Abstract
The biosynthetic pathway for the synthesis of the compatible solute α-mannosylglycerate (MG) in the thermophilic bacterium Thermus thermophilus HB27 was identified based on the activities of recombinant mannosyl-3-phosphoglycerate synthase (MPGS) (EC 2.4.1.217) and mannosyl-3-phosphoglycerate phosphatase (MPGP) (EC 3.1.3.70). The sequences of homologous genes from the archaeon Pyrococcus horikoshii were used to identify MPGS and MPGP genes in T. thermophilus HB27 genome. Both genes were separately cloned and overexpressed in Escherichia coli, yielding 3 to 4 mg of pure recombinant protein per liter of culture. The molecular masses were 43.6 and 28.1 kDa for MPGS and MPGP, respectively. The recombinant MPGS catalyzed the synthesis of α-mannosyl-3-phosphoglycerate (MPG) from GDP-mannose and d-3-phosphoglycerate, while the recombinant MPGP catalyzed the dephosphorylation of MPG to MG. The recombinant MPGS had optimal activity at 80 to 90°C and a pH optimum near 7.0; MPGP had maximal activity between 90 and 95°C and at pH 6.0. The activities of both enzymes were strictly dependent on divalent cations; Mn2+ was most effective for MPGS, while Mn2+, Co2+, Mg2+, and to a lesser extent Ni2+ activated MPGP. The organization of MG biosynthetic genes in T. thermophilus HB27 is different from the P. horikoshii operon-like structure, since the genes involved in the conversion of fructose-6-phosphate to GDP-mannose are not found immediately downstream of the contiguous MPGS and MPGP genes. The biosynthesis of MG in the thermophilic bacterium T. thermophilus HB27, proceeding through a phosphorylated intermediate, is similar to the system found in hyperthermophilic archaea.
The compatible solute mannosylglycerate (MG) was originally identified in red algae of the order Ceramiales (4) but has never been encountered in mesophilic bacteria or archaea. MG is, however, a common compatible solute of halotolerant or slightly halophilic thermophilic and hyperthermophilic prokaryotes. The organisms that accumulate MG include the slightly halophilic euryarchaeotes of the genera Pyrococcus and Thermococcus (22, 23), the crenarchaeote Aeropyrum pernix, and the species Archaeoglobus veneficus and Archaeoglobus profundus (12). Among thermophilic bacteria, MG has been identified in Thermus thermophilus, Rhodothermus marinus, and Rubrobacter xylanophilus (28, 33). The role of MG as a compatible solute under salt stress has been confirmed in several of these organisms, and there is increasing evidence that this organic solute also has a role in stabilizing proteins from thermal denaturation, since it is one of the most efficient thermoprotectants known in vitro (3, 30).
The biosynthetic route for the synthesis of MG has been examined in Rhodothermus marinus and Pyrococcus horikoshii. The synthesis of MG proceeds via two alternate routes in R. marinus. In one pathway, GDP-mannose is condensed with d-glycerate to produce MG in a single glycosyl transfer reaction catalyzed by MG synthase (26). In the other pathway, mannosyl-3-phosphoglycerate synthase (MPGS) catalyzes the conversion of GDP-mannose and d-3-phosphoglycerate into mannosyl-3-phosphoglycerate (MPG), which is subsequently converted to MG by mannosyl-3-phosphoglycerate phosphatase (MPGP). In contrast to the case in R. marinus, the synthesis of MG in P. horikoshii proceeds only via the two-step pathway involving the phosphorylated intermediate (9).
Strains of most species of the genus Thermus are ubiquitous inhabitants of neutral and alkaline continental hot springs, where the concentration of salt is very low. On the other hand, only T. thermophilus has been isolated from shallow marine and abyssal hydrothermal vents, where the concentration of sodium reaches that in seawater. Furthermore, all known strains of the species T. thermophilus are halotolerant and are capable of growing in media containing about 5 to 6% NaCl (8). Unlike in R. marinus, Pyrococcus furiosus, P. horikoshii, Thermococcus celer, and Thermococcus stetteri, where MG is the major compatible solute during growth on yeast extract-based media, trehalose is the major compatible solute of the T. thermophilus strains examined. However, MG is a secondary compatible solute that accumulates concomitantly with the salinity of the growth medium (28).
The identification of the genes and the investigation of biosynthetic pathways involved in the synthesis of MG are considered essential to our knowledge of the physiological role of MG in thermal and osmotic stress responses of thermophilic bacteria and hyperthermophilic archaea, as well as the puzzling distribution of this compatible solute among prokaryotes. The annotation of genes based on sequence similarity to homologous proteins is far from being a reliable approach to the identification of the correct enzyme activity, and it is therefore important that sequence information should be supported by functional data (10). In this regard, we identified, cloned, and functionally overexpressed the MPGS and MPGP gene sequences from Thermus thermophilus HB27 and characterized the recombinant enzymes leading to the synthesis of MG.
MATERIALS AND METHODS
Strains, identification and cloning of the genes encoding MPGS and MPGP from T. thermophilus, and functional overexpression in Escherichia coli.
T. thermophilus strain HB27 (DSM 7039) was obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany. For the attempt to identify the genes responsible for MG biosynthesis in T. thermophilus HB27 we used the sequences of mgs from R. marinus and the MPGS and MPGP genes from P. horikoshii for BLAST (1) searches in the T. thermophilus HB27 genome sequence database (Göttingen Genomics Laboratory [www.g2l.bio.uni-goettingen.de]). Two open reading frames (ORFs) with high homology to the MPGS and MPGP genes were detected, and based on the sequences provided, the full MPGS gene was amplified from T. thermophilus HB27 genomic DNA by using forward primer TT1 (5′-GCGGAATTCATGCGTCTGGAGATTCCCAAC-3′) and reverse primer TT2 (5′-GCGAAGCTTTCATGGCACCCGGAAGCGGG-3′); the complete MPGP gene was amplified with forward primer TT3 (5′-GCGGAATTCATGATCGTCTTCACCGACCTG-3′) and reverse primer TT4 (5′-GCGCTGCAGTCAGGGCCCGCTCCCTCCTCG-3′). Forward primers were constructed with additional EcoRI recognition sequences (underlined) immediately upstream of the start codon (boldface), and reverse primers were constructed by adding HindIII and PstI sites (underlined) to TT2 and TT4, respectively, and directly behind the stop codon (boldface). PCR amplifications were carried out in a Perkin-Elmer GeneAmp PCR system 2400 instrument with reaction mixtures (50 μl) containing 100 ng of T. thermophilus HB27 DNA, 200 ng of each primer, 10 mM Tris-HCl (pH 9.0), 1.5 mM MgCl2, 50 mM KCl, 2.5 U of Taq DNA polymerase, and a 0.2 mM concentration of each deoxynucleoside triphosphate. The mixtures were preincubated for 5 min at 94°C and then subjected to 25 cycles of denaturation at 94°C for 1 min, annealing at 60°C for 1 min, and primer extension at 72°C for 1 min. The extension reaction in the last cycle was prolonged for 7 min. Amplification products were visualized on 1% agarose gels and purified by band excision (Novagen). Both PCR products were purified after digestion of the MPGS gene with EcoRI and HindIII and digestion of the MPGP gene with EcoRI and PstI and were ligated into the corresponding sites of expression vector pKK223-3 (Amersham Biosciences) to obtain plasmids pTTS and pTTP, respectively. Most DNA manipulations followed standard molecular techniques and procedures (32). Both constructs were sequenced on both strands (AGOWA, Berlin, Germany).
E. coli XL1-Blue and BL21-Rosetta strains (Novagen) were used as hosts for overexpression. The nucleotide sequences of the MPGS and MPGP genes exhibit a high number of CCC codons for proline, which are rarely used by E. coli. To avoid translational constraints or misincorporation upon high-level expression (17), the BL21-Rosetta strain carrying a plasmid with extra tRNA genes for the codons rarely used in E. coli was used as the host. Clones bearing pTTS or pTTP were grown in 1 liter of YT medium at pH 7.0 and 37°C. Ampicillin was added to a final concentration of 100 μg/ml for selection of plasmids pTTS and pTTP. Chloramphenicol was added to a final concentration of 40 μg/ml for selection of the plasmid with the tRNA genes. E. coli clones were grown to the mid-exponential growth phase (optical density at 600 nm of 0.8) at 37°C, induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside), and grown further for 12 h at 30°C.
Cells were harvested by centrifugation (7,000 × g, 10 min, 4°C). The cell pellet was resuspended in Tris-HCl (20 mM, pH 7.6) containing MgCl2 (5 mM) and (per milliliter of the suspension) DNase I (10 μg) and the protease inhibitors phenylmethylsulfonyl fluoride (80 μg), leupeptin (20 μg), and antipain (20 μg) (all from Sigma-Aldrich). Cells were disrupted by sonication, followed by centrifugation (18,000 × g, 30 min, 4°C) to remove debris. MPGS and MPGP activities were confirmed in E. coli cell extracts by incubating 10 μl of supernatant with the mixtures described below. Cell extracts from E. coli containing the plasmid pKK223-3 without insert were used as negative controls. E. coli cell extracts containing the recombinant enzymes were incubated for 20 min at 70°C to denature the majority of the host proteins and centrifuged (25,000 × g, 30 min, 4°C). The supernatants were filtered through 0.22-μm-pore-size filters (Schleicher & Schuell) and used for purification of the enzymes. The protein contents of all samples were determined by the Bradford assay (5).
Enzyme assays during purification of the proteins.
To detect MPGS activity in E. coli cell extracts during the purification of the recombinant enzyme from E. coli cell extracts, a reaction mixture (100 μl) containing 25 μl of the eluted fraction, 2.5 mM GDP-mannose, 2.5 mM d-3-phosphoglycerate (sodium salt), and 20 mM MgCl2 in 25 mM Tris-HCl (pH 7.6) was used. The reaction mixture was incubated at 70°C for 15 min and cooled on ice, followed by incubation for 15 min with 2 U of alkaline phosphatase (Sigma-Aldrich) at 37°C. The products formed before and after the addition of alkaline phosphatase were visualized by thin-layer chromatography (TLC) (9). Standards of MG, MPG, d-mannose, guanosine, d-3-phosphoglycerate, GDP-mannose, and GDP were used for comparative purposes.
To detect MPGP activity in E. coli cell extracts and during the fast protein liquid chromatography purification of the recombinant enzyme, a reaction mixture (50 μl) containing 15 μl of the eluted fraction, MPG (obtained from synthesis with purified MPGS), 25 mM Tris-HCl (pH 7.6), and 20 mM MgCl2 was used. The reaction mixture was incubated at 70°C for 15 min and cooled on ice. The products formed were analyzed by TLC as described above with appropriate standards.
Purification of MPGS.
The MPGS activity assay was performed as described above, and the enzyme was purified with two sequential Q-Sepharose fast-flow columns (Hi-Load 16/10) equilibrated with 20 mM Tris-HCl, pH 7.6. Elution was carried out at a constant flow of 3 ml/min with linear NaCl gradients (0.0 to 1.0 M) in the same buffer, and MPGS activity was located by TLC after incubation of fractions with substrates as described above. Active fractions were concentrated, and the purity of the samples was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Purification of MPGP.
MPGP-containing supernatant obtained after heat denaturation was applied to a Q-Sepharose fast-flow column as described for the recombinant MPGS. Elution was carried out at a constant flow rate of 3 ml/min with a linear NaCl gradient (0.0 to 1.0 M). Active fractions were concentrated, dialyzed against 20 mM Tris-HCl (pH 7.6), loaded onto a Mono Q column and eluted by a linear gradient of NaCl (0.0 to 1.0 M) at a constant flow rate of 1 ml/min. Active fractions were concentrated, and the purity of the samples was evaluated by SDS-PAGE.
Characterization of the recombinant enzymes.
For the characterization of MPGS substrate specificity, several sugar nucleotides were tested as possible donors of the sugar moiety, namely, GDP-mannose, ADP-mannose, UDP-mannose, ADP-glucose, GDP-glucose, UDP-glucose, and UDP-galactose. The three-carbon compounds glycerol, d-3-phosphoglycerate, d-2-phosphoglycerate, l-glycerol-3-phosphate, 2,3-bisphospho-d-glycerate, and phosphoenolpyruvate (all from Sigma-Aldrich) were tested as sugar acceptors. The reaction mixtures, containing a 2.5 mM concentration of each substrate in 25 mM Tris-HCl (pH 7.6) and 25 mM MgCl2, were incubated at 70°C for 30 min, followed by dephosphorylation at 37°C for 15 min with 2 U of alkaline phosphatase. Samples were tested for the formation of products by TLC (9).
MPG was synthesized by using recombinant MPGS from T. thermophilus HB27 (20 μg/ml). The reaction mixture contained 10 mM GDP-mannose and 10 mM d-3-phosphoglycerate in 25 mM Bis-Tris propane (pH 8.0) and 0.1 mM MnCl2. The synthesis proceeded for 30 min at 80°C and was stopped in an ethanol-ice bath. Two volumes of cold acetone were added, and the sample was further incubated on ice to precipitate the enzyme. After centrifugation, acetone was evaporated at 80°C. The MPG produced was used as the substrate for MPGP assays. The concentration of MPG was measured by specific dephosphorylation of a 5-μl aliquot at 90°C for 5 min in a reaction mixture containing 5 μg of pure MPGP, 25 mM Tris-HCl (pH 7.0), and 10 mM MgCl2 in a total volume of 50 μl. The reaction was stopped on ice, and the volume was adjusted to 300 μl with water for phosphate quantification (2). The reaction mixture was also spotted on a Silica Gel 60 TLC plate (Merck) to confirm that the dephosphorylation of MPG was complete. The solvent system used was composed of n-propanol and ammonia (1:1 by volume), which separated the MPG formed from residual GDP-mannose in the reaction mixture. Visualization of the compounds was achieved with α-naphthol-sulfuric acid solution (9).
Several sugar phosphates, i.e., MPG (synthesized with pure MPGS as described above), mannose-6-phosphate (mannose-6P), glucose-6P, fructose-6P, and trehalose-6P, as well as GDP (all from Sigma-Aldrich), were examined as possible substrates for MPGP in a mixture containing a 2.5 mM concentration of each substrate, 50 mM Tris-HCl (pH 7.6), and 25 mM MgCl2 by incubation of the reaction mixtures at 70°C for 30 min followed by TLC analysis.
The protocol used to examine the temperature profile, pH dependence, effect of cations, and thermal stability of the recombinant enzymes was as follows. The activity of MPGS was calculated from the synthesis of MPG from GDP-mannose and d-3-phosphoglycerate. Reactions were initiated at a specific temperature by adding an exact amount of protein and were stopped at different times by cooling on an ethanol-ice bath. Cold acetone was added to denature protein and evaporated as described above. Samples were then incubated with excess T. thermophilus MPGP to ensure rapid and complete dephosphorylation of the MPG formed. The free phosphate was quantified (2). The complete dephosphorylation of MPG was verified by TLC.
The activity of MPGP was based on the same method of release of inorganic phosphate from MPG. Reactions were commenced at a specific temperature by adding an exact amount of protein and stopped at different times by cooling on an ethanol-ice bath. The reaction mixture contained 2 mM MPG in 25 mM Bis-Tris propane buffer, pH 7.0. The formation of free phosphate was quantified (2). The temperature profiles for activity of MPGS and MPGP were determined between 25 and 103°C. The effects of pH on MPGS and MPGP activities were determined in 25 mM MES (morpholineethanesulfonic acid) buffer (pH 4.5 to 6.0), 25 mM Tris-HCl buffer (pH 5.5 to 7.0), and Bis-Tris propane buffer (pH 6.5 to 9.0) at 80 and 90°C, respectively. All pH values were measured at room temperature; the actual pH at 80 and 90°C was calculated by using the conversion factor ΔpKa/ΔT (°C) = −0.011 for MES, −0.030 for Tris-HCl, and −0.015 for Bis-Tris propane. The thermal stabilities of both enzymes were determined at 70, 80, 90, and 100°C as follows. MPGS (10 μl of a solution of 1.0 mg/ml) was incubated in 25 mM Bis-Tris propane (pH 8.0) for all temperatures. MPGP (10 μl of a solution of 1.0 mg/ml) was incubated in 25 mM Bis-Tris propane (pH 7.0) at all temperatures but 80°C. At appropriate times, samples were withdrawn and immediately examined for residual activities at 80°C.
The protocol used to determine the kinetic parameters of MPGS was as follows. The reactions were initiated by the addition of 0.2 μg of MPGS to the 25 mM Bis-Tris propane (pH 8.0) solutions containing either GDP-mannose (0.1 to 5.0 mM) plus d-3-phosphoglycerate (5 mM) or GDP-mannose (5 mM) plus d-3-phosphoglycerate (0.1 to 5.0 mM) and an excess of MPGP (2 μg) to ensure complete dephosphorylation of MPG. The reactions were stopped at different times by cooling on ice-ethanol bath, and the phosphate released was determined as described above. The reaction used to determine the kinetic parameters of MPGP was initiated by the addition of 0.2 μg of MPGP to the 25 mM Bis-Tris propane (pH 7.0) solutions containing MPG (0.1 to 2.0 mM) and stopped at different times by cooling on ice-ethanol bath. Phosphate released was quantified. All samples for MPGS and MPGP reactions were preheated for 2 min, and all reactions were initiated by the addition of the enzyme preparation. Kinetic parameters for all substrates were determined at 80°C. All experiments were performed in duplicate. Values for Vmax and Km were determined from Hanes plots. The protein content of all samples was determined by the Bradford assay (5).
Nucleotide sequence accession numbers.
A 3.0-kb sequence containing the MPGS and MPGP genes, as well as flanking sequences, has been deposited in GenBank under accession number AY193871. The DNA sequences corresponding to putative phosphomannose mutase (PMM), putative phosphomannose isomerase (PMI), and putative mannose-1P-guanylyltransferase/PMI (M1P-GT/PMI) have been deposited under accession numbers AY194229, AY194230, AY194231, and AY194232, respectively.
RESULTS
Identification of the genes encoding MPGS and MPGP in T. thermophilus HB27.
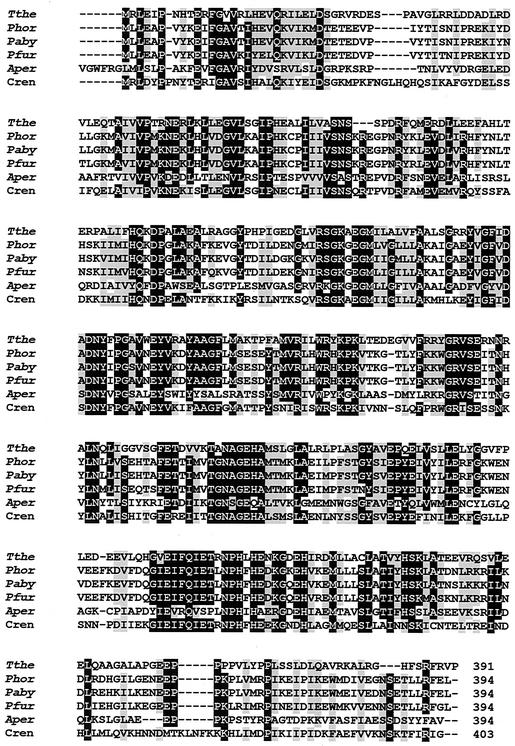
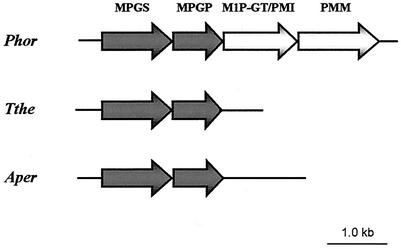
Based on the amino acid sequences of MPGS and MPGP from P. horikoshii, BLAST searches were performed at the T. thermophilus HB27 genome database, resulting in the identification of two ORFs displaying high homology to the MPGS and MPGP genes of the hyperthermophilic archaeon. These ORFs were identified as the genes encoding MPGS and MPGP by their functional overexpression in E. coli. The MPGS gene contains 1,176 bp coding for a polypeptide with 391 amino acids and a calculated molecular mass of 43.6 kDa. This protein has 46% amino acid identity with MPGS from P. horikoshii, 43% identity with MPGS from Pyrococcus abyssi, 42% identity with MPGS from P. furiosus, 36% identity with the putative MPGS from A. pernix, and 38% identity with a putative MPGS found in an environmental sequence belonging to an uncultured crenarchaeote (Fig. 1). The MPGP gene contains 780 bp encoding a protein with 259 amino acids and a molecular mass of 28.1 kDa exhibiting 28% amino acid identity with MPGP from P. horikoshii (GenPept accession number BAA30022), 25% amino acid identity with MPGP from P. abyssi (GenPept accession number CAB50139), 32% amino acid identity with MPGP from P. furiosus (GenPept accession number AAL80714), and 26% amino acid identity with putative MPGP from A. pernix (GenPept accession number BAA79870). In contrast to the adjacent MPGS and MPGP gene sequences in these organisms, a sequence coding for putative MPGP was not found downstream of the MPGS gene in the uncultured crenarchaeote fragment (GenBank accession number AJ496176). A search on the complete genome of T. thermophilus HB27 (A. Henne, personal communication) did not reveal the presence of a gene corresponding to mgs from R. marinus (GenBank accession number AF173987). The genome of T. thermophilus HB27, unlike those of P. horikoshii (19), P. furiosus (31), and P. abyssi (National Centre for Sequencing, Evry, France [www.genoscope.cns.fr]), does not possess two genes immediately downstream from the MPGS and MPGP genes encoding a putative PMM or a putative bifunctional M1P-GT/PMI (Fig. 2). However, putative genes encoding PMM (GenBank accession numbers AY194229 and AY194230) and a gene encoding a putative type I PMI (GenBank accession number AY194231) are found in the T. thermophilus HB27 genome. Another gene encoding a protein with 34% amino acid identity to the putative bifunctional M1P-GT/PMI of P. horikoshii is also found in the genome of T. thermophilus HB27 (GenBank accession number AY194232), but sequence analysis showed that the isomerase domain is truncated.
FIG. 1.
CLUSTALX (36) alignment of MPGS amino acid sequences from T. thermophilus HB27 (Tthe) (GenBank accession number AY193871), P. horikoshii (Phor) (GenPept accession number BAA30023), P. abyssi (Paby) (GenPept accession number CAB50138), P. furiosus (Pfur) (GenPept accession number AAL80715), A. pernix (Aper) (GenPept accession number BAA79872), and an uncultured crenarchaeote (Cren) (GenPept accession number CAD42692). Identical amino acids are shaded in black, and conserved residues are shaded in gray.
FIG. 2.
Schematic comparison of the organization of the genes encoding MPGS and MPGP in the genomes of T. thermophilus HB27 (Tthe) (Göttingen Genomics Laboratory), A. pernix (Aper) (20), and P. horikoshii (Phor) (19). Arrows represent genes. MPGS, EC 2.4.1.217; MPGP, EC 3.1.3.70; M1P-GT/PMI, EC 2.7.7.22/5.3.1.8; PMM, EC 5.4.2.8.
Cloning of the MPGS and MPGP genes, functional overexpression in E. coli, and purification of recombinant enzymes.
PCR amplification of the MPGS and MPGP genes from genomic DNA of T. thermophilus yielded bands with the expected sizes in agarose gels. Activity assays carried out with E. coli cell extracts after expression of E. coli clones containing plasmids pTTS and pTTP, revealed MPG synthesis by MPGS-expressing clones and MPG dephosphorylation by MPGP-expressing clones. SDS-PAGE analysis of crude extracts of XL1-Blue clones showed weak expression bands compared to results for the negative control cell extracts bearing empty vectors (results not shown). The analysis of T. thermophilus HB27 MPGS and MPGP gene nucleotide sequences showed the presence of rare CCC proline-encoding codons, occurring at 3.8 and 6.2%, respectively, whereas in E. coli they occur at only 0.5% (17). Higher levels of expression were achieved with BL21-Rosetta clones coexpressing the recombinant enzyme and tRNA for the rare codons.
Yields of 3 to 4 mg of protein from MPGS and MPGP per liter of culture were estimated. Heat treatment of cell extracts at 70°C for 20 min resulted in extensive purification of the 44-kDa (MPGS) and 28-kDa (MPGP) proteins. The recombinant MPGS and MPGP preparations were pure as judged by SDS-PAGE. Bands corresponding to MPGS and MPGP were not detected in cell extracts from the E. coli BL-21 Rosetta strain containing the empty plasmid (results not shown).
Catalytic properties of MPGS from T. thermophilus.
MPGS showed a highly specific substrate specificity, since only the combination of GDP-mannose with d-3-phosphoglycerate resulted in the formation of MPG (results not shown). The identification of the reaction product of MPGS as MPG was achieved by standard nuclear magnetic resonance spectroscopy (9). The α configuration of MPG was confirmed from the measurement of the coupling constant between the anomeric carbon and the directly bound proton (J = 171.8 Hz).
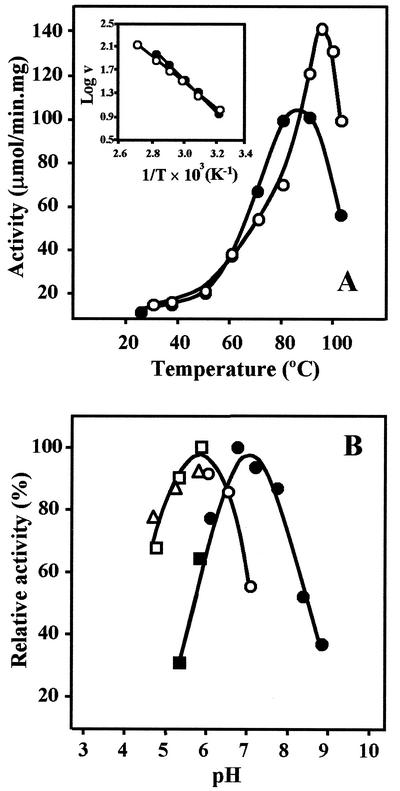
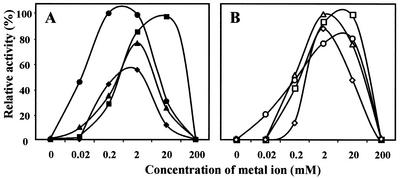
At 25°C the activity of the enzyme was 10% of the maximal activity reached between 80 and 90°C (Fig. 3). At 103°C MPGS still had 55% of the total activity. The activation energy for MPGS determined from Arrhenius plots was 48.4 kJ/mol. The half-lives determined at 80, 90, and 100°C were 189, 22, and 10 min, respectively. Within the pH range examined (5.0 to 9.0), the activity of the enzyme at 80°C was maximal near pH 7.0 (Fig. 3). MPGS exhibited Michaelis-Menten kinetics, and the Km values for the substrates were determined from double-reciprocal plots. The activity of MPGS was strictly dependent on divalent cations in the following order of efficiency: Mn2+ > Mg2+ > Co2+ > Ni2+ (Fig. 4). The maximum activation of the enzyme by Mn2+ was between 0.2 and 2.0 mM. On the other hand, Mg2+ had a similar effect on the activation of MPGS, but this took place at about 20.0 mM. At this concentration Mn2+ inhibited the enzyme. Other divalent cations tested, such as Ca2+, Sr2+, Cu2+, Ba2+, and Zn2+, did not stimulate MPGS activity at any concentration. NaCl and KCl, in the concentration range of 50 to 300 mM, inhibited enzyme activity (results not shown). All results are summarized in Table 1.
FIG. 3.
(A) Temperature dependence of the activity of recombinant MPGS (•) and the recombinant MPGP (○). The enzyme activities were determined at between 25 and 103°C. The inset shows the Arrhenius plot of the data at between 40 and 90°C. (B) pH dependence of the recombinant MPGS (solid symbols) and the recombinant MPGP (open symbols) of T. thermophilus HB27. The enzyme activities were determined at between pH 4.5 and 9.0 in MES (▵), Tris-HCl (▪ and □) and Bis-Tris propane (• and ○).
FIG. 4.
Metal ion dependence of the in vitro activity of MPGS (A) and MPGP (B). The activity assays were performed in the presence of various concentrations of manganese ions (• and ○), magnesium ions (▪ and □), nickel ions (♦ and ⋄), and cobalt ions (▴ and ▵).
TABLE 1.
Biochemical properties of the enzymes and kinetic parameters for the substrates involved in the synthesis of MG in T. thermophilus HB27
| Property (Unit) | Value for:
|
|
|---|---|---|
| MPGS | MPGP | |
| Optimum temp for activity (°C) | 80-90 | 95 |
| Optimum pH for activity | ∼7 | ∼6 |
| Activation energy (kJ/mol) | 48.4 | 41.7 |
| Half-life (min) at: | ||
| 80°C | 189 | NDa |
| 90°C | 22 | 25 |
| 100°C | 10 | 2 |
| Km (mM) for: | ||
| GDP-mannose | 0.33 | |
| 3-Phosphoglycerate | 0.13 | |
| MPG | 0.52 | |
| Vmax (μmol/min · mg) | 122 | 172 |
ND, not determined.
Catalytic properties of MPGP.
MPGP also exhibited high substrate specificity, since MPG was the only substrate dephosphorylated by the enzyme (results not shown). At 30°C the enzyme had 15% of the activity reached at 95°C (Fig. 3). At 103°C MPGP retained 70% of the activity. The activation energy for MPGP was 41.7 kJ/mol. Half-lives determined at 90 and 100°C were 25 and 2 min, respectively. The optimum pH for activity of the enzyme was near 6.0 (Fig. 3). MPGP exhibited Michaelis-Menten kinetics, and the Km for the substrate is shown in Table 1. This enzyme was also strictly dependent on divalent cations, but MPGP was not as selective as MPGS, since Co2+, Mg2+, Mn2+, and, to a lesser extent, Ni2+ activated the enzyme to similar degrees at a concentration of about 2.0 mM (Fig. 4). Other cations, i.e., Ca2+, Sr2+, Ba2+, Cu2+, and Zn2+, had no effect on MPGP activity at any concentration. All results are summarized in Table 1.
DISCUSSION
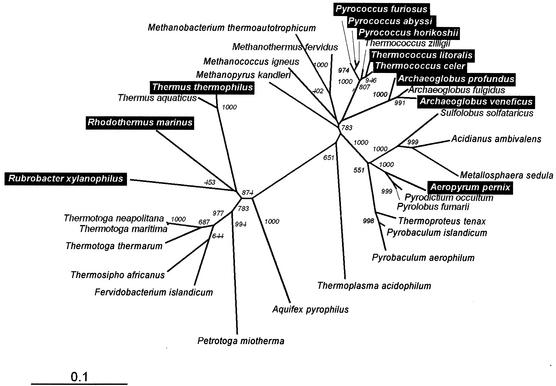
Experimental data show that MG accumulation is scattered among phylogenetically unrelated groups of prokaryotes who share only the ability to grow at very high temperatures under saline conditions (Fig. 5). Several hyperthermophilic archaea of the genus Pyrococcus, the slightly halophilic species of Thermococcus, and A. pernix accumulate MG (34). The thermophilic bacteria T. thermophilus (classified within the Deinococcus-Thermus phylum) (8), R. marinus (a member of the phylum Cytophaga-Flavobacterium-Bacteroides) (35), and R. xylanophilus (representing the deepest branch within the phylum Actinobacteria) (6) also accumulate MG. However, other hyperthermophilic bacteria and archaea do not accumulate this compatible solute. Organisms such as Thermotoga maritima, which accumulate primarily di-myo-inositol-phosphate and its derivatives, or the type strain of the archaeon Archaeoglobus fulgidus, which accumulates primarily diglycerol phosphate as compatible solutes (24, 25), whose genomes have been sequenced, lack genes for the synthesis of MG (21, 27).
FIG. 5.
Unrooted phylogenetic tree based on 16S rRNA sequences for the type strains of each of the prokaryotes in which MG has been found (black boxes) (9, 22, 23, 25, 28, 34). The remaining organisms present in the tree were also investigated for the presence of compatible solutes but do not accumulate MG (22, 24, 25, 33). The ClustalX program (36) was used for sequence alignments and to generate the phylogenetic tree. The significance of the branching order was evaluated by bootstrap analysis of 1,000 computer-generated trees. Bootstrap values are indicated. Bar, 0.1 change per site.
Two biosynthetic pathways for MG have been identified in R. marinus, but only the two-step pathway has been identified in P. horikoshii (9, 26). The identification of the sequences of the genes from P. horikoshii provided us with the information to search for homologs in other organisms that accumulate MG whose genomes have been sequenced and in the unpublished T. thermophilus HB27 genome. Of some 100 complete genomes examined, we found genes coding for MPGS and MPGP only in T. thermophilus HB27, a thermophilic and slightly halophilic bacterium that accumulates trehalose and MG under osmotic stress (28). An MG synthase homolog gene from R. marinus was not encountered in T. thermophilus HB27. Thus, at present R. marinus is the only organism that possesses two biosynthetic pathways for the synthesis of MG, one of which is restricted to this organism (26).
Genes involved in MG biosynthesis in P. horikoshii are arranged in an operon-like structure containing four genes coding for the enzymes catalyzing the conversion of the central metabolite fructose-6P to GDP-mannose and further to MG. The organization of the genes in T. thermophilus HB27 is similar to that in Pyrococcus spp. and A. pernix in that the MPGP gene is found immediately downstream of the MPGS gene. However, in strain HB27 the genes encoding PMM and M1P-GT/PMI are not found downstream of the MPGP gene as they are in the Pyrococcus spp. Thus, the gene organization in T. thermophilus HB27 is most similar to the one found in the crenarchaeote A. pernix. It could be that the Pyrococcus operon-like arrangement and cotranscription of genes responsible for the pathway from fructose-6P provide a functional advantage and efficient synthesis of MG in an organism where it is the major compatible solute. However, the physiological implications of a simpler genetic structure for MG synthesis in T. thermophilus HB27 are not clear, because R. marinus, in which MG and the unique MG derivative mannosylglyceramide are the major compatible solutes, also possesses a gene arrangement like that of T. thermophilus. (N. Borges and H. Santos, unpublished results).
Putative genes coding for enzymes involved in the conversion of the glycolytic intermediate fructose-6P into GDP-mannose could be found in the T. thermophilus HB27 genome, but they were located elsewhere in the chromosome. One ORF encoding a putative type I PMI was found elsewhere in the genome and could be responsible for the conversion of fructose-6P into mannose-6P. Another ORF was identified as a putative type II PMI, and like type II PMIs, it is an M1P-GT/PMI bifunctional enzyme (16). However, this ORF does not contain a complete isomerase domain because of a C-terminal truncation of more than 100 amino acids and is probably not functional. Other putative type II PMIs present in different organisms exhibit similar truncations; among them are the M1P-GT/PMIs from Deinococcus radiodurans, Thermotoga maritima, and Synechocystis (18, 27, 38), but activities for these have not been studied so far. The reaction catalyzing the conversion of mannose-6P into mannose-1P, the specific substrate for GDP-mannose synthesis, could be carried out in T. thermophilus HB27 by either one or both of two putative PMMs found in the genome. The M1P-GT domain of the truncated M1P-GT/PMI could then catalyze the conversion of mannose-1P and GDP into GDP-mannose, since no other gene for this activity appears to be present in the genome of this organism. Moreover, this organism appears to lack a M1P-GT homolog that uses GTP (EC 2.7.7.13) instead of GDP (EC 2.7.7.22) for the formation of GDP-mannose.
The syntheses of the compatible solutes glucosylglycerol, galactosylglycerol, trehalose, and sucrose share, with the synthesis of MG, the formation of a phosphorylated intermediate (7, 11, 14, 37). Despite this common feature, the MPGSs of T. thermophilus, Pyrococcus spp., A. pernix, and an uncultured crenarchaeote share insignificant sequence similarity with these osmolyte-phosphate synthases and were classified in a separate glycosyltransferase family, designated GT55 (http://afmb.cnrs-mrs.fr/∼cazy/CAZY/index.html).
MPGS and MPGP from T. thermophilus HB27 share most in vitro biochemical and kinetic properties with their counterparts in P. horikoshii: the maximal activities of the enzymes are between 80 and 100°C, the activities of both enzymes are almost undetectable below 40°C but they are still active at above 100°C, and the pH dependences and the substrate specificities are the same. The Vmax and Km values for the substrates of the bacterial and the archaeal enzymes are also very similar. The thermostability of T. thermophilus MPGS at 100°C is only slightly lower than the thermostability measured at 98°C for P. horikoshii MPGS. The thermostability of T. thermophilus HB27 MPGP, however, is considerably lower than that of the homofunctional enzyme from P. horikoshii (9). The property that distinguishes MPGSs and MPGPs from T. thermophilus and P. horikoshii is the absolute requirement of the bacterial enzymes for divalent cations for activity. Manganese was most effective for activity of MPGS, and at the lowest concentration it could be partially replaced by higher concentrations of Co2+, Ni2+, or Mg2+. On the other hand, MPGS from P. horikoshii displayed 46% of the maximal activity in the absence of divalent cations, and 15 mM Mg2+ had the maximum effect on the enzyme activity. The cation dependence of T. thermophilus MPGS is unique with respect to other enzymes involved in the synthesis of sugar or sugar derivative compatible solutes, such as glucosylglycerol-phosphate synthase from Synechocystis sp. strain PCC 6803, trehalose-phosphate synthase from Mycobacterium smegmatis, and sucrose-phosphate synthase from Synechocystis. These enzymes, like P. horikoshii MPGS, are active in the absence of cations, but Mg2+ stimulates their activity (7, 14, 29). The absolute cation dependence of the bacterial MPGP for activity also represents the main difference between the bacterial and the archaeal enzyme.
T. thermophilus, unlike P. horikoshii, which grows at temperatures around the boiling point of water (13), has an optimum temperature of only around 70°C and a maximum growth temperature of 80 to 83°C (8). Thus, the relative thermostabilities of the T. thermophilus MPGS and MPGP are very high compared to those of the P. horikoshii enzymes.
Knowledge of the physiological role of MG in T. thermophilus will provide a more detailed understanding of osmoadaptation in thermophilic and hyperthermophilic organisms, and this bacterium, which is suitable for genetic manipulations (15), is the organism of choice to accomplish these objectives. Characterization of this pathway involved in the synthesis of MG in T. thermophilus is an essential step towards this goal.
Acknowledgments
This research was funded by the European Commission 5th Framework Programme, project QLK3-CT-2000-00640, and FCT/FEDER projects PRAXIS/P/BIO/12082/1998 and POCTI/35715/BIO/2000. N. Empadinhas acknowledges a Ph.D. grant from PRAXIS XXI (BD/21665/99).
We thank Joey Marugg (Nestlé Research Center, Lausanne, Switzerland) for advice on molecular biology.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Ames, B. N. 1966. Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 8:115-118. [Google Scholar]
- 3.Borges, N., A. Ramos, N. D. Raven, R. J. Sharp, and H. Santos. 2002. Comparative study of the thermostabilizing properties of mannosylglycerate and other compatible solutes on model enzymes. Extremophiles 6:209-216. [DOI] [PubMed] [Google Scholar]
- 4.Bouveng, H., B. Lindberg, and B. Wickberg. 1955. Low-molecular carbohydrates in algae. Structure of the glyceric acid mannoside from red algae. Acta Chem. Scand. 9:807-809. [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Carreto, L., E. Moore, M. F. Nobre, R. Wait, P. W. Riley, R. J. Sharp, and M. S. da Costa. 1996. Rubrobacter xylanophilus sp. nov., a new thermophilic species isolated from a thermally polluted effluent. Int. J. Syst. Bacteriol. 46:460-465. [Google Scholar]
- 7.Curatti, L., E. Folco, P. Desplats, G. Abratti, V. Limones, L. Herrera-Estrella, and G. Salerno. 1998. Sucrose-phosphate synthase from Synechocystis sp. strain PCC 6803: identification of the spsA gene and characterization of the enzyme expressed in Escherichia coli. J. Bacteriol. 180:6776-6779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.da Costa, M. S., M. F. Nobre, and F. A. Rainey. 2001. The genus Thermus, p. 404-414. In D. R. Boone and R. W. Castenholtz (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer, New York, N.Y.
- 9.Empadinhas, N., J. D. Marugg, N. Borges, H. Santos, and M. S. da Costa. 2001. Pathway for the synthesis of mannosylglycerate in the hyperthermophilic archaeon Pyrococcus horikoshii. Biochemical and genetic characterization of key enzymes. J. Biol. Chem. 276:43580-43588. [DOI] [PubMed] [Google Scholar]
- 10.Gerlt, J. A., and P. C. Babbitt. 2000. Can sequence determine function? Genome Biol. 1:5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Giaever, H. M., O. B. Styrvold, I. Kaasen, and A. R. Strøm. 1988. Biochemical and genetic characterization of osmoregulatory trehalose synthesis in Escherichia coli. J. Bacteriol. 170:2841-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gonçalves, L. G., R. Huber, M. S. da Costa, and H. Santos. 2003. A variant of the hyperthermophile Archaeoglobus fulgidus adapted to grow at high salinity. FEMS Microbiol. Lett. 218:239-244. [DOI] [PubMed] [Google Scholar]
- 13.Gonzalez, J. M., Y. Masuchi, F. T. Robb, J. W. Ammerman, D. L. Maeder, M. Yanagibayashi, J. Tamaoka, and C. Kato. 1998. Pyrococcus horikoshii sp. nov., a hyperthermophilic archaeon isolated from a hydrothermal vent at the Okinawa Trough. Extremophiles 2:123-130. [DOI] [PubMed] [Google Scholar]
- 14.Hagemann, M., U. Effmert, T. Kerstan, A. Schoor, and N. Erdmann. 2001. Biochemical characterization of glucosylglycerol-phosphate synthase of Synechocystis sp. strain PCC 6803: comparison of crude, purified, and recombinant enzymes. Curr. Microbiol. 43:278-283. [DOI] [PubMed] [Google Scholar]
- 15.Hidaka, Y., M. Hasegawa, T. Nakahara, and T. Hoshino. 1994. The entire population of Thermus thermophilus cells is always competent at any growth phase. Biosci. Biotechnol. Biochem. 58:1338-1339. [DOI] [PubMed] [Google Scholar]
- 16.Jensen, S. O., and P. R. Reeves. 1998. Domain organization in phosphomannose isomerases (types I and II). Biochim. Biophys. Acta 1382:5-7. [DOI] [PubMed] [Google Scholar]
- 17.Kane, J. F. 1995. Effects of rare codon clusters on high-level expression of heterologous proteins in Escherichia coli. Curr. Opin. Biotechnol. 6:494-500. [DOI] [PubMed] [Google Scholar]
- 18.Kaneko, T., S. Sato, H. Kotani, A. Tanaka, E. Asamizu, Y. Nakamura, N. Miyajima, M. Hirosawa, M. Sugiura, S. Sasamoto, T. Kimura, T. Hosouchi, A. Matsuno, A. Muraki, N. Nakazaki, K. Naruo, S. Okumura, S. Shimpo, C. Takeuchi, T. Wada, A. Watanabe, M. Yamada, M. Yasuda, and S. Tabata. 1996. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 3:109-136. [DOI] [PubMed] [Google Scholar]
- 19.Kawarabayasi, Y., M. Sawada, H. Horikawa, Y. Haikawa, Y. Hino, S. Yamamoto, M. Sekine, S. Baba, H. Kosugi, A. Hosoyama, Y. Nagai, M. Sakai, K. Ogura, R. Otsuka, H. Nakazawa, M. Takamiya, Y. Ohfuku, T. Funahashi, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K. Aoki, and H. Kikuchi. 1998. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 5:55-76. [DOI] [PubMed] [Google Scholar]
- 20.Kawarabayasi, Y., Y. Hino, H. Horikawa, S. Yamazaki, Y. Haikawa, K. Jin-no, M. Takahashi, M. Sekine, S. Baba, A. Ankai, H. Kosugi, A. Hosoyama, S. Fukui, Y. Nagai, K. Nishijima, H. Nakazawa, M. Takamiya, S. Masuda, T. Funahashi, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, H. Kikuchi, et al. 1999. Complete genome sequence of an aerobic hyper-thermophilic crenarchaeon, Aeropyrum pernix K1. DNA Res. 6:83-101, 145-152. [DOI] [PubMed]
- 21.Klenk, H. P., R. A. Clayton, J. F. Tomb, O. White, K. E. Nelson, K. A. Ketchum, R. J. Dodson, M. Gwinn, E. K. Hickey, J. D. Peterson, D. L. Richardson, A. R. Kerlavage, D. E. Graham, N. C. Kyrpides, R. D. Fleischmann, J. Quackenbush, N. H. Lee, G. G. Sutton, S. Gill, E. F. Kirkness, B. A. Dougherty, K. McKenney, M. D. Adams, B. Loftus, J. C. Venter, et al. 1997. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390:364-370. [DOI] [PubMed] [Google Scholar]
- 22.Lamosa, P., L. O. Martins, M. S. da Costa, and H. Santos. 1998. Effects of temperature, salinity, and medium composition on compatible solute accumulation by Thermococcus spp. Appl. Environ. Microbiol. 64:3591-3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martins, L. O., and H. Santos. 1995. Accumulation of mannosylglycerate and di-myo-inositol-phosphate by Pyrococcus furiosus in response to salinity and temperature Appl. Environ. Microbiol. 61:3299-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martins, L. O., L. S. Carreto, M. S. da Costa, and H. Santos. 1996. New compatible solutes related to di-myo-inositol-phosphate in members of the order Thermotogales. J. Bacteriol. 178:5644-5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martins, L. O., R. Huber, H. Huber, K. O. Stetter, M. S. da Costa, and H. Santos. 1997. Organic solutes in hyperthermophilic archaea. Appl. Environ. Microbiol. 63:896-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martins, L. O., N. Empadinhas, J. D. Marugg, C. Miguel, C. Ferreira, M. S. da Costa, and H. Santos. 1999. Biosynthesis of mannosylglycerate in the thermophilic bacterium Rhodothermus marinus. Biochemical and genetic characterization of a mannosylglycerate synthase. J. Biol. Chem. 274:35407-35414. [DOI] [PubMed] [Google Scholar]
- 27.Nelson, K. E., R. A. Clayton, S. R. Gill, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, W. C. Nelson, K. A. Ketchum, L. McDonald, T. R. Utterback, J. A. Malek, K. D. Linher, M. M. Garrett, A. M. Stewart, M. D. Cotton, M. S. Pratt, C. A. Phillips, D. Richardson, J. Heidelberg, G. G. Sutton, R. D. Fleischmann, J. A. Eisen, O. White, S. L. Salzberg, H. O. Smith, J. C. Venter, and C. M. Fraser. 1999. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature 399:323-329. [DOI] [PubMed] [Google Scholar]
- 28.Nunes, O. C., C. M. Manaia, M. S. da Costa, and H. Santos. 1995. Compatible solutes in the thermophilic bacteria Rhodothermus marinus and “Thermus thermophilus.” Appl. Environ. Microbiol. 61:2351-2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan, Y. T., R. R. Drake, and A. D. Elbein. 1996. Trehalose-P synthase of mycobacteria: its substrate specificity is affected by polyanions. Glycobiology 6:453-461. [DOI] [PubMed] [Google Scholar]
- 30.Ramos, A., N. D. H. Raven, R. J. Sharp, S. Bartolucci, M. Rossi, R. Cannio, J. Lebbink, J. Van der Oost, W. M. De Vos, and H. Santos. 1997. Stabilization of enzymes against thermal stress and freeze-drying by mannosylglycerate. Appl. Environ. Microbiol. 63:4020-4025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robb, F. T., D. L. Maeder, J. R. Brown, J. DiRuggiero, M. D. Stump, R. K. Yeh, R. B. Weiss, and D. M. Dunn. 2001. Genomic sequence of hyperthermophile, Pyrococcus furiosus: implications for physiology and enzymology. Methods Enzymol. 330:134-157. [DOI] [PubMed] [Google Scholar]
- 32.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 33.Santos, H., and M. S. da Costa. 2001. Organic solutes from thermophiles and hyperthermophiles. Methods Enzymol. 334:302-315. [DOI] [PubMed] [Google Scholar]
- 34.Santos, H., and M. S. da Costa. 2002. Compatible solutes of organisms that live in hot saline environments. Environ. Microbiol. 4:501-509. [DOI] [PubMed] [Google Scholar]
- 35.Silva, Z., C. Horta, M. S. da Costa, A. P. Chung, and F. A. Rainey. 2000. Polyphasic evidence for the reclassification of Rhodothermus obamensis Sako et al. 1996 as a member of the species Rhodothermus marinus Alfredsson et al. 1988. Int. J. Syst. E vol. Microbiol. 50:1457-1461. [DOI] [PubMed] [Google Scholar]
- 36.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomson, K-S. 1983. Purification of UDP galactose:sn-glycerol-3-phosphate-d-galactosyltransferase from Poterioochromonas malhamensis. Biochim. Biophys. Acta 759:154-159. [Google Scholar]
- 38.White, O., J. A. Eisen, J. F. Heidelberg, E. K. Hickey, J. D. Peterson, R. J. Dodson, D. H. Haft, M. L. Gwinn, W. C. Nelson, D. L. Richardson, K. S. Moffat, H. Qin, L. Jiang, W. Pamphile, M. Crosby, M. Shen, J. J. Vamathevan, P. Lam, L. McDonald, T. Utterback, C. Zalewski, K. S. Makarova, L. Aravind, M. J. Daly, C. M. Fraser, et al. 1999. Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286:1571-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]