Abstract
Selectively regulating gene expression is an essential molecular tool that is lacking for many pathogenic gram-positive bacteria. In this report, we describe the evaluation of a series of promoters regulated by the bacteriophage P1 temperature-sensitive C1 repressor in Enterococcus faecium, Enterococcus faecalis, and Staphylococcus aureus. Using the lacZ gene to monitor gene expression, we examined the strength, basal expression, and induced expression of synthetic promoters carrying C1 operator sites. The promoters exhibited extremely low basal expression and, under inducing conditions, gave high levels of expression (100- to 1,000-fold induction). We demonstrate that the promoter system could be modulated by temperature and showed rapid induction and that the mechanism of regulation occurred at the level of transcription. Controlled expression with the same constructs was also demonstrated in the gram-negative bacterium Escherichia coli. However, low basal expression and the ability to achieve derepression were dependent on both the number of mismatches in the C1 operator sites and the promoter driving c1 expression. Since the promoters were designed to contain conserved promoter elements from gram-positive species and were constructed in a broad-host-range plasmid, this system will provide a new opportunity for controlled gene expression in a variety of gram-positive bacteria.
The overuse of antibiotics has contributed to the emergence and increasing prevalence of antibiotic-resistant bacteria. Gram-positive cocci such as Staphylococcus aureus and enterococci are the leading cause of hospital-acquired infections (39). S. aureus causes a variety of infections ranging from localized skin suppuration to life-threatening septicemia. Alarmingly, S. aureus isolates resistant to vancomycin, the last effective antibiotic, are emerging worldwide (22). Enterococcus species are a leading cause of urinary tract infection, nosocomial infection, and surgical-wound infection (39). Enterococcus faecalis is responsible for the majority of enterococcal infections (26, 41) and, for the time being, usually remains sensitive to at least one antibiotic. In contrast Enterococcus faecium, which causes fewer infections, is more likely to be resistant to all antibiotics.
The recent emergence of antibiotic-resistant gram-positive bacteria has highlighted the need for genetic studies addressing the mechanism of bacterial pathogenesis. Regulated promoters are essential for the functional analysis of genes through expression studies (1) and reverse genetics. However, only a few regulated promoters are available for use in enterococci and streptococci. The tetracycline-regulated promoter system has been shown to function in Streptococcus pneumoniae (47), Bacillus subtilis (11), and S. aureus (1, 25). In addition, the xylose-inducible promoter system has been used for B. subtilis and staphylococci (27, 52, 56). However, the levels of regulation achieved with these systems are below those obtained for gram-negative bacteria (33), and tight basal expression is achieved at the expense of reduced inducibility (11, 56). The most studied regulated promoter from gram-positive bacteria is the nisA promoter, derived from the Lactococcus lactis nisin gene cluster. For regulated expression, the system requires coexpression of histidine protein kinase NisK and response regulator NisR (6). Induction is achieved by the addition of subinhibitory levels of the lantibiotic nisin. Controlled gene expression over a 1,000-fold range in L. lactis has been demonstrated (4). However, more-modest regulation has been demonstrated in heterologous hosts (6) such as E. faecalis (20-fold), Streptococcus agalactiae (10-fold), and Streptococcus pyogenes (60-fold).
One of the reasons why there are fewer regulated promoters for gram-positive species may be the more stringent control of promoter usage in gram-positive species than in gram-negative species. Multiple conserved regions, in addition to the −35 and −10 hexamers, have been identified in promoters from gram-positive species (14, 20, 50, 51). Consequently, well-characterized promoters from gram-negative species such as Ptac and Ptrc are inactive in gram-positive hosts even though they contain consensus −35 and −10 hexamers (38).
The temperate bacteriophage P1 can infect and lysogenize many gram-negative species (55). Stable lysogeny is maintained by the action of the components of the tripartite immunity system (17). The C1 repressor protein acts as a central regulator by controlling the expression of a variety of genes (3, 7, 18, 19) by binding to C1 asymmetric operator (7) sites (consensus sequence, ATTGCTCTAATAAATTT). A bacteriophage P1-derived promoter in conjunction with the temperature-sensitive C1 repressor (40) has been used to regulate gene expression in gram-negative bacteria (45). In this report we demonstrate that the P1 temperature-sensitive C1 repressor can be used to control gene expression by using synthetic promoters in the pathogenic gram-positive species E. faecium, E. faecalis, and S. aureus. We compare the strengths of the promoters in different species and show that the promoters exhibit extremely low basal expression and that the control of regulation occurs at the level of transcription. Since the promoter system was constructed in a broad-host-range plasmid and contained conserved promoter elements, the system will provide a new opportunity for controlled gene expression in gram-positive bacteria.
MATERIALS AND METHODS
Strains and media.
The bacterial strains used in this study were E. coli DH5α (φ80dlacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 deoR Δ(lacZYA-argF)U169; obtained from Gibco-BRL), S. aureus RN4220 (restriction-deficient strain kindly provided by Jean Lee, Channing Laboratory, Boston, Mass.), E. faecalis ATCC 47077 (designation OG1RF), and E. faecium ATCC 12952. The following growth media (Difco) were used: Luria-Bertani broth for E. coli, tryptic soy broth for S. aureus, brain heart infusion broth for E. faecalis, and Todd-Hewitt broth for E. faecium.
Reporter plasmid construction.
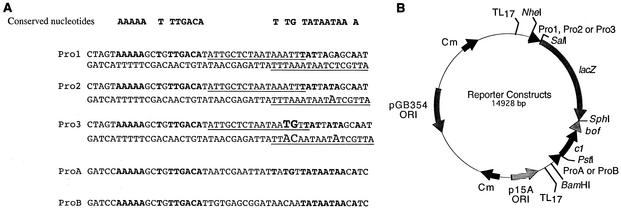
The reporter plasmids were constructed in the gram-negative and gram-positive species shuttle vector pAM401 (53). The lacZ gene was amplified by PCR with pBHRlacZ (45) as the template and the upstream primer 5′-AGGACGGTCGACTAAGGAGGTGAAAAGTATGGTCGTTTTACAAGCTCG and downstream primer 5′-TCCTCCGCATGCTCCCCCCTGCCCGGTTAT, which contained SalI and SphI restriction sites (underlined) for cloning into the SalI and SphI sites of pAM401. The upstream primer also contained a ribosome binding site (RBS; 5′-TAAGGAGG) positioned 8 bp upstream of a start codon (boldface) to initiate translation. The C1-regulated promoters (Fig. 1A; Pro1, -2, and -3) were obtained by annealing complementary oligonucleotides that contained partial and full SalI overhangs (5′ and 3′, respectively). The promoters were cloned (in the same orientation as lacZ) into the SalI site of the lacZ construct, thereby re-creating the 3′ SalI site only. To increase the number of cloning sites, the oligonucleotides also contained a SpeI site at the 5′ end. To reduce readthrough from cryptic promoters into the 5′ end of the expression cassette, the TL17 transcriptional terminators (54) were cloned into the SpeI site. To prevent “runaway” transcription, the terminators were also cloned at the 3′ end of the expression cassette (EcoRV site). To control gene expression, the coding sequences for the c1 repressor and bof modulator were inserted initially into the cloning vector pBluescript II SK(+) (Stratagene). The forward PCR primers used to amplify c1 and bof contained both an RBS and restriction endonuclease site. To incorporate both of these features, c1 and bof were amplified by a seminested-PCR strategy. c1 was amplified by using the thermosensitive P1 mutant as the template (40) (kindly provided by Michael Yarmolinsky, National Institutes of Health, Bethesda, Md.) with the forward primer 5′-TAAGGAGGTGAAAAGTATGATAAATTATGTCTACGGC and the reverse primer 5′-CTAGCTGAATTCCTATTGCGCGCTTTCGGGGTTG. After 10 amplification cycles, an aliquot (1 μl) was reamplified with the forward nested primer 5′-CGCAGTGAATTCTAAGGAGGTGAAAAGTATG and the same reverse primer. The forward primers each contained an RBS upstream of the start codon (boldface), and the reverse primer and the latter forward primer contained EcoRI restriction sites (underlined sequences) for cloning into the corresponding sites of pBluescript II SK(+). Similarly, the forward primer 5′-TAAGGAGGTGAAAAGTATGAAAAAGCGATACTACACAG, reverse primer5′-GTAGTAGCATGCGGTGAGCAAACAGCCAT, and nested forward primer 5′-GCTAGGAAGCTTTAAGGAGGTGAAAAGTATG were used to amplify bof with bacteriophage P1 DNA as the template. The bof primers contained HindIII and SphI sites (underlined). However, bof was cloned 3′ of c1 into the HindIII and HincII sites of pBluescript II SK(+). To drive expression of c1 and bof, complementary oligonucleotides containing promoter elements (Fig. 1A; ProA and -B) were cloned upstream of c1 and bof into the BamHI and PstI sites of pBluescript II SK(+). The promoter c1.bof fragments with BamHI/SphI overhangs were then cloned into the corresponding sites of pDAS101, pDAS111, and pDAS121 to create the final reporter constructs (Fig. 1B and Table 1).
FIG. 1.
Construction of the temperature-sensitive C1-regulated promoter system. (A) Topography and sequences of the promoters. Determined based on compilation analysis, the conserved promoter nucleotides from gram-positive bacteria are in boldface (14, 20). The synthetic promoters (Pro1, -2, and -3) consist of two partially overlapping C1 operators (top and bottom strands; underlined sequences). Pro1 carries two C1 operator sites that match the 17-bp consensus (7, 18), while Pro2 and Pro3 deviate from the consensus by 1 and 5 nucleotides, respectively (large font). Pro2 differs from Pro1 by a single nucleotide in the −10 hexamer (G versus the consensus T). Pro3 differs from Pro2 by 2 nucleotide changes in the spacer region (AT versus the consensus TG). ProA and ProB, which drive c1 expression, differ in the nucleotide spacer sequence between the −35 and −10 hexamers. (B) Map of the reporter plasmid and its relevant features. The lacZ reporter gene was placed under the transcriptional control of a C1-regulated promoter (Pro1, -2, or -3; arrows denote direction). To control gene expression and to aid the binding of the repressor to its operator site, the temperature-sensitive C1 repressor and Bof modulator were cloned 3′ of lacZ and placed under the transcriptional control of either ProA or -B. The reporter construct contains the p15A origin of replication, the origin of replication derived from pGB354, and the chloramphenicol (Cm) resistance markers from pACYC184 and pGB354 (53).
TABLE 1.
Plasmids used in this study
| Plasmid | Description and relevant characteristics | Origin or reference |
|---|---|---|
| pAM401 | Gram-positive and -negative species shuttle vector | 53 |
| pDAS100 | pAM401 containing lacZ and transcriptional terminators | This study |
| pDAS101 | pDAS100 with Pro1 driving lacZ | This study |
| pDAS102 | pDAS101 with ProA driving c1 and bof | This study |
| pDAS103 | pDAS101 with ProB driving c1 and bof | This study |
| pDAS111 | pDAS100 with Pro2 driving lacZ | This study |
| pDAS112 | pDAS111 with ProA driving c1 and bof | This study |
| pDAS113 | pDAS111 with ProB driving c1 and bof | This study |
| pDAS121 | pDAS100 with Pro3 driving lacZ | This study |
| pDAS122 | pDAS121 with ProA driving c1 and bof | This study |
| pDAS123 | pDAS121 with ProB driving c1 and bof | This study |
Transformation.
E. coli was transformed as described by Sambrook et al. (42). E. faecalis and E. faecium were electroporated as described by Friesenegger et al. (9) except cells were resuspended at 1/100 of their original culture volume. S. aureus was electroporated by the method described by Lee (31). Chloramphenicol was used to select for plasmids at the following concentrations: 25 μg/ml, E. coli; 20 μg/ml, E. faecalis; 5 μg/ml, E. faecium; 15 μg/ml, S. aureus.
RNA extraction and slot blot hybridizations.
RNA was extracted from E. faecium, E. faecalis, and S. aureus by using the Qiagen RNeasy kit according to the manufacturer's instructions with the following modification: to break open the bacterial cells, the samples were vortexed continuously for 10 min in the presence of acid-washed glass beads (212 to 300 μm). RNA (up to 10 μg) was vacuum blotted onto Duralon UV membranes (Stratagene) with a slot blot apparatus (42). Two identical RNA blots were prepared for each species. Both membranes were probed with a 35S-tailed (Roche) oligonucleotide complementary to either lacZ (5′-CGCTCAGGTCAAATTCAGACGGCAAACGA) or a conserved region of 16S rRNA (5′-CCAACATCTCACGACACGAGCTGACGACAA). Hybridization was performed in a mixture containing 1× Denhardt's solution, 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 50 μg of poly(A)/ml, 500 μg of salmon sperm/ml, 10% dextran sulfate, and 45% formamide at 37°C. Washing was performed at 37°C at a final stringency of 0.5× SSC and 0.1% sodium dodecyl sulfate (SDS). The membranes were visualized with a phosphorimager.
Reporter gene assays.
β-Galactosidase (β-Gal) activity was assayed as described by Miller (35) except that the cells were permeabilized with 4 drops of chloroform and 2 drops of 0.1% SDS.
Nucleotide sequence accession number.
The sequence of the reporter plasmid pDAS112 has been deposited in GenBank under accession no. AY230218.
RESULTS AND DISCUSSION
Rationale for and characteristics of the reporter gene constructs.
Controlled expression using a temperature-sensitive C1-regulated promoter system has been demonstrated previously for gram-negative bacteria (44, 45) by using a promoter responsible for driving ban gene expression in bacteriophage P1 (18). The ban promoter consisted of two overlapping C1 operator sites, and, although it did not contain consensus −35 and −10 hexamers, it was highly active in enteric gram-negative bacteria (43, 45). However, reporter gene analysis indicated that this promoter was inactive in the gram-positive species E. faecalis and S. aureus (data not shown). Functional promoters derived from gram-negative species are often inactive in gram-positive species (21, 30, 36, 38), which may reflect the fact that several conserved promoter elements (14, 20, 50, 51), in addition to the −35 and −10 hexamers, are required for activity. Consequently, compilation analysis of promoters from gram-positive species (14, 20) allowed three promoters (Pro1, -2, and -3) with conserved elements to be designed. The conserved elements consisted of the −35 and −10 hexamers, an A tract and T 5′ of the −35 hexamer, a TG dinucleotide 5′ of the −10 hexamer, and two A nucleotides 3′ of the −10 hexamer (Fig. 1A). The promoters differed by a single nucleotide within the −10 hexamer (Pro1 versus Pro2) or by the addition of TG nucleotides (Pro2 versus Pro3; Fig. 1A). The promoters were also designed to contain two partially overlapping C1 operator sites (7). Placement of the C1 operators downstream of the −10 hexamer resulted in only partial repression in the presence of C1 in E. coli (data not shown); consequently the operator on the top strand was placed between the −35 and −10 hexamers while the operator on the bottom strand completely covered the −10 hexamer. In the presence of a repressor, this placement was expected to more effectively prevent transcription by exclusion of the RNA polymerase and/or by the masking of the promoter elements (18, 29). However, as a result of the optimization of promoter elements, Pro3 and Pro2 carried five and one mismatch to the consensus C1 operator sequence, respectively. Nevertheless, these operator sites were expected to be effective since functional C1 binding sites containing mismatches to the consensus sequence have been identified throughout the P1 prophage (3, 43).
The amount of repressor produced is crucial to the effectiveness of a promoter system; small amounts of repressor can result in partial repression, while too much repressor results in the inability to achieve derepression (45, 46). Therefore, the c1 gene was placed under the transcriptional control of two promoters (ProA and ProB; Fig. 1A) which have consensus −35 and −10 hexamers but which differ in their spacer sequences. Since variations in the spacer sequence have been shown to alter promoter strength by up to 400-fold (24), this was expected to provide differing amounts of C1 repressor. To enhance binding of the C1 repressor to its operator (49), the bof gene was cloned 3′ of the c1 gene. The Bof protein does not bind to C1 alone but binds to C1 operators by forming a C1-Bof-operator DNA ternary complex (48). To ensure efficient translation, the primers amplifying lacZ incorporated a contrived RBS derived from gram-positive species (6). This resulted in a 200-fold increase in β-Gal activity in E. faecalis (data not shown) compared to that with the lacZ RBS (GGAGG[N]6ATG) used for reporter gene studies of gram-negative bacteria (45); consequently, the former RBS was also incorporated into the forward primers amplifying c1 and bof. At the permissive temperature, C1 binds to its operator sites and prevents transcription from the reporter gene, while at the nonpermissive temperature, C1 is thermally unstable, thereby allowing transcription to proceed.
The reporter plasmids were constructed in pAM401, which contains a p15A replicon derived from pACYC184 and a pGB354 replicon derived from the broad-host-range gram-positive-bacterium plasmid pIP501 (53). Consequently, the plasmid can be used for studies of enteric gram-negative bacteria, Streptococcus species (8, 28), Enterococcus species (9), Streptococcus gordonii (unpublished results), L. lactis (10), Lactobacillus casei (12), and Pediococcus species (13). The copy number of the pAM401 parental plasmid (pGB354) in streptococci has been reported to be approximately 50 copies/cell (2).
Previous work using the parent plasmid pAM401 has demonstrated transformation efficiencies of approximately 105 for E. faecalis and E. faecium (9). Although the transformation efficiencies were not measured in this report, it is expected that similar transformation efficiencies can be obtained for the larger reporter constructs since plasmids up to 25 kb can be electroporated without any loss in transformation efficiency (34, 37). In our experiments, the relative transformation efficiency was slightly lower for S. aureus than for E. faecalis and E. faecium (data not shown).
Plasmid maintenance studies were performed to test the stability of pDAS122 in S. aureus, pDAS113 in E. faecium, and pDAS113 in E. faecalis. Cultures (in triplicate) harboring the plasmids were grown overnight in selective (chloramphenicol) liquid media and then plated on nonselective or selective solid media. The percentages of chloramphenicol-resistant colonies obtained for E. faecalis, E. faecium, and S. aureus were 89, 77, and 61%, respectively. Therefore, strains carrying the reporter constructs were always grown under selective pressure.
Analysis of β-Gal activity from the temperature-sensitive C1-regulated promoters in E. coli.
Since E. coli is the preferred host for cloning and propagation of plasmids, it is important to know whether the regulated promoter is efficiently repressed in this host; this may be essential if, for example, the gene of interest encodes a toxic protein. Therefore, to demonstrate the functionality of the promoter system, β-Gal activity was measured initially in E. coli. β-Gal activity was measured by using three C1-regulated promoters driving lacZ at the permissive (31°C) and nonpermissive (42°C) temperatures. In the absence of C1, the activities of all three promoters were high, with Pro2 and Pro3 producing approximately 5- to 10-fold more Miller units than Pro1 (Table 2). This was most likely due to the nucleotide change from G to the consensus T within the −10 hexamers in Pro2 and Pro3 (Fig. 1A). Pro2 and Pro3 exhibited similar activities, indicating that the TG dinucleotide had little effect on promoter strength in E. coli. In the presence of C1 and at low temperature, β-Gal activity was significantly reduced, indicating that C1 can efficiently repress transcription from these promoters. In particular, the basal expression of Pro2 was much lower than that of Pro3, which was probably a reflection of the number of mismatches in the C1 operator sites (one and five mismatches, respectively) and hence the ability to more effectively repress transcription. Interestingly, the basal expression of Pro2 was also lower than that of the control vector, which contained a promoterless lacZ gene (pDAS100). This may be explained by the observation that, in E. coli, repressor-bound operators can prevent the formation of active complexes between RNA polymerase and promoters and also terminate ongoing transcription (5). It also suggests that the terminators used in the plasmid construction were not 100% effective, leading to some promoter readthrough from the plasmid backbone. Little difference was observed in the basal levels of expression from the C1-regulated promoters when C1 was expressed from ProA or ProB, suggesting that adequate amounts of C1 were produced from both constructs to effectively repress transcription. At the nonpermissive temperature, β-Gal activity from the C1-regulated promoters significantly increased, although levels were still below levels obtained in the absence of C1 (Table 2). Nevertheless, the range of regulation was similar to that for the bacteriophage P1-derived C1-regulated promoter system in E. coli described previously (44).
TABLE 2.
Basal and induced activities from lacZ fusions to C1-regulated promoters in E. coli DH5αa
| Construct | Promoter driving:
|
Avg activity in Miller units (±SD)
|
||
|---|---|---|---|---|
| lacZ | c1 | Basal (31°C) | Induced (42°C) | |
| pDAS100 | 4.2 (0.5) | 6.0 (0.7) | ||
| pDAS101 | Pro1 | 1,117.5 (223.4) | 2,197.1 (77.9) | |
| pDAS102 | Pro1 | ProA | ND | ND |
| pDAS103 | Pro1 | ProB | ND | ND |
| pDAS111 | Pro2 | 15,478.9 (675.7) | 10,899.2 (531.1) | |
| pDAS112 | Pro2 | ProA | <0.25 | 15.9 (1.2) |
| pDAS113 | Pro2 | ProB | <0.25 | 5.7 (0.1) |
| pDAS121 | Pro3 | 9,119.0 (272.4) | 9,575.1 (666.2) | |
| pDAS122 | Pro3 | ProA | 2.8 (0.1) | 2,386.1 (504.8) |
| pDAS123 | Pro3 | ProB | 2.0 (0.3) | 213.1 (11.1) |
Overnight cultures were diluted 1:100 and grown to an optical density at 600 nm (OD600) of approximately 0.1 at 31°C. The cultures were then divided equally and incubated at 31 or 42°C for 95 min prior to being assayed for β-Gal activity (OD600 of approximately 0.6). The control strain carried a plasmid (pDAS100) containing a promoterless lacZ gene. Values are for multiple cultures (n = 3) assayed in triplicate. A value of <0.25 indicates activity below the limits of detection for the assay. ND, not determined.
Analysis of β-Gal expression using the temperature-sensitive C1-regulated promoters in E. faecium, E. faecalis, and S. aureus.
The C1-regulated promoters in E. faecium, E. faecalis, and S. aureus were analyzed (Table 3). In the absence of C1, the activity of Pro1 was low to undetectable; therefore, constructs containing this promoter were not examined further. However, Pro2 expression was high, indicating that the 1-nucleotide difference between Pro1 and Pro2, in contrast to what was found for E. coli, was essential for activity in these species. The addition of the TG dinucleotide (Pro3) further increased the strength of the promoter. In the presence of C1 at the permissive temperature, the basal activity of Pro2 was reduced to the background level displayed by the control strain carrying the promoterless lacZ construct. This indicated that the Pro2 promoter was completely repressed in the presence of C1. In E. faecalis and S. aureus, the basal level of expression was below the limits of detection when c1 was expressed from either ProA or ProB. In E. faecium, however, basal activity was slightly higher when ProB, instead of ProA, was used to drive c1 expression, suggesting that the ability to repress transcription was dependent on the levels of C1 expressed.
TABLE 3.
Basal and induced activities from lacZ fusions to C1-regulated promoters in E. faecium, E. faecalis, and S. aureusa
| Species and construct | Promoter driving:
|
Avg activity in Miller units (±SD)
|
||
|---|---|---|---|---|
| lacZ | c1 | Basal (31°C) | Induced (42°C) | |
| E. faecium | ||||
| pDAS100 | 1.3 (0.1) | 0.6 (0.03) | ||
| pDAS101 | Pro1 | 1.7 (0.4) | 1.4 (0.35) | |
| pDAS102 | Pro1 | ProA | ND | ND |
| pDAS103 | Pro1 | ProB | ND | ND |
| pDAS111 | Pro2 | 1,769.9 (89.6) | 3,849.2 (131.6) | |
| pDAS112 | Pro2 | ProA | 1.8 (0.3) | 1.6 (0.1) |
| pDAS113 | Pro2 | ProB | 3.4 (0.2) | 640.3 (14.5) |
| pDAS121 | Pro3 | 2,344.6 (165.1) | 2,564.3 (387.7) | |
| pDAS122 | Pro3 | ProA | 227.6 (10.8) | 699.0 (57.2) |
| pDAS123 | Pro3 | ProB | 825.1 (16.3) | 1,528.1 (65.4) |
| E. faecalis | ||||
| pDAS100 | <0.25 | <0.25 | ||
| pDAS101 | Pro1 | <0.25 | <0.25 | |
| pDAS102 | Pro1 | ProA | ND | ND |
| pDAS103 | Pro1 | ProB | ND | ND |
| pDAS111 | Pro2 | 1,139.1 (23.6) | 3,068.3 (119.7) | |
| pDAS112 | Pro2 | ProA | <0.25 | <0.25 |
| pDAS113 | Pro2 | ProB | <0.25 | 269.0 (49.5) |
| pDAS121 | Pro3 | 2,332.4 (54.6) | 4,860.7 (149.8) | |
| pDAS122 | Pro3 | ProA | 2.83 (1.4) | 1,181.2 (59.5) |
| pDAS123 | Pro3 | ProB | 884.0 (145.4) | 1,120.5 (29.1) |
| S. aureus | ||||
| pDAS100 | <0.25 | <0.25 | ||
| pDAS101 | Pro1 | <0.25 | <0.25 | |
| pDAS102 | Pro1 | ProA | ND | ND |
| pDAS103 | Pro1 | ProB | ND | ND |
| pDAS111 | Pro2 | 76.1 (7.9) | 183.4 (35.5) | |
| pDAS112 | Pro2 | ProA | <0.25 | <0.25 |
| pDAS113 | Pro2 | ProB | <0.25 | 4.6 (0.74) |
| pDAS121 | Pro3 | 129.5 (16.8) | 257.8 (55.1) | |
| pDAS122 | Pro3 | ProA | <0.25 | 26.4 (5.8) |
| pDAS123 | Pro3 | ProB | 54.6 (3.8) | 138.4 (9.1) |
Overnight cultures were diluted 1:100 and grown to an optical density at 600 nm (OD600) of approximately 0.1 at 31°C. The cultures were then divided equally and incubated at 31 or 42°C for 120 min (E. faecium), 95 min (E. faecalis), or 75 min (S. aureus) prior to being assayed for β-Gal activity (OD600 of approximately 0.6). The control strain carried a plasmid (pDAS100) containing a promoterless lacZ gene. Values are for multiple cultures (n = 3) assayed in triplicate. ND, not determined.
Basal expression exhibited by Pro3 was generally higher than that exhibited by Pro2 and was more dependent on the promoter driving c1 expression and, presumably, the concentration of repressor present. The higher basal expression may be a reflection of the increased number of mismatches in the C1 operator sites compared to that for Pro2 (five compared to one), leading to less-efficient binding of the C1 repressor (18). Moreover, since the Pro3 promoter was generally stronger than the Pro2 promoter, the higher expression may also reflect the increased ability of RNA polymerase to compete with the repressor for binding to the unoccupied promoters. Nevertheless, low basal expression was still observed in S. aureus and E. faecalis when c1 was expressed from the ProA promoter.
Under inducing conditions when either the Pro2 or Pro3 promoter was used, striking differences in the levels of induced expression were achieved depending on whether ProA or ProB was used to drive c1 expression and which species was tested (Table 3). Induction was not observed when the Pro2 promoter in combination with the ProA promoter was used for any of the species (pDAS112). In contrast, high induced activity was obtained when Pro2 in combination with ProB was used to drive c1 expression in E. faecalis, E. faecium, and, to a lesser extent, S. aureus. Using the Pro3 promoter in combination with ProA to drive c1 expression (pDAS122) resulted in regulated expression in E. faecalis and S. aureus only, while using Pro3 in combination with ProB (pDAS123) did not result in regulated expression for any of the species due to the high basal expression. Therefore, use of only some of the constructs resulted in regulated expression. Moreover, the results suggested that differences in C1 expression correlated with the ability to achieve derepression. Partial derepression when the repressor is in excess has been demonstrated previously for regulated promoter systems in gram-negative bacteria (33, 45). Indeed, ProA might be expected to be more active than ProB, resulting in higher levels of C1 expressed, since it contains more conserved nucleotides. However, low levels of basal activity and elevated induced expression were obtained in S. aureus and E. faecalis by using the Pro3 promoter in combination with ProA to drive c1 expression. This suggests that induced expression depends on both the interaction between the repressor and operator site and the amount of repressor present. C1 has also been shown to be more thermally stable when tightly bound to DNA than it is in its unbound form, which can be dissociated only by further temperature increases (16). It should be noted that induced expression was achieved in E. coli with these constructs irrespective of the promoters utilized (Table 2). Nevertheless, these results demonstrated that a temperature-sensitive C1-regulated promoter can be effectively repressed to levels comparable to those for the control vectors yet yield high levels of induced expression. Induction/repression ratios for E. faecium, E. faecalis, and S. aureus were approximately 200, and at least 1,000, and 100, respectively. Consequently, these data represent the first heterologous regulated promoter system to be described for E. faecium and reflect a range of regulation in E. faecalis which is similar to those for the promoter systems described for E. coli (15). The level of regulation achieved for S. aureus is comparable, if not better, than those for previously described promoter systems (25, 56). In addition, since different combinations of promoters were evaluated, constructs can be selected depending on whether tight basal or highly induced expression is preferred.
However, note that the level of β-Gal activity detected varied by species and was highest in E. coli, followed by E. faecium, E. faecalis, and S. aureus. In particular, β-Gal activity was low in S. aureus, which may be due to weak promoter activity, inefficient translation of the lacZ message, or poor stability of the LacZ protein. Alternatively, it may reflect inefficient permeabilization of the S. aureus cells and hence a lower sensitivity of the β-Gal assay. However, since the promoters were designed on the basis of conserved elements from gram-positive species and since the relative lacZ message levels were similar, it is likely that the low expression levels were due to weak translation and/or detection rather than poor promoter activity.
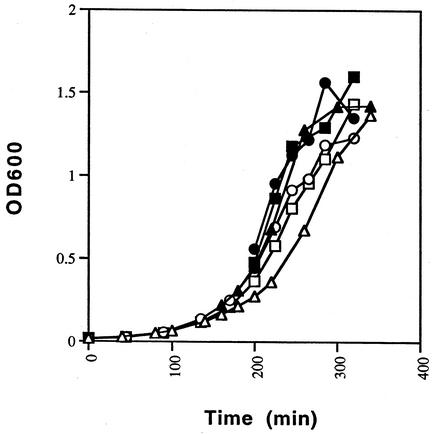
To investigate the growth characteristics of E. faecalis, E. faecium, and S. aureus under the same conditions that were used for the promoter analysis, cell growth was monitored spectrophotometrically at 31 and 42°C (Fig. 2). When the culture temperature was changed from 31 to 42°C, cell growth rates increased. The increase in growth rate may result in differences in the levels of expression for the different constructs that are independent of the temperature-sensitive C1 repressor and hence the regulated promoter system. For example, in the absence of the repressor, activities from the promoter constructs (pDAS111 and pDAS121) would be expected to be similar at both temperatures. However, except for pDAS121 in E. faecium, the activities were approximately twofold higher at 42°C than at 31°C (Table 3). It is reasonable to speculate that these differences can be attributed to the increased growth rate at 42°C; although Miller units (β-Gal activity) account for the number of cells assayed, they do not take into consideration the growth rates of the different species. Therefore, the actual level of induced expression and hence the range of regulation may be slightly lower than the reported values.
FIG. 2.
Time course analysis of cell growth at 31 and 42°C. Overnight cultures were diluted 1:100 and grown at 31°C to early log phase. The cultures (S. aureus [squares], E. faecalis [triangles], and E. faecium [circles]) were then divided equally and incubated at 31°C (open symbols) or 42°C (solid symbols). Cell growth was monitored spectrophotometrically by measuring optical density at 600 nm (OD600).
Transcriptional regulation of lacZ expression.
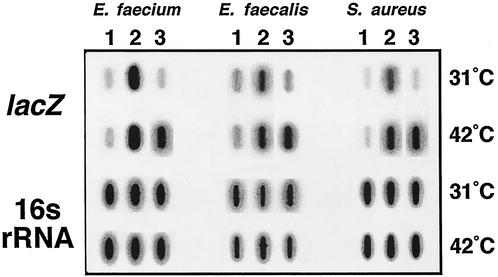
To analyze the regulation of lacZ expression at the transcriptional level, slot blot analysis was performed (32). Since promoters were located in both orientations in the plasmid, hybridizations were performed with an oligonucleotide complementary to lacZ as a probe. RNA was prepared from cultures carrying (i) promoterless lacZ control constructs, (ii) reporter constructs lacking the c1 repressor, and (iii) reporter constructs under repressed and derepressed conditions (Fig. 3). The blots were also hybridized with a complementary oligonucleotide homologous to a conserved region of 16S rRNA to verify equal loading of the RNA. Levels of lacZ expression from the promoterless lacZ control constructs and the constructs lacking c1 were low and high, respectively, as expected (Fig. 3). Furthermore, the levels of lacZ transcripts produced from the control vectors and reporter constructs under repressed conditions were similar, indicating that C1 can efficiently repress transcription. In contrast, at elevated temperatures, lacZ expression from the reporter constructs was significantly increased. The results are therefore in agreement with enzymatic assays and confirmed that the regulation of lacZ expression occurred primarily at the level of transcription.
FIG. 3.
Slot blot analysis of lacZ expression in E. faecium, E. faecalis, and S. aureus. Overnight cultures were diluted 1:100 and grown to an optical density at 600 nm of approximately 0.1. Cultures were then divided equally and incubated at 31 or 42°C for 120, 95, and 75 min, respectively. Cultures carried either a promoterless lacZ construct (pDAS100; lane 1); a reporter construct lacking the c1 repressor (pDAS111 for E. faecium and E. faecalis and pDAS121 for S. aureus; lane 2), or a C1-regulated reporter construct (pDAS113 for E. faecium and E. faecalis and pDAS122 for S. aureus; lane 3). RNA was extracted from the cultures, blotted onto the membrane, and hybridized to either a lacZ or 16S rRNA complementary oligonucleotide probe.
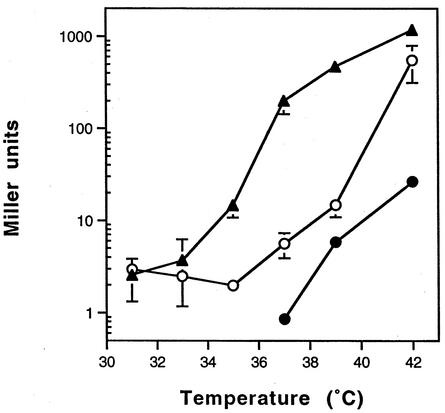
Modulation of β-Gal activity in E. faecium, E. faecalis, and S. aureus.
The ability to obtain different levels of expression by partial induction of the promoter is an important feature of a controlled expression system. Therefore, to assess the ability to modulate the temperature-sensitive C1-regulated promoter system in E. faecium, E. faecalis, and S. aureus, β-Gal activity was measured at different temperatures. The results indicated that, by varying temperature, it was possible to modulate expression (Fig. 4). However, the degree to which the promoter could be modulated was dependent on the host. For example, in E. faecalis there was a steady increase in β-Gal activity as the temperature increased. In contrast, the level of β-Gal expressed in E. faecium remained relatively unchanged until 39°C. For all three species, maximal induction was achieved at the highest temperature tested (42°C), which is in agreement with previous results indicating C1 instability at 42°C and above (16). Since enterococci can tolerate temperatures of 45°C (23), higher induced activities may be observed by a further increase in temperature.
FIG. 4.
Modulation of expression from the temperature-sensitive C1-regulated promoter in S. aureus (•), E. faecium (○), and E. faecalis (▴). Overnight cultures carrying the reporter construct were diluted 1:100 and grown at 31°C. The culture was then divided equally and incubated for 75 (S. aureus), 120 (E. faecium), or 95 min (E. faecalis) at the designated temperatures prior to assaying for β-Gal activity (optical density at 600 nm at the time of harvesting, approximately 0.6). Values (± standard deviations) are averages of triplicate cultures assayed in triplicate. The reporter constructs used for S. aureus, E. faecalis, and E. faecium were pDAS122, pDAS122, and pDAS113, respectively.
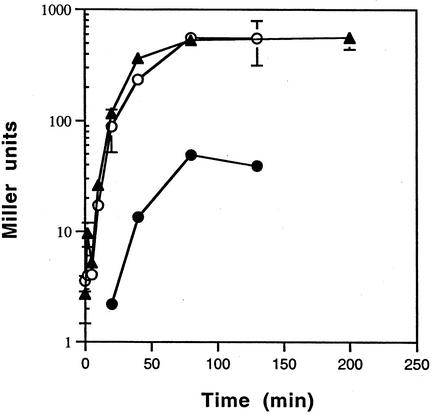
To examine the kinetics of induction, the cultures were grown at low temperature and then induced at the elevated temperatures. At the times indicated in Fig. 5, cultures were harvested and β-Gal activity was measured. The kinetics of induction for E. faecium, E. faecalis, and S. aureus were similar and indicated that the temperature-sensitive C1-regulated promoter has a fast rate of induction. In addition, the results indicated that incubation under inducing conditions need only be maintained for 80 min to achieve maximal induction (Fig. 5).
FIG. 5.
Time course analysis of temperature induction of lacZ expression in S. aureus (•), E. faecium (○), and E. faecalis (▴). Overnight cultures carrying the reporter constructs were diluted 1:100 and grown at 31°C to early log phase. Aliquots of the culture were then incubated at 42°C for the indicated times prior to harvesting for β-Gal activity (optical density at 600 nm was approximately 0.6). Values reported (± standard deviations) are averages of duplicate cultures assayed in triplicate. The reporter constructs used for S. aureus, E. faecalis, and E. faecium were pDAS122, pDAS122, and pDAS113, respectively.
In conclusion, we have demonstrated that the bacteriophage P1 temperature-sensitive C1 repressor can be used to control gene expression in clinically relevant gram-positive bacteria. For all three species investigated, the promoters were shown to be tightly repressed, an essential characteristic of a regulated promoter system. In E. faecalis, the level of regulation was 1,000-fold, bringing a level of efficiency comparable to those for promoter systems currently used in gram-negative bacteria. Furthermore, significant regulation was obtained in E. faecium, a species for which no heterologous regulated promoter systems have been described.
The C1-regulated promoters and promoters driving c1 expression were designed based on conserved gram-positive species promoter elements and therefore should be active in a wide variety of bacteria. The vectors were also constructed in a broad-host-range vector capable of replication in gram-positive species as well as enteric gram-negative species. Tight basal expression and controlled induction with the same reporter plasmid were demonstrated in both E. coli and gram-positive species; these are features that may have many applications. Furthermore, as temperature is the inducer, the promoter system is not dependent on exogenously supplied inducers. For these reasons, we expect the temperature-sensitive regulated promoter system to be useful for genetic studies of both pathogenic gram-negative and gram-positive species.
Acknowledgments
This work was supported by Hexal Gentech Forschungs GmbH.
DNA sequencing data were obtained by the Biotechnology Resource Laboratory of the Medical University of South Carolina.
REFERENCES
- 1.Bateman, B. T., N. P. Donegan, T. M. Jarry, M. Palma, and A. L. Cheung. 2001. Evaluation of a tetracycline-inducible promoter in Staphylococcus aureus in vitro and in vivo and its application in demonstrating the role of sigB in microcolony formation. Infect. Immun. 69:7851-7857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behnke, D., M. S. Gilmore, and J. Ferretti. 1982. pGB301 vector plasmid family and its use for molecular cloning in streptococci, p. 239-242. In D. Schlessinger (ed.), Microbiology. American Society for Microbiology, Washington, D.C.
- 3.Citron, M., M. Velleman, and H. Schuster. 1989. Three additional operators, Op21, Op68, and Op88, of bacteriophage P1. Evidence for control of the P1 dam methylase by Op68. J. Biol. Chem. 264:3611-3617. [PubMed] [Google Scholar]
- 4.de Ruyter, P. G., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deuschle, U., R. Gentz, and H. Bujard. 1986. lac repressor blocks transcribing RNA polymerase and terminates transcription. Proc. Natl. Acad. Sci. USA 83:4134-4137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eichenbaum, Z., M. J. Federle, D. Marra, W. M. de Vos, O. P. Kuipers, M. Kleerebezem, and J. R. Scott. 1998. Use of the lactococcal nisA promoter to regulate gene expression in gram-positive bacteria: comparison of induction level and promoter strength. Appl. Environ. Microbiol. 64:2763-2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eliason, J. L., and N. Sternberg. 1987. Characterization of the binding sites of c1 repressor of bacteriophage P1. Evidence for multiple asymmetric sites. J. Mol. Biol. 198:281-293. [DOI] [PubMed] [Google Scholar]
- 8.Engel, H. W., N. Soedirman, J. A. Rost, W. J. van Leeuwen, and J. D. van Embden. 1980. Transferability of macrolide, lincomycin, and streptogramin resistances between group A, B, and D streptococci, Streptococcus pneumoniae, and Staphylococcus aureus. J. Bacteriol. 142:407-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friesenegger, A., S. Fiedler, L. A. Devriese, and R. Wirth. 1991. Genetic transformation of various species of Enterococcus by electroporation. FEMS Microbiol. Lett. 63:323-327. [DOI] [PubMed] [Google Scholar]
- 10.Garvey, P., G. F. Fitzgerald, and C. Hill. 1995. Cloning and DNA sequence analysis of two abortive infection phage resistance determinants from the lactococcal plasmid pNP40. Appl. Environ. Microbiol. 61:4321-4328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geissendorfer, M., and W. Hillen. 1990. Regulated expression of heterologous genes in Bacillus subtilis using the Tn10 encoded tet regulatory elements. Appl. Microbiol. Biotechnol. 33:657-663. [DOI] [PubMed] [Google Scholar]
- 12.Gibson, E. M., N. M. Chace, S. B. London, and J. London. 1979. Transfer of plasmid-mediated antibiotic resistance from streptococci to lactobacilli. J. Bacteriol. 137:614-619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez, C. F., and B. S. Kunka. 1983. Plasmid transfer in Pediococcus spp.: intergeneric and intrageneric transfer of pIP501. Appl. Environ. Microbiol. 46:81-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graves, M. C., and J. C. Rabinowitz. 1986. In vivo and in vitro transcription of the Clostridium pasteurianum ferredoxin gene. Evidence for “extended” promoter elements in gram-positive organisms. J. Biol. Chem. 261:11409-11415. [PubMed] [Google Scholar]
- 15.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heinrich, J., H. D. Riedel, B. R. Baumstark, M. Kimura, and H. Schuster. 1989. The c1 repressor of bacteriophage P1 operator-repressor interaction of wild-type and mutant repressor proteins. Nucleic Acids Res. 17:7681-7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heinrich, J., M. Velleman, and H. Schuster. 1995. The tripartite immunity system of phages P1 and P7. FEMS Microbiol. Rev. 17:121-126. [DOI] [PubMed] [Google Scholar]
- 18.Heinzel, T., M. Velleman, and H. Schuster. 1989. ban operon of bacteriophage P1. Mutational analysis of the c1 repressor-controlled operator. J. Mol. Biol. 205:127-135. [DOI] [PubMed] [Google Scholar]
- 19.Heinzel, T., M. Velleman, and H. Schuster. 1990. The c1 repressor inactivator protein coi of bacteriophage P1. Cloning and expression of coi and its interference with c1 repressor function. J. Biol. Chem. 265:17928-17934. [PubMed] [Google Scholar]
- 20.Helmann, J. D. 1995. Compilation and analysis of Bacillus subtilis σA-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 23:2351-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henkin, T. M., and A. L. Sonenshein. 1987. Mutations of the Escherichia coli lacUV5 promoter resulting in increased expression in Bacillus subtilis. Mol. Gen. Genet. 209:467-474. [DOI] [PubMed] [Google Scholar]
- 22.Hiramatsu, K., H. Hanaki, T. Ino, K. Yabuta, T. Oguri, and F. C. Tenover. 1997. Methicillin-resistant Staphylococcus aureus clinical strain with reduced vancomycin susceptibility. J. Antimicrob. Chemother. 40:135-136. [DOI] [PubMed] [Google Scholar]
- 23.Huycke, M. M., D. F. Sahm, and M. S. Gilmore. 1998. Multiple-drug resistant enterococci: the nature of the problem and an agenda for the future. Emerg. Infect. Dis. 4:239-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jensen, P. R., and K. Hammer. 1998. The sequence of spacers between the consensus sequences modulates the strength of prokaryotic promoters. Appl. Environ. Microbiol. 64:82-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ji, Y., A. Marra, M. Rosenberg, and G. Woodnutt. 1999. Regulated antisense RNA eliminates alpha-toxin virulence in Staphylococcus aureus infection. J. Bacteriol. 181:6585-6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones, R. N., S. A. Marshall, M. A. Pfaller, W. W. Wilke, R. J. Hollis, M. E. Erwin, M. B. Edmond, and R. P. Wenzel. 1997. Nosocomial enterococcal bloodstream infections in the SCOPE program: antimicrobial resistance, species occurrence, molecular testing results, and laboratory testing accuracy. SCOPE Hospital Study Group. Diagn. Microbiol. Infect. Dis. 29:95-102. [DOI] [PubMed] [Google Scholar]
- 27.Kim, L., A. Mogk, and W. Schumann. 1996. A xylose-inducible Bacillus subtilis integration vector and its application. Gene 181:71-76. [DOI] [PubMed] [Google Scholar]
- 28.Kreikemeyer, B., and P. G. Jerlstrom. 1999. An Escherichia coli-Enterococcus faecalis shuttle vector as a tool for the construction of a group B Streptococcus heterologous mutant expressing the beta antigen (Bac) of the C protein complex. FEMS Microbiol. Lett. 180:255-262. [DOI] [PubMed] [Google Scholar]
- 29.Lanzer, M., and H. Bujard. 1988. Promoters largely determine the efficiency of repressor action. Proc. Natl. Acad. Sci. USA 85:8973-8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee, G., C. Talkington, and J. Pero. 1980. Nucleotide sequence of a promoter recognized by Bacillus subtilis RNA polymerase. Mol. Gen. Genet. 180:57-65. [DOI] [PubMed] [Google Scholar]
- 31.Lee, J. C. 1995. Electroporation protocols for microorganisms, p. 209-215. In J. A. Nickoloff (ed.), Methods in molecular biology, vol. 47. Humana Press, Inc., Totowa, N.J.
- 32.Leonhardt, H., and J. C. Alonso. 1988. Construction of a shuttle vector for inducible gene expression in Escherichia coli and Bacillus subtilis. J. Gen. Microbiol. 134:605-609. [DOI] [PubMed] [Google Scholar]
- 33.Lutz, R., and H. Bujard. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McIntyre, D. A., and S. K. Harlander. 1989. Genetic transformation of intact Lactococcus lactis subsp. lactis by high-voltage electroporation. Appl. Environ. Microbiol. 55:604-610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Moran, C. P., Jr., N. Lang, S. F. LeGrice, G. Lee, M. Stephens, A. L. Sonenshein, J. Pero, and R. Losick. 1982. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol. Gen. Genet. 186:339-346. [DOI] [PubMed] [Google Scholar]
- 37.Powell, I. B., M. G. Achen, A. J. Hillier, and B. E. Davidson. 1988. A simple and rapid method for genetic transformation of lactic streptococci by electroporation. Appl. Environ. Microbiol. 54:655-660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poyart, C., and P. Trieu-Cuot. 1997. A broad-host-range mobilizable shuttle vector for the construction of transcriptional fusions to β-galactosidase in gram-positive bacteria. FEMS Microbiol. Lett. 156:193-198. [DOI] [PubMed] [Google Scholar]
- 39.Rehm, S. J. 2002. Two new treatment options for infections due to drug-resistant gram-positive cocci. Cleve. Clin. J. Med. 69:397-401, 405-413. [DOI] [PubMed] [Google Scholar]
- 40.Rosner, J. L. 1972. Formation, induction, and curing of bacteriophage P1 lysogens. Virology 48:679-680. [DOI] [PubMed] [Google Scholar]
- 41.Ruoff, K. L., L. de la Maza, M. J. Murtagh, J. D. Spargo, and M. J. Ferraro. 1990. Species identities of enterococci isolated from clinical specimens. J. Clin. Microbiol. 28:435-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N. Y.
- 43.Schaefer, T. S., and J. B. Hays. 1991. Bacteriophage P1 Bof protein is an indirect positive effector of transcription of the phage bac-1 ban gene in some circumstances and a direct negative effector in other circumstances. J. Bacteriol. 173:6469-6474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schofield, D. A., C. Westwater, J. W. Dolan, J. S. Norris, and M. G. Schmidt. 2002. Tight regulation and modulation via a C1-regulated promoter in Escherichia coli and Pseudomonas aeruginosa. Curr. Microbiol. 44:425-430. [DOI] [PubMed] [Google Scholar]
- 45.Schofield, D. A., C. Westwater, J. W. Dolan, M. G. Schmidt, and J. S. Norris. 2001. Controlled expression in Klebsiella pneumoniae and Shigella flexneri using a bacteriophage P1-derived C1-regulated promoter system. J. Bacteriol. 183:6947-6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stark, M. J. 1987. Multicopy expression vectors carrying the lac repressor gene for regulated high-level expression of genes in Escherichia coli. Gene 51:255-267. [DOI] [PubMed] [Google Scholar]
- 47.Stieger, M., B. Wohlgensinger, M. Kamber, L. Rolf, and W. Keck. 1999. Integrational plasmids for the tetracycline-regulated expression of genes in Streptococcus pneumoniae. Gene 226:243-251. [DOI] [PubMed] [Google Scholar]
- 48.Velleman, M., T. Heinzel, and H. Schuster. 1992. The Bof protein of bacteriophage P1 exerts its modulating function by formation of a ternary complex with operator DNA and C1 repressor. J. Biol. Chem. 267:12174-12181. [PubMed] [Google Scholar]
- 49.Velleman, M., M. Heirich, A. Gunther, and H. Schuster. 1990. A bacteriophage P1-encoded modulator protein affects the P1 c1 repression system. J. Biol. Chem. 265:18511-18517. [PubMed] [Google Scholar]
- 50.Voskuil, M. I., and G. H. Chambliss. 1998. The −16 region of Bacillus subtilis and other gram-positive bacterial promoters. Nucleic Acids Res. 26:3584-3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Voskuil, M. I., K. Voepel, and G. H. Chambliss. 1995. The −16 region, a vital sequence for the utilization of a promoter in Bacillus subtilis and Escherichia coli. Mol. Microbiol. 17:271-279. [DOI] [PubMed] [Google Scholar]
- 52.Wieland, K. P., B. Wieland, and F. Gotz. 1995. A promoter-screening plasmid and xylose-inducible, glucose-repressible expression vectors for Staphylococcus carnosus. Gene 158:91-96. [DOI] [PubMed] [Google Scholar]
- 53.Wirth, R., F. Y. An, and D. B. Clewell. 1986. Highly efficient protoplast transformation system for Streptococcus faecalis and a new Escherichia coli-S. faecalis shuttle vector. J. Bacteriol. 165:831-836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wright, J. J., A. Kumar, and R. S. Hayward. 1992. Hypersymmetry in a transcriptional terminator of Escherichia coli confers increased efficiency as well as bidirectionality. EMBO J. 11:1957-1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yarmolinsky, M. B., and N. Sternberg. 1988. Bacteriophage P1, p. 291-438. In R. Calendar (ed.), The bacteriophages, vol. 1. Plenum Publishing Corp., New York, N.Y.
- 56.Zhang, L., F. Fan, L. M. Palmer, M. A. Lonetto, C. Petit, L. L. Voelker, A. St John, B. Bankosky, M. Rosenberg, and D. McDevitt. 2000. Regulated gene expression in Staphylococcus aureus for identifying conditional lethal phenotypes and antibiotic mode of action. Gene 255:297-305. [DOI] [PubMed] [Google Scholar]