Abstract
A heterologous metabolism of polyhydroxyalkanoate (PHA) biosynthesis and degradation was established in Escherichia coli by introducing the Ralstonia eutropha PHA biosynthesis operon along with the R. eutropha intracellular PHA depolymerase gene. By with this metabolically engineered E. coli, enantiomerically pure (R)-3-hydroxybutyric acid (R3HB) could be efficiently produced from glucose. By employing a two-plasmid system, developed as the PHA biosynthesis operon on a medium-copy-number plasmid and the PHA depolymerase gene on a high-copy-number plasmid, R3HB could be produced with a yield of 49.5% (85.6% of the maximum theoretical yield) from glucose. By integration of the PHA biosynthesis genes into the chromosome of E. coli and by introducing a plasmid containing the PHA depolymerase gene, R3HB could be produced without plasmid instability in the absence of antibiotics. This strategy can be used for the production of various enantiomerically pure (R)-hydroxycarboxylic acids from renewable resources.
Polyhydroxyalkanoates (PHAs) are a group of completely biodegradable polyesters that are synthesized and accumulated by various bacteria (1, 5, 9, 27). More than 140 kinds of carboxylic acids hydroxylated at the 3-, 4-, 5-, or 6-position, all in the (R)-configuration if they possess a chiral center on the position of hydroxyl group, can be incorporated into PHAs by employing different bacteria under various culture conditions (5, 9, 28). Therefore, it was reasoned that various enantiomerically pure (R)-(−)-hydroxycarboxylic acids (RHAs) might be conveniently prepared by depolymerizing biosynthesized PHAs. These RHAs contain two functional groups that are convenient to modify for the synthesis of various chiral compounds, especially fine chemicals such as antibiotics, vitamins, perfumes, and pheromones (3, 9, 21, 23). For example, (R)-(−)-3-hydroxybutyric acid (R3HB) is an important precursor of 4-acetoxyazetidinone, which in turn is used to make carbapenem antibiotics, which have close to a billion-dollar market. Poly-(R)-(−)-3-hydroxybutyrate (PHB) is the most ubiquitous member of the PHAs. Methods for producing R3HB by chemical digestion of PHB have been reported (13, 23, 24). In these methods, however, organic solvents were used in large amounts, and the production efficiency was rather low due to complicated processes.
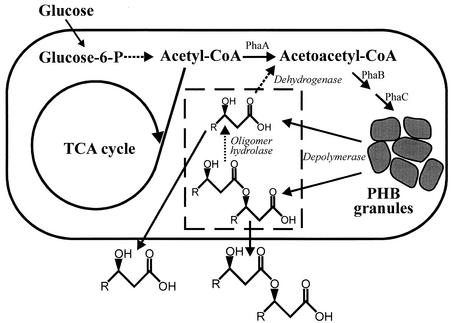
The metabolism for the synthesis and degradation of PHB plays an important role in many bacteria for the reservation and reutilization of excess carbon and energy sources and reducing power (1). The metabolic pathway for the synthesis and degradation of PHB in Ralstonia eutropha was already proposed (1, 5, 11, 18, 22, 25). PHB is synthesized from acetyl-coenzyme A (CoA) by three sequential enzymatic reactions catalyzed by β-ketothiolase, acetoacetyl-CoA reductase, and PHA synthase (18, 22, 25). When the conditions are met, PHB is depolymerized to R3HB by intracellular PHA depolymerase and oligomer hydrolase (1, 5, 11). Then the R3HB dehydrogenase converts R3HB to acetoacetate, which is further metabolized in the cell. By employing this cyclic nature of PHB synthesis and degradation, we recently demonstrated that R3HB could be efficiently produced in naturally PHA-producing bacteria by providing appropriate environmental conditions (lowering the pH, for example) that support high activity of intracellular PHA depolymerase and no activity of R3HB dehydrogenase (11).
It has previously been demonstrated that recombinant E. coli strains harboring the Ralstonia eutropha PHA biosynthesis genes (18, 22, 25) were able to accumulate a large amount of PHB, up to 90% of dry cell weight (4, 10, 25). Recently, Saegusa et al. (19) cloned the intracellular PHA depolymerase gene from R. eutropha. It was therefore reasoned that R3HB and other RHAs may be efficiently produced by establishing a heterologous PHA biosynthesis and degradation pathway in Escherichia coli, as depicted in Fig. 1.
FIG. 1.
Poly-(R)-(−)-3-hydroxybutyrate (PHB) metabolism and the metabolic engineering strategy for the production of (R)-(−)-3-hydroxybutyric acid in E. coli. Abbreviations: PhaA, β-ketothiolase; PhaB, acetoacetyl-CoA reductase; PhaC, PHA synthase.
In this article, we report the development of metabolically engineered E. coli strains harboring a heterologous PHA synthesis and degradation pathway and a novel strategy for the production of enantiomerically pure RHAs by these strains.
MATERIALS AND METHODS
Bacterial strains, plasmids, PCR primers, and growth condition.
The bacterial strains, plasmids, and PCR primers used in this study are listed in Table 1. R. eutropha H16 was grown at 30°C in nutrient broth (NB; Difco laboratories, Detroit, Mich.). Recombinant E. coli XL1 Blue transformed with various plasmids were grown at 37°C in Luria-Bertani medium (LB; 10 g of Bacto-tryptone, 5 g of Bacto-yeast extract, and 10 g of sodium chloride per liter) supplemented with 20 g of glucose per liter or in a chemically defined medium (R medium) (4) supplemented with 20 g of glucose per liter and 20 mg of thiamine per liter. When specified, ampicillin (100 μg/ml) and/or chloramphenicol (50 μg/ml) was added for the maintenance of plasmids.
TABLE 1.
Strains, plasmids, and PCR primers
| Strain, plasmid, or primer | Relevant features | Source or referencea |
|---|---|---|
| Strains | ||
| E. coli XL1-Blue | supE44 hsdR17 recA1 endA1 gyrA96 thi relA1 lac F′[proAB+ lacIqlacZ ΔM15 Tn10(tetr)] | Stratagene |
| E. coli B | Wild type; ATCC 11303 | ATCC |
| E. coli B-PHA+ | B (ATCC 11303) pta::phaCAB, Kmr | This study |
| R. eutropha H16 | Wild type; ATCC 17699 | ATCC |
| Plasmids | ||
| pACYC184 | Cloning vector; Cmr Tcr | NEB |
| pUC19 | Cloning vector; Apr | NEB |
| pSYL105 | Expression vector containing R. eutropha phaCRe, phaARe; and phaBRe; hok/sok locusb; Apr | 10 |
| p5184 | pACYC184 carrying XbaI fragment of pSYL105 containing phaCRe, phaARe, and phaBRe as an operon and hok/sok locus | This study |
| pSYL105Red | pSYL105 containing phaZiRe | This study |
| pTrcEBG | pTrc99A containing red operon of bacteriophage λ | 8 |
| pUC19Red | pUC19 containing phaZiRe | This study |
| pUCRed_stb | pUC19Red containing hok/sok locus | This study |
| pUC19pta | pUC19 containing segments of E. coli pta gene | This study |
| pUC19pta-PHA | pUC19pta containing R. eutropha PHA biosynthesis operon | This study |
| Primersc | ||
| phaZ-XB/F | 5′-GCTCTAGAGGATCCTTGTTTTCCGCAGCAACAGAT-3′ | Genotech |
| phaZ-BH/R | 5′-GCGGATCCAAGCTTACCTGGTGGCCGAGGC-3′ | Genotech |
| pta-P1 | 5′-GCGAATTCTTTAAAGACGCGCGCATTTCTAAACT-3′ | Genotech |
| pta-P2 | 5′-GCGGTACCGAGCTCCGGGTTGATCGCACAGTCA-3′ | Genotech |
| pta-P3 | 5′-GGCGAGCTCGCGCATGCCCGACCGCTGAACAGCTG-3′ | Genotech |
| pta-P4 | 5′-GCAAGCTTTTTAAAGCGCAGTTAAGCAAGATAATC-3′ | Genotech |
Stratagene, Stratagene Cloning Systems, La Jolla, Calif.; ATCC, American Type Culture Collection, Rockville, Md., NEB, New England Biolabs, Beverly, Mass.
parB (hok/sok) locus of plasmid R1.
Primers were synthesized at Genotech, Daejeon, Korea.
Batch cultures were carried out in a 2.5-liter jar fermentor (KoBiotech Co., Incheon, Korea) containing 1.2 liters of R medium supplemented with 20 g of glucose per liter and 20 mg of thiamine per liter at 37°C and 500 rpm without pH control. For the production of poly(3-hydroxybutyrate-co-3-hydroxyvalerate) copolymers, cells were cultured in R medium containing 10 g of glucose per liter and 1 g of propionic acid per liter (26). Plasmid stability was measured as described previously (12) by comparing the number of colonies appearing on antibiotic- and non-antibiotic-containing plates after serially subculturing the recombinant E. coli cells in LB medium.
Genetic manipulation.
Chromosomal DNA of R. eutropha was prepared by Marmur's method (15). PCR was carried out with an automatic thermal cycler (Takara Shuzo Co., Kyoto, Japan). Isolation and purification of plasmid DNA, restriction digestion, ligation, gel electrophoresis, and transformation were carried out as described by Sambrook et al. (20).
Integration of the R. eutropha PHA biosynthesis genes into the chromosome of E. coli B was carried out as previously reported (8, 29). The E. coli phosphotransacetylase (pta) gene was selected for the site of integration of the R. eutropha PHA biosynthesis genes. Plasmid pTrcEBG (8) containing the red operon of bacteriophage λ was transformed into E. coli B. Transformants were prepared as electroporation-competent cells after induction with 1 mM isopropylthiogalactopyranoside (IPTG) for the expression of the red operon. The pta gene was amplified as two fragments from the chromosomal DNA of E. coli B by PCR with two pairs of primer sets, pta-P1 plus pta-P2 and pta-P3 plus pta-P4 (Table 1). Plasmid pUC19pta was prepared by inserting these two PCR products digested with EcoRI-KpnI and SphI-HindIII, respectively, into pUC19.
To prepare the R. eutropha PHA biosynthesis operon, pSYL105 (10) was digested with EcoRI, made blunt ended, and digested with BamHI. Plasmid pUC19pta-PHA was constructed by inserting the PHA biosynthesis operon into the SmaI and BamHI sites of pUC19pta. Finally, the DNA cassette containing the PHA biosynthesis operon and the homologous regions of the pta gene was prepared by digesting pUC19pta-PHA with DraI and mixed with electroporation-competent cells. After electroporation, the desired transformants were selected by identifying PHB-accumulating colonies on LB plates containing 20 g of glucose per liter at 37°C. From the selected transformants, pTrcEBG was removed by cultivating cells in the absence of ampicillin.
Analytical procedures.
Cell concentration (grams of dry cell weight per liter of culture broth) and PHB concentration were determined as previously described (4). The PHB content was defined as the percentage of the ratio of PHB concentration to the cell concentration. Residual cell concentration was calculated by subtracting PHB concentration from the cell concentration. The RHA concentration was measured by high-performance liquid chromatography (HPLC) as previously described (11). When PHB was degraded by an intracellular PHA depolymerase, mainly dimers of R3HB were produced and excreted into the medium along with R3HB monomers.
Dimers of R3HB were hydrolyzed to R3HB monomers by mild heat treatment under alkaline condition as follows. To a 1.5-ml microcentrifuge tube containing 500 μl of cell-free supernatant, 500 μl of 10 N NaOH was added. The tightly sealed tube was incubated at 95°C for 2 h. After cooling to room temperature, the reaction mixture was diluted with an appropriate amount (2- to 50-fold) of distilled water. The resulting solution was filtered and analyzed by HPLC. The monomer yield was defined as the percentage of the ratio of the amount of R3HB formed after heat treatment to the total amount of glucose consumed. The theoretical yield of R3HB from glucose is 57.8%.
RESULTS AND DISCUSSION
Metabolic engineering of E. coli.
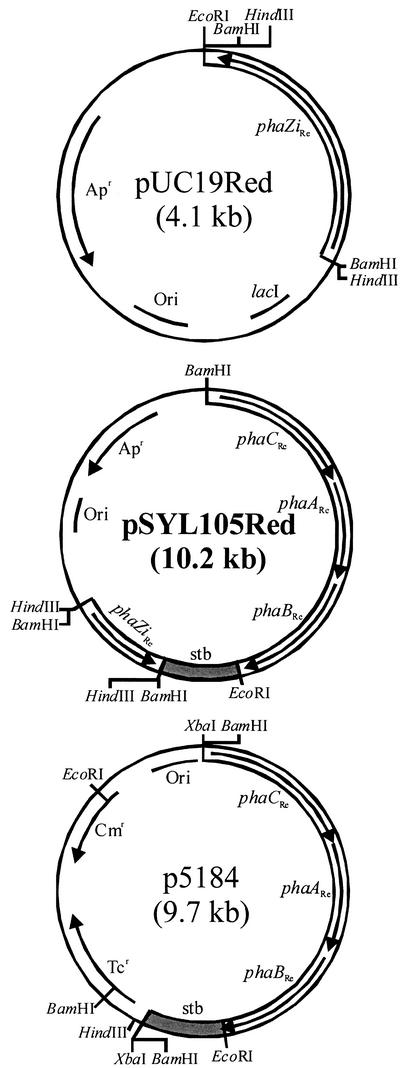
To establish a heterologous metabolism of PHA biosynthesis and degradation in E. coli, plasmids that contain the genes encoding the PHA biosynthetic enzymes and an intracellular PHA depolymerase were constructed as follows. The R. eutropha intracellular PHA depolymerase gene (phaZiRe), including its native promoter region, was amplified from R. eutropha chromosomal DNA by PCR with primers phaZ-XB/F and phaZ-BH/R. Plasmid pUC19Red was constructed by inserting the BamHI-digested PCR product into the BamHI site of plasmid pUC19 (Fig. 2). The 1.4-kbp HindIII-digested fragment of pUC19Red containing the phaZiRe gene was cloned into the HindIII site of pSYL105 (10) harboring the R. eutropha PHA biosynthesis genes to construct pSYL105Red (Fig. 2). These plasmids thus contain the genes coding for four enzymes responsible for PHA biosynthesis and degradation under the control of their native promoters and ribosome-binding sites: β-ketothiolase (phaARe), NADPH-dependent acetoacetyl-CoA reductase (phaBRe), and PHA synthase (phaCRe) as an operon and an intracellular PHA depolymerase (phaZiRe).
FIG. 2.
Plasmids used in this study. Apr, ampicillin resistance gene; Cmr, chloramphenicol resistance gene; lacI, lactose operon repressor; ori, origin of replication; phaA, β-ketothiolase; phaB, acetoacetyl-CoA reductase; phaC, PHA synthase; phaZi, intracellular PHA depolymerase; stb, the hok/sok locus of plasmid R1; Tcr, tetracycline resistance gene.
Production of R3HB.
To examine the possibility of R3HB production by this strategy, flask cultures of E. coli XL1-Blue harboring pSYL105Red were carried out. The recombinant E. coli strains produced R3HB monomers and dimers and excreted them into the medium. These results suggest that the heterologous PHA biosynthesis and degradation metabolism had been successfully established in E. coli. The final cell and PHB concentrations, the R3HB concentration after alkaline heat treatment, and the yield of R3HB determined after 51 h of cultivation were 1.3 g/liter, 0.45 g/liter, 8.3 g/liter, and 41.5%, respectively, when a complex medium was used, and 1.4 g/liter, 0.12 g/liter, 9.6 g/liter, and 48.0%, respectively, when a chemically defined medium was used. Throughout the culture, the PHB concentration was higher in complex medium than in chemically defined medium. However, more R3HB could be produced while less PHB remained in cells when a chemically defined medium was used, which is beneficial from the recovery point of view. Therefore, batch cultures were carried out in a chemically defined medium.
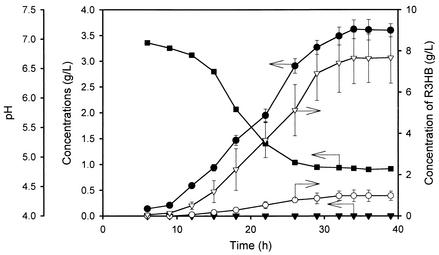
The results obtained by batch culture of E. coli XL1-Blue (pSYL105Red) are shown in Fig. 3A. Rapid production of R3HB started at the beginning of culture when a chemically defined medium was used. During the exponential growth phase (6 to 15 h), R3HB was produced mainly as a monomer up to 5.2 g/liter. The R3HB concentration after alkaline digestion was 6.4 g/liter at 15 h. During the stationary phase (after 15 h), the R3HB concentration decreased while the total R3HB concentration (after alkaline heat treatment) continued to increase. This phenomenon is likely to be due to the formation R3HB dimers from the monomers by esterification. Generally, the carboxylic acid group of R3HB can be readily esterified with alcohol groups to form esters (17). Since R3HB molecules possess both alcohol and carboxylic acid functional groups, the R3HB monomers produced can serve as reactants for reversible intermolecular esterification to form dimers. When R3HB was excreted mainly as monomers by the depolymerization of PHB during the exponential growth phase, the ratio of R3HB monomers to dimers was high enough for the monomers to undergo the esterification reaction. Nonetheless, our desired R3HB product could be successfully obtained by alkaline heat treatment.
FIG. 3.
Time profiles of cell concentration (•), PHB concentration (▾), pH (▪), and R3HB concentration before (○) and after (▿) alkaline heat treatment during batch culture of recombinant E. coli harboring pSYL105Red (A) or p5184 and pUC19Red (B) in a chemically defined medium containing 20 g of glucose per liter.
Two-plasmid system.
Even though R3HB could be produced by employing a single plasmid, there was some PHB remaining in the cell, which lowered the final monomer yield. It was reasoned that a higher intracellular PHA depolymerase activity would be required for the complete depolymerization of PHB. Therefore, a two-plasmid system in which the depolymerase gene is on a high-copy-number plasmid while the biosynthesis genes are on a medium-copy-number plasmid was developed to examine the possibility of increasing the monomer yield. For the expression of the PHA biosynthesis genes, plasmid p5184 (Fig. 2) was constructed by inserting the 5.5-kbp XbaI-digested fragment of pSYL105 containing the R. eutropha PHA biosynthesis operon and the hok/sok locus (6) to the XbaI site of medium-copy-number plasmid pACYC184. For the expression of the phaZiRe gene, high-copy-number plasmid pUC19Red (Fig. 2) was used.
In the batch culture of recombinant E. coli harboring both p5184 and pUC19Red (Fig. 3B), cell growth was rather slow, and the final cell concentration was lower than that obtained with the single-plasmid system. This seemed to be due to the greater metabolic burden exerted by the presence of two plasmids compared with one plasmid. However, all PHB synthesized was immediately depolymerized to R3HB monomers and dimers without intracellular accumulation. Even though the copy number of the PHB biosynthesis genes was lower than that with the single-plasmid system, the maximum R3HB concentration after alkaline heat treatment obtained was 9.9 g/liter, which is slightly higher than that obtained with the single-plasmid system. The monomer yield was 49.5%, which is 85.6% of the maximum theoretical yield. It is notable that R3HB monomers were not dimerized, which is different from what was observed with the single-plasmid system. This seems to be because the R3HB concentration of less than 2 g/liter is not high enough to trigger the esterification reaction.
As described above, all PHB was immediately depolymerized when the two-plasmid system was used, while some PHB remained in the cell when the single-plasmid system was used. This phenomenon can be explained as follows. We have previously reported that the crystallinity of PHB produced in recombinant E. coli is much higher than that of PHB produced in a natural PHB producer (7). PHB produced with a single-plasmid system seems to become partially crystalline because of its rapid accumulation before depolymerization. The intracellular PHA depolymerase cannot depolymerize crystalline PHB (19). Therefore, if PHB once accumulates in E. coli due to the higher PHA biosynthetic capacity compared with depolymerization capacity, some old PHB would become more crystalline and would not be a good substrate of the intracellular PHB depolymerase. When the two-plasmid system was used, the intracellular R3HB depolymerase activity was high enough to immediately degrade PHB produced by PHA biosynthesis enzymes present in lower copy numbers.
Integration of PHA biosynthesis genes into E. coli chromosome.
One of the problems most frequently encountered during the industrial production of bioproducts by employing recombinant strains is plasmid instability. Addition of antibiotics to stabilize plasmids is not desirable, especially for the production of bulk products in a large-scale fermentor. The use of the hok/sok locus of plasmid R1 (6) is a good method to stabilize a plasmid by postsegregationally killing the plasmid-free cells, but it cannot be applied to the two-plasmid system. In the two-plasmid system that we used, only plasmid p5184 contains the hok/sok locus. In order to examine the stability of plasmids, E. coli XL1-Blue harboring p5184 and pUC19Red was serially subcultured for 30 generations without the addition of antibiotics. Plasmid p5184 containing the hok/sok locus was stably maintained for 30 generations. However, 85% of the cells lost pUC19Red after six generations (data not shown). Therefore, segregational instability of plasmid was found to be a problem in the absence of antibiotics.
To overcome this problem of plasmid instability, the PHA biosynthesis operon was integrated into the chromosome of E. coli B by homologous recombination (29) to make E. coli B-PHA+. The phosphotransacetylase (pta) gene was selected for the site of integration because this can increase the flux towards the PHB biosynthesis pathway by eliminating the competing pathway consuming acetyl-CoA to form acetic acid. E. coli B-PHA+ was transformed with pUCRed_stb, which was constructed by cloning the hok/sok locus into pUC19Red. By batch fermentation of E. coli B-PHA+ harboring pUCRed_stb in the absence of antibiotics, R3HB was produced to a high level even though there was only a single copy of the PHA biosynthesis operon (Fig. 4). The plasmid was stably maintained until the end of fermentation. The final R3HB concentration and the yield obtained were 7.7 g/liter and 38.5%, respectively, after alkaline heat treatment.
FIG. 4.
Time profiles of cell concentration (•), PHB concentration (▾), pH (▪), and R3HB concentration before (○) and after (▿) alkaline heat treatment during batch culture of recombinant E. coli B-PHA+ harboring pUCRed_stb in a chemically defined medium containing 20 g of glucose per liter.
To see if other RHAs could be produced by this strategy, production of R3HB and (R)-3-hydroxyvaleric acid (R3HV) by first making cells synthesize poly(3-hydroxybutyrate-co-3-hydroxyvalerate) was examined. By flask culture of E. coli B-PHA+(pUCRed_stb) in a chemically defined medium containing 10 g of glucose per liter and 1 g of propionic acid per liter, the final concentrations of R3HB and R3HV after alkaline heat treatment obtained in 48 h were 2.8 and 0.4 g/liter, respectively.
As demonstrated above, R3HB can be efficiently produced by employing metabolically engineered E. coli harboring the genes involved in the biosynthesis and degradation of PHA. By using a two-plasmid system, R3HB was most efficiently produced, with a yield of 49.5% from glucose. The problem of using antibiotics for the plasmid maintenance could be overcome by integrating the PHA biosynthesis operon into the E. coli chromosome and using the hok/sok locus of plasmid R1 in a plasmid harboring the PHA depolymerase gene. Various PHAs other than PHB can be produced in E. coli by engineering the metabolic pathways and by providing different substrates (2, 14, 16). More than 140 different monomer units can be incorporated into PHAs (28). Production of these RHAs will be possible by employing the strategy reported here and with the appropriate pairs of PHA biosynthesis and degradation genes having different substrate specificities. This was demonstrated by producing R3HB and R3HV by culturing a metabolically engineered E. coli in a medium containing glucose and propionic acid. This is a good example of “green chemistry” in that high-value fine chemicals are produced from renewable resources such as glucose and fatty acids.
Acknowledgments
This work was supported by the Ministry of Commerce, Industry and Energy, the Brain Korea 21 program of the Ministry of Education, and the National Research Laboratory program of the Ministry of Science and Technology. Further support from the Center for Ultramicrochemical Process Systems is appreciated.
REFERENCES
- 1.Anderson, A. J., and E. A. Dawes. 1990. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol. Rev. 54:450-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antonio, R. V., A. Steinbüchel, and B. H. A. Rehm. 2000. Analysis of in vivo substrate specificity of the PHA synthase from Ralstonia eutropha: formation of novel copolyesters in recombinant Escherichia coli. FEMS Microbiol. Lett. 182:111-117. [DOI] [PubMed] [Google Scholar]
- 3.Chiba, T., and T. Nakai. 1985. A synthetic approach to (+)-thienamycin from methylene (R)-3-hydroxybutanoate. Chem. Lett. 1985:651-654. [Google Scholar]
- 4.Choi, J., S. Y. Lee, and K. Han. 1998. Cloning of the Alcaligenes latus polyhydroxyalkanoate biosynthesis genes and use of these genes for the enhanced production of poly(3-hydroxybutyrate) in Escherichia coli. Appl. Environ. Microbiol. 64:4897-4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doi, Y. (ed.). 1990. Microbial polyesters. VCH, New York, N.Y.
- 6.Gerdes, K. 1988. The parB (hok/sok) locus of plasmid R1: a general purpose plasmid stabilization system. Bio/Technology 6:1402-1405. [Google Scholar]
- 7.Hahn, S. K., Y. K. Chang, and S. Y. Lee. 1995. Recovery and characterization of poly(3-hydroxybutyric acid) synthesized in Alcaligenes eutrophus and recombinant Escherichia coli. Appl. Environ. Microbiol. 61:34-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jeong, K. J., and S. Y. Lee. 2002. Excretion of β-endorphin into culture medium by with outer membrane protein F as a function partner in recombinant Escherichia coli. Appl. Environ. Microbiol. 68:4979-4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee, S. Y. 1996. Bacterial polyhydroxyalkanoates. Biotechnol. Bioeng. 49:1-14. [DOI] [PubMed] [Google Scholar]
- 10.Lee, S. Y., K. M. Lee, H. N. Chang, and A. Steinbüchel. 1994. Comparison of recombinant Escherichia coli strains for synthesis and accumulation of poly-(3-hydroxybutyric acid), and morphological changes. Biotechnol. Bioeng. 44:1337-1347. [DOI] [PubMed] [Google Scholar]
- 11.Lee, S. Y., Y. Lee, and F. Wang. 1999. Chiral compounds from bacterial polyesters: sugars to plastics to fine chemicals. Biotechnol. Bioeng. 65:363-368. [DOI] [PubMed] [Google Scholar]
- 12.Lee, S. Y., L. D. Mermelstein, and E. T. Papoutsakis. 1993. Determination of plasmid copy number and stability in Clostridium acetobutylicum ATCC 824. FEMS Microbiol. Lett. 108:319-324. [DOI] [PubMed] [Google Scholar]
- 13.Lee, Y., S. H. Park, I. T. Lim, K. Han, and S. Y. Lee. 2000. Preparation of alkyl (R)-(−)-3-hydroxybutyrate by acidic alcoholysis of poly-(R)-(−)-3-hydroxybutyrate. Enzyme Microb. Technol. 27:33-36. [DOI] [PubMed] [Google Scholar]
- 14.Madison, L. L., and G. W. Huisman. 1999. Metabolic engineering of poly(3-hydroxyalkanoates): from DNA to plastic. Microbiol. Mol. Biol. Rev. 63:21-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marmur, J. 1961. A procedure for the isolation of deoxyribonucleic acids from microorganisms. J. Mol. Biol. 3:208-218. [Google Scholar]
- 16.Matsusaki, H., H. Abe, and Y. Doi. 2000. Biosynthesis and properties of poly(3-hydroxybutyrate-co-3-hydroxyalkanoates) by recombinant strains of Pseudomonas sp. 61-3. Biomacromolecules 1:17-22. [DOI] [PubMed] [Google Scholar]
- 17.Miltenberger, K., and H. Aktiengesellschaft. 1985-1996. Hydroxycarboxylic acid, aliphatic, p. 507-517. In H.-J. Arpe, E. Biekert, H. T. Davis, W. Gerharz, H. Gerrens, W. Keim, J. L. McGuire, A. Mitsutani, H. Pilat, Sir C. Reece, D. P. Sheetz, H. E. Simmons, E. Weise, R. Wirtz, and H.-R. Wüthrich (ed.), Ullmann's encyclopedia of industrial chemistry, 5th ed. Wiley-VCH, Weinheim, Germany.
- 18.Peoples, O. P., and A. J. Sinskey. 1989. Poly-β-hydroxybutyrate biosynthesis in Alcaligenes eutrophus H16. Identification and characterization of the PHB polymerase gene (phbC). J. Biol. Chem. 264:15298-15303. [PubMed] [Google Scholar]
- 19.Saegusa, H., M. Shiraki, C. Kanai, and T. Saito. 2001. Cloning of an intracellular poly[d(−)-3-hydroxybutyrate] depolymerase gene from Ralstonia eutropha H16 and characterization of the gene product. J. Bacteriol. 183:94-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 21.Schnurrenberger, P., E. Hungerbühler, and D. Seebach. 1987. Total synthesis of (+)-colletodiol from (S,S)-tartarate and (R)-3-hydroxybutanoate. Liebigs Ann. Chem. 1987:733-744. [Google Scholar]
- 22.Schubert, P., A. Steinbüchel, and H. G. Schlegel. 1988. Cloning of the Alcaligenes eutrophus poly-β-hydroxybutyrate synthetic pathway and synthesis of poly-β-hydroxybutyrate in Escherichia coli. J. Bacteriol. 170:5837-5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seebach, D., A. K. Beck, R. Breitschuh, and K. Job. 1992. Direct degradation of the biopolymer poly[(R)-3-hydroxybutyric acid] to (R)-3-hydroxybutanoic acid and its methyl ester. Org. Synth. 71:39-47. [Google Scholar]
- 24.Seebach, D., and M. F. Zuger. 1982. Über die depolymerisierung von poly-(R)-3-hydroxy-buttersaureester (PHB). Helv. Chim. Acta 65:495-503. (In German.) [Google Scholar]
- 25.Slater, S. C., W. H. Voige, and D. Dennis. 1988. Cloning and expression in Escherichia coli of the Alcaligenes eutrophus H16 poly-β-hydroxybutyrate biosynthetic pathway. J. Bacteriol. 170:4431-4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Slater, S., T. Gallaher, and D. Dennis. 1992. Production of poly-(3-hydroxybutyrate-co-3-hydroxyvalerate) in a recombinant Escherichia coli strain. Appl. Environ. Microbiol. 58:1089-1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steinbüchel, A., and B. Fuchtenbusch. 1998. Bacterial and other biological systems for polyester production. Trends Biotechnol. 16:419-427. [DOI] [PubMed] [Google Scholar]
- 28.Steinbüchel, A., and H. E. Valentin. 1995. Diversity of bacterial polyhydroxyalkanoic acids. FEMS Microbiol. Lett. 128:219-228. [Google Scholar]
- 29.Yu, D., H. M. Ellis, E.-C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]