. Molecular cloning, sequencing, and expression of the genes encoding adenosylcobalamin-dependent diol dehydrase of Klebsiella oxytoca. J. Biol. Chem. 270:7142-7148. [DOI] [PubMed] [Google Scholar]
- 46.Tobimatsu, T., T. Sakai, Y. Hashida, N. Mizoguchi, S. Miyoshi, and T. Toraya. 1997. Heterologous expression, purification, and properties of diol dehydratase, an adenosylcobalamin-dependent enzyme of Klebsiella oxytoca. Arch. Biochem. Biophys. 347:132-140. [DOI] [PubMed] [Google Scholar]
- 47.Toraya, T., and K. Mori. 1999. A reactivating factor for coenzyme B12-dependent diol dehydratase. J. Biol. Chem. 274:3372-3377. [DOI] [PubMed] [Google Scholar]
- 48.Toraya, T. 2000. Radical catalysis of B12 enzymes: structure, mechanism, inactivation, and reactivation of diol and glycerol dehydratases. Cell Mol. Life Sci. 77:106-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Toraya, T., S. Kuno, and S. Fukui. 1980. Distribution of coenzyme B12-dependent diol dehydratase and glycerol dehydratase in selected genera of Enterobacteriaceae and Propionibacteriaceae. J. Bacteriol. 141:1439-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toraya, T., T. Shirakashi, T. Kosuga, and S. Fukui. 1976. Substrate specificity of coenzyme B12-dependent diol dehydrase: glycerol as both a good substrate and a potent inactivator. Biochem. Biophys. Res. Commun. 69:475-480. [DOI] [PubMed] [Google Scholar]
- 51.Yamanishi, M., M. Yunoki, T. Tobimatsu, H. Sato, J. Matsui, A. Dokiya, Y. Iuchi, K. Oe, K. Suto, N. Shibata, Y. Morimoto, N. Yasuoka, and T. Toraya. 2002. The crystal structure of coenzyme B12-dependent glycerol dehydratase in complex with cobalamin and propane-1,2-diol. Eur. J. Biochem. 269:4484-4494. [DOI] [PubMed] [Google Scholar]