Abstract
A loss of FAS (CD95) function has been proposed to constitute an important step in early mucosa-associated lymphoid tissue (MALT) lymphoma development and FAS mutations have been recognized in malignant lymphomas, in particular at extranodal sites. Since primary gastric lymphomas frequently exhibit resistance to FAS-mediated apoptosis, we investigated whether FAS is mutated in 18 gastric MALT lymphomas and 28 diffuse large B-cell lymphomas (DLBCL). We detected seven mutations in five lymphomas, one MALT lymphoma and four DLBCL; two DLBCL had two mutations. The MALT lymphoma exhibited a point mutation in the splice donor region of intron 3. Three DLBCL had missense mutations in exon 2, which encodes a signal peptide and a portion of the extracellular FAS ligand-binding domain. One DLBCL carried a point mutation in the splice donor region of intron 8, which would result in exon skipping. Two DLBCL harbored a missense mutation in exon 9, which encodes the intracellular death domain. The two death domain mutations inhibited FAS ligand-induced apoptosis in a dominant-negative mode, when transiently expressed in human T47D breast carcinoma and Jurkat T cells. A signal peptide and an extracellular domain mutation, however, failed to inhibit apoptosis in these transfection assays. They are likely to reduce apoptosis in lymphoma cells solely by a loss of function. In summary, our data show that FAS mutations are rare in primary gastric MALT lymphomas (5.6%) but occur in a subset of primary gastric DLBCL (14.3%) and suggest that these mutations contribute to the pathogenesis of gastric lymphomas by rendering lymphocytes resistant to apoptosis.
Most extranodal lymphomas arise in the gastrointestinal tract and the stomach is the most frequent site involved.1,2 The great majority of primary gastric lymphomas are of B-cell lineage and derive from mucosa-associated lymphoid tissue (MALT), which is acquired in the course of a Helicobacter pylori gastritis.1,2 The proliferation of gastric MALT lymphoma cells depends on the presence of T cells specifically activated by H. pylori antigens. The importance of this stimulation is impressively demonstrated by remission of more than half of the lymphomas with antibiotic H. pylori eradication therapy.1,2 MALT lymphomas are indolent (low-grade) malignant lymphomas, but they may undergo transformation to aggressive (high-grade) diffuse large B-cell lymphoma (DLBCL).3
A dysregulation of apoptosis seems to be important for the survival of MALT lymphoma cells. This is illustrated by the frequent alteration of apoptosis genes. The t(11;18)(q21;q21) is present in 24% of gastric MALT lymphomas.4–6 It involves apoptosis inhibitor-2 (API2) and the MALT lymphoma-associated translocation (MALT1) gene.7 API2 is an inhibitor of caspases, proteases that function as executioners of apoptosis. MALT1 encodes a paracaspase that activates the transcription factor nuclear factor-κB, which confers survival signals.8 Importantly, MALT lymphomas with a t(11;18)(q21;q21) are resistant to H. pylori eradication and are associated with advanced gastric tumors.9 Unexpectedly, the t(11;18)(q21;q21) is usually not present in gastric DLBCL.4,5 MALT1 expression can also be increased by gene amplification or a t(14;18)(q32;q21), which juxtaposes the gene to the immunoglobulin heavy-chain locus.10,11
The p53 is a nuclear protein that functions as a transcription factor and mediates cell cycle arrest or apoptosis in response to cellular stress exerted, eg, by hypoxia or DNA damage. Loss of function mutations of p53, withconcomitant mutation in one allele and loss of the other, have been identified in 7 of 60 (11.7%) cases of gastric MALT− and DLBCL.12
The t(1;14)(p22;q32) and t(1;2)(p22;p12) are exclusive to MALT lymphoma but occur rarely.2,13,14 The translocations juxtapose the B-cell lymphoma/leukemia (bcl-10) gene to an immunoglobulin gene locus and thus deregulate its expression. Bcl-10 contains a caspase recruiting domain (CARD), a motif that functions in apoptosis signaling and mediates protein-protein interactions. A truncated bcl-10 mutant isolated from a gastric lymphoma with t(1;14)(p22;q32) exhibited a loss of the pro-apoptotic activity of wild-type bcl-10 and acquisition of a transforming potential.15
Malignant lymphomas, including gastric lymphomas, are frequently resistant to apoptosis induction via the death receptor FAS (CD95, Apo-1, TNFRSF6).16,17 FAS is a member of the tumor necrosis factor receptor superfamily that transmits an apoptosis signal on interaction with FAS ligand (FASL).18–21 FAS is widely expressed, whereas the expression of FASL in the immune system is more restricted and largely confined to cytotoxic T lymphocytes and natural killer cells.20,21 FAS is important for lymphocyte homeostasis. It is required for the elimination of autoreactive B lymphocytes, the clonal selection in germinal centers and the elimination of excess effector lymphocytes during immune responses.20–22 Germline FAS mutations cause an autoimmune lymphoproliferative syndrome (ALPS) in mice and humans, characterized by massive lymphadenopathy, splenomegaly, and autoimmune manifestations.23–25 A loss of FAS regulatory function has been proposed to constitute an important step in early MALT-lymphoma development,17 and somatic FAS mutations are frequent in some subtypes of malignant lymphomas, in particular at extranodal sites.17,26–28 Therefore, a potential mechanism that may account for FASL resistance in gastric lymphomas seems to be mutation of FAS. We have therefore searched for FAS gene alterations in primary gastric MALT− and DLBCL to clarify whether FAS mutations play a role in reducing apoptosis, thereby providing a survival advantage to lymphoma cells.
Materials and Methods
Lymphoma Samples
Forty-six lymphomas were studied. They were classified as primary gastric lymphoma on histopathology and clinical staging and comprised 18 MALT lymphomas and 28 DLBCL. Staging was done according to Musshoff, modified by Radaszkiewicz et al.29 28 lymphomas were stage I (10 MALT and 18 DLBCL), 16 were stage II (7 MALT and 9 DLBCL), and two cases were stage III (1 MALT and 1 DLBCL). Histological diagnoses were established according to the World Health Organization classification of tumors of hematopoetic and lymphoid tissues.30
Polymerase Chain Reaction
Genomic DNA was isolated from paraffin-embedded tissues with QIAamp DNA MINI kit (Qiagen, Hilden, Germany). Only tissue blocks with lymphoma cells comprising at least 70% of the total cellularity were used. FAS exons 2–9, which encode the entire mature molecule, and adjacent intron sequences were amplified by polymerase chain reaction (PCR) using the “inward” primers listed in Table 1. For samples that were difficult to amplify, a nested PCR reaction was used with the “outward” and “inward” primers listed in Table 1. Exons 2–8 were amplified in single PCR reactions, while exon 9 was subdivided into three segments (Table 1). PCR was performed with either an Omni-Gene (Thermo Hybaid, Needham Heights, MA) or a Biometra thermocycler (Whatman Biometra, Göttingen, Germany) in final volumes of 50 μl, containing 10 mmol/L Tris-HCl (pH 8.3), 50 mmol/L KCl, 1.5 mmol/L MgCl2 (or 2.5 mmol/L, 3.0 mmol/L, 3.5 mmol/L MgCl2, depending on the primer pair used), 200 μmol/L of each dNTP, 0.5 μmol/L of each primer, 0.5 μg DNA, and 2.5 U Taq polymerase AmpliTaq Gold (PerkinElmer, Foster City, CA). The amplification protocol consisted of 36 cycles with denaturation at 95°C, annealing at 54°C (exon 6, 7, and 8), 58°C (exon 3, 5, and 9), or 62°C (exon 2 and 4), and elongation at 72°C for 1 minute. An initial denaturation step at 94°C and a final incubation at 72°C for 10 minutes each were included.
Table 1.
PCR Primers Used for Amplification of Exons 2–9 and the Enhancer Region of FAS
| Inward primer | Outward primer | |
|---|---|---|
| Exon-Primer* | ||
| 2-F | GTGGGTTACACTGGTTTACACGTTGC | GATACTGCCAATTTTGG |
| 2-FM | GTGGGTTACACTCGTTTACACGTTGC | |
| 2-R | CATGATTACTATGTGCTACTCCTAACTG | GTCATGATTACTATGTGCT |
| 3-F | TCCTGTTCAAACACTTGCTCC | TATACTTCCCACCCTGTT |
| 3-FM | TCCTGTTCAGACACTTGCTCC | |
| 3-R | GTTCTACATGAAATTCCAAGATTGG | GTAGGCCCCAATTTCAAA |
| 4-F | AGCCGCGATAACTAATAGTTTCC | CCTGCCCACCATTTTCATA |
| 4-FM | AGCCGCGATAACTGATAGTTTCC | |
| 4-R | TCTCAGTCAGTGTTACTTCCCTAGG | TTGGAGGCAAAGCAGGA |
| 5-F | TTGCCAGAGATGCAAAGATG | TGAAGGAATACGTTTGC |
| 5-FM | TTGCCAGAGCTGCAAAGATG | |
| 5-R | GGGGGAAAGGAGAATATAACC | CTTCACATCTTTCCATG |
| 6-F | TTCATATAAAATGTCCAATGTTCC | AACCAATCACTCTTGATT |
| 6-FM | TTCATATAATATGTCCAATGTTCC | |
| 6-R | CTGCAGTTTGAACAAAGCAA | CTTCCCCCAAGTTATTTC |
| 7-F | TCTCACATGCATTCTACAAGGC | TGTTCTCACATGCATTC |
| 7-FM | TCTCACATGCAGTCTACAAGGC | |
| 7-R | TTCTTTTCAAGGAAAGCTGATACC | GGAAGTAACAAAAAGCCA |
| 8-F | TATTTTTATTTGTCTTTCTCTGCTTCC | GGAAAAATTAGAAGTTCAC |
| 8-FM | TATTTTTATTTGTCTGTCTCTGCTTCC | |
| 8-R | TTGGCCTATTACTCTAAAGGATGC | TTTACTCTGAAATTGGC |
| 9-1F* | CATTTAGAAAAACAAATTTTCAGAC | CATGGTTTTCACTAATGGG |
| 9-1FM | CATTTAGAAAAACAGATTTTCAGAC | |
| 9-1R | GGACATTGTCATTCTTGATCTC | TGCCAATTACGAAGCAGT |
| 9-2F | CAAGTTAAAGGCTTTGTTCG | ATTGCTGGAGTCATGACA |
| 9-2FM | CAAGTTAATGGCTTTGTTCG | |
| 9-2R | CTGCAAGAGTACAAAGATTGG | CCTTGAGGATGATAGTCTG |
| 9-3F | GCGTATGACACATTGATTAAAGATC | TGGCATCAACTTCATGGA |
| 9-3R | CAGAACTGAATTTGTTGTTTTTCAC | CCCAGTAAAAAACCAAGCA |
| 9-3RM | CAGAACTTGAATTTGCTGTTTTTCAC | |
| Enhancer Region Primer | ||
| −734GC | TCCCTTTTCAGAGCCCTATGG | |
| −623 | GACTTGCGGGGCATTTGAC |
The first numeral denotes the exon number, a second numeral overlapping primer pairs within an exon.
F, forward primer; R, reverse primer; FM, forward mutated; RM, reverse mutated. Mutated nucleotides are written in bold letters and are underlined.
Single-Strand Conformation Polymorphism Analysis
PCR products were analyzed by single-strand conformation polymorphism (SSCP) to screen for mutations in the FAS gene. Four microliters of a 50 μl PCR reaction were mixed with 36 μl SSCP solution [10% sodium dodecyl sulfate (SDS); 0.5 mol/L ethylene diaminetetraacetic acid (EDTA), pH 8.0; 1 mol/L Tris-HCL, pH 8.0] and then 6 μl of this mixture were combined with 6 μl denaturation buffer (95% formamide; 10 mmol/L EDTA, pH 8.0; 0.1% bromophenol blue; 0.1% xylene cyanol). Afterward, samples were denatured at 95°C for 10 minutes, chilled quickly in an ice/100% ethanol bath and loaded on an 8% or 10% (in case of exon 7) vertical polyacrylamide gel containing 10% (v/v) glycerol. The gel was run at 6 watts for 16 hours in 1 × Tris-borate-EDTA buffer (pH 8.0) at 4°C. DNA bands were visualized by silver staining (Silver Stain Plus; Bio-Rad, Vienna, Austria). The mutation detection rate of the SSCP gels was determined by analyzing PCR products generated with 10 mutated FAS primers (“mutated primers”, Table 1) and found to be 90%.
DNA Sequencing
DNA samples that exhibited an aberrant band pattern in at least two SSCP gels, generated with independently repeated PCR products, were subcloned into a TA-cloning vector (Invitrogen, Groningen, The Netherlands) and 8 to 22 clones per sample were sequenced with the SequiTherm EXCEL II DNA sequencing kit (Epicenter, Madison, WI) on a LI-COR 4000 (MWG-Biotech, Ebersberg, Germany) sequencer. Only sequence alterations that were present in at least three clones were regarded as a mutation.
Loss of Heterozygosity Analysis
To determine whether a loss of the normal CD 95 allele had occurred in lymphomas with a CD 95 mutation two known polymorphisms at position −670 in the enhancer region (primers “−734 GC” and “−623” listed in Table 1) and position 416 (codon 58) in exon 3 were used.31,32 The SSCP band patterns of lymphomas were compared to control tissues not involved by lymphoma, obtained either from the same patient or unrelated individuals. A reduction in intensity of a SSCP band in a lymphoma sample as compared to controls was regarded as indicative of an allele loss.
Site-Directed Mutagenesis and Cloning Procedures
The five mutations identified in three gastric DLBCL (cases 1, 2, and 3 in Table 2) were introduced in FAS expression vectors by site-directed mutagenesis to assess their biological activity in transfection assays. A FAS wild-type cDNA cloned in pBluescript II KS (pBS-APO14.2; kindly provided by Peter H. Krammer, German Cancer Research Center, Heidelberg) served as template. Mutations were generated with the QuikChange kit (Stratagene, La Jolla, CA), following the instructions of the manufacturer. Case 2 and 3 (Table 2) carried two mutations each, which were generated consecutively by first introducing one mutation and than repeating the mutagenesis protocol to insert the second mutation. The PCR parameters for site-directed mutagenesis consisted of an initial denaturation at 95°C for 30 seconds, followed by 12 cycles each of denaturation at 95°C for 30 seconds, annealing at 55°C for 1 minute and elongation at 68°C for 11 minutes. The oligonucleotide primers used to construct the mutants were the following (positions of the mutated nucleotides are in bold and underlined): CD 95 mutant 1 (case 1 in Table 2), 5′-forward-GGTGTCAATGAAGCCAAAATAAATGAGATCAAGAATGAC-3′; 5′-reverse-GTCATTCTTGATCTCATTTATTTTGGCTTCATTGACACC-3′; CD 95 mutant 2 (case 2 in Table 2), 5′-forward-CAGCAGAACAGAAAGTTTAACTGCTTCGTAATTGG-3′; 5′reverse–CCAATTACGAAGCAGTTAAACTTTCTGTTCTGCTG-3′. 5′-forward-GAGGAAGACTGTTAGTATAGTTGAGAC TCAGAACTTGG-3′; 5′-reverse-CCAAGTTCTGAGTCTCAACTATAGTAACAGTCTTCCTC-3′; CD 95 mutant 3 (case 3 in Table 2), 5′-forward-CCTCTGGTTCTTACGTTTGTTGCTAGATTATCG-3; 5′-reverse-CGATAATCTAGCAACAAACGTAAGAACCAGAGG-3′ and 5′-forward-AAGTGACTGACATCA ACCCCAAGGGATTGGAATTG-3′; 5′-reverse-CAATTCCAATCCCTTGGGGTTGATGTCAGTCACTT-3′. The generation of the desired mutations was confirmed by DNA sequencing and the mutated FAS cDNAs (mutants 1, 2, and 3) as well as the wild-type sequence were released from pBluescript II KS with the restriction enzyme NotI and ligated into the NotI site of the mammalian expression vector pcDNA3.1/V5-HIS-C (Invitrogen). This vector contains a human cytomegalovirus immediate-early promoter and a C-terminal tag encoding the V5 epitope, which allows verification of expression of a cloned gene with an anti-V5 tag antibody. The integrity of the FAS cDNAs and the proper reading frames were confirmed by DNA sequencing of wild-type and mutated FAS expression vectors in forward and reverse orientation with the BigDye terminator cycle sequencing kit (Applied Biosystems, Warrington, UK) on an ABI Prism 310 DNA sequencer (Applied Biosystems).
Table 2.
FAS Mutations Identified in Five Primary Gastric Lymphomas
| Case no. | Histology | Stage | Nucleotide change | Localization | Codon* | Predicted effect |
|---|---|---|---|---|---|---|
| 1 | DLBCL | El2 | GAT→AAT | Exon 9 | 244 | Asp→Asn |
| 2 | DLBCL | Ell1 | ACA→ATA | Exon 2 | 27 | Thr→lle |
| CAA→TAA | Exon 9 | 260 | Gln→STOP | |||
| 3 | DLBCL | El2 | TCT→TTT | Exon 2 | −3, signal peptide | Ser→Phe |
| TCC→CCC | Exon 2 | 16 | Ser→Pro | |||
| 4 | DLBCL | El2 | IVS + 2 T→A | Intron 8 | - | Splice defect |
| 5 | MALT | El1 | IVS + 17 G→A | Intron 3 | - | Splice defect? |
Numbering of codons is according to Itoh et al.34
DLBCL, diffuse large B-cell lymphoma; MALT, mucosa-associated lymphoid tissue lymphoma; IVS, intervening sequence.
Cell Culture
The human breast cancer cell line T47D (ATCC, Manassas, VA) and the human T-cell leukemia cell line Jurkat (a gift of M. Eisenbauer, Institute of Tumorbiology-Cancer Research, Vienna) were used to assay the biological activities of the mutant CD 95 constructs 1, 2, and 3. Jurkat cells were grown in Roswell Park Memorial Institute medium (RPMI −1640; GIBCO, Paisley, UK) and T47D cells in Dulbecco’s modified Eagle’s medium (DMEM; GIBCO). Both culture media contained 10% fetal calf serum (FCS; GIBCO), 1% penicillin-streptomycin (GIBCO) and 1% GlutamaxR (GIBCO). Cells were incubated at 37°C in a humidified atmosphere containing 5% CO2.
Transient Transfection of T47D Cells
T47D cells were plated into 100-mm dishes and grown overnight to 40% to 50% confluence, when cells were co-transfected with a FAS expression vector (FAS wild-type or mutant 1, 2, or 3) and a luciferase expression vector (pGL3; Promega, Mannheim, Germany) using LipofectAMINE reagent (Invitrogen). The following amount of expression vector was applied per culture dish: 8 μg FAS wild-type and 2 μg pGL3, or 8 μg mutant CD 95 construct 1, 2, or 3 and 2 μg pGL3 or 8 μg pCDNA3.1/V5-HIS-C and 2 μg pGL3. 10 μg of DNA (1 μg/μl distilled water) and 30 μl LipofectAMINE were each transferred into 800 μl DMEM and then both solutions were combined. LipofectAMINE-DNA complexes were allowed to form for 30 minutes and were then applied to T47D, which had been rinsed once with DMEM, without FCS and phenol red. Cells were incubated for 2 hours in a humidified incubator at 37°C and then the LipofectAMINE-DNA complexes were removed and DMEM supplemented with 10% FCS/1% GlutamaxR was added. On the next day cells were split into 12-well dishes at 20% to 30% confluence, supplied with fresh culture medium and on the following day (48 hours after transfection) exposed to 30 ng/ml human soluble FASL (sFASL; Alexis Biochemicals, Montreal, Canada). Control cells were cultured without the addition of sFASL.
Nucleofection of Jurkat Cells
Transient co-nucleofection of Jurkat cells with the same expression vector combinations as described above for T47D cells was performed with a Nucleofector electroporation device (Amaxa Biosystems, Köln, Germany). For nucleofection 2 × 106 Jurkat cells were harvested by centrifugation and the cell pellet was resuspended in 100 μl of Nucleofector Solution (Amaxa Biosystems) and 2 μg DNA (1 μg pGL3 and 1 μg FAS wild-type or mutant 1, 2, or 3 or pCDNA3.1/V5-HIS-C expression vector) were added. Nucleofection was performed with Nucleofector settings optimized for Jurkat cells (program S-18). On the next day cells were split into 12-well dishes (2 × 105 cells per well) with a renewal of culture medium and exposed to 10 ng/ml sFASL. Control cells were cultured without the addition of sFASL.
Luciferase Assay
Cell death induction by sFASL in T47D and Jurkat cells, co-transfected with FAS- (wild-type or mutant 1, 2, or 3) and luciferase expression vectors, was determined by using the luciferase activity of cell lysates as a reporter of cell viability.33 The luciferase activity of transfected cells was determined after 24 hours of sFASL treatment. Cell pellets were lysed in reporter lysis buffer (Promega), and to achieve complete cell lysis, the cell suspensions were immediately frozen at −80°C and then thawed rapidly. Subsequently the lysates were centrifuged at 12 000 × g for 1 minute at room temperature and the supernatant was assayed for luciferase activity. Therefore, 20 μl of supernatant were mixed with 100 μl luciferase assay reagent (Promega), and the luciferase activity was measured with a luminometer (Lumat LB 9507; EG&G Berthold, Bad Wildbad, Germany). The luciferase activity was recorded as relative light units (RLU). The RLU values of sFASL-treated samples were expressed as percentage of untreated control samples. Experiments were repeated three times and each experiment was done in triplicates.
Statistical Analysis
Statistical analysis was performed with two-sided Student′s t-test.
RNA Extraction and Reverse Transcriptase-PCR
Total cellular RNA was isolated from transfected T47D and Jurkat cells using the RNeasy Mini kit (Qiagen) and treated with deoxyribonuclease I (Invitrogen) to eliminate residual DNA contamination. First-strand cDNA synthesis was primed with oligo-dT and catalyzed by Super Script II Rnase H reverse transcriptase (Invitrogen). A 550-bp DNA fragment encompassing a FAS cDNA segment and an adjacent transcribed vector sequence was amplified by PCR. The two oligonucleotide primers used were homologous to a sequence in the FAS cDNA (forward 5′-CCAACCTTAAATCCTGAAAC-3′) and a vector sequence (reverse 5′-TAGAAGGCACAGTCGAGG-3′). The amplification conditions on a Biometra Thermocycler were as follows: initial denaturation at 95°C for 12 minutes, followed by 28 cycles of denaturation at 95°C, annealing at 45°C and extension at 72°C for 1 minute each. Negative controls for the PCR consisted of omission of the reverse transcriptase in the 1. strand cDNA reaction or omitting the addition of DNA. PCR products were analyzed on 2% Seakem (Cambrex, Apen, Germany) agarose gels and sequenced.
Western Blot Analysis
To confirm the expression of the transfected CD 95 wild-type and mutant cDNAs a Western blot analysis was performed. Transfected cells were lysed in SDS sample buffer (25 mmol/L Tris, pH 6.8; 3% SDS; 10% glycerol; 36 mmol/L dithiothreitol, 0.925 mmol/L EDTA) with proteinase inhibitors [20 μmol/L N-tosyl-l-phenylalanine chloromethyl ketone, 20 μmol/L tosyl-l-lysine chloromethyl ketone (Boehringer Ingelheim, Ingelheim, Germany) and 10 ng/ml benzamidinchloride (ς-Aldrich, Vienna, Austria)]. The viscous lysates were sonicated and the protein content was measured with Bradford reagent (Bio-Rad Laboratories, Vienna, Austria). Equal amounts of total protein (20 μg/lane) were loaded onto 10% SDS-polyacrylamide gel electrophoresis gels. Proteins were electrophoresed with a miniprotean cell (Bio-Rad Laboratories) at 80 V for 2 to 3 hours and blotted onto PVDF membranes (Hybond P; Amersham, Buckinghamshire, UK) overnight at 4°C. To confirm equal protein transfer, membranes were stained with Poinceau S. The incubation with primary anti-V5 tag antibody (1:500; Invitrogen) was done overnight at 4°C and was followed by incubation with the secondary antibody (peroxidase-conjugated goat anti-mouse IgG; dilution 1:2.000; Calbiochem, La Jolla, CA) for 1 to 2 hours at room temperature. The ECL Kit (Amersham) was used for blot development and chemiluminescence was visualized on Xomat UV films (Kodak, Vienna, Austria).
Results
FAS Gene Mutation Analysis
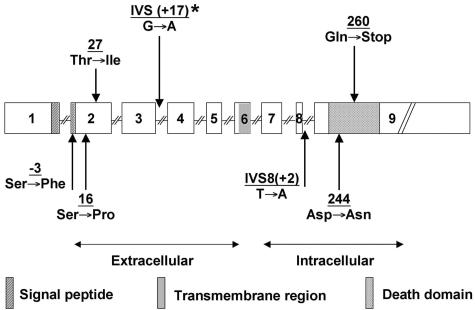
Five (10.9%) out of 46 gastric lymphomas analyzed harbored mutations in the FAS gene. In two of the lymphomas two different mutations were co-existent (Table 2, Figure 1). Only one (5.6%) of the 18 MALT lymphomas carried a FAS mutation, whereas 4 (14.3%) of 28 DLBCL were mutated. All mutations were point mutations. Three missense mutations affected exon 2. One of them was located in the amino-terminal signal peptide, two localized to the extracellular domain, which interacts with FASL and is important for the formation of functional FAS trimers.20,21 Two mutations were identified in exon 9 and affected the intracellular death domain, an 80 amino acid motif that is essential for apoptosis signaling.34 One of the death domain mutations (case 1 in Table 2) altered codon 244, a residue that is highly conserved and known to have a profound influence on protein structure and function.35 The second death domain mutation introduced a stop codon at position 260, which would result in a protein truncated within the death domain. Two mutations were located in the splice donor region of intron 3 at position + 17 and intron 8 at the invariable position + 2.
Figure 1.
Scheme of the human FAS gene locus, illustrating nature and location of seven mutations identified in five primary gastric lymphomas (1 MALT lymphoma and 4 DLBCL). *, MALT lymphoma.
All four DLBCL with a FAS mutation were without a histologically detectable low-grade component. It is thus unclear whether they represent transformed MALT lymphomas or have arisen de novo. The mutated lymphomas were either stage EI (4 cases) or EII (1 case; Table 2).
In three of the five lymphomas (cases 1, 2, and 4; Table 2) with FAS mutations corresponding normal tissue was available for analysis. In all three cases the mutation was absent in normal tissue (data not shown), indicating that the mutations had arisen somatically.
We further investigated whether the normal FAS allele was retained in the lymphomas by exploiting two biallelic polymorphisms located in the promoter region and in exon 3 of the FAS gene.31,32 All but one (Case 5, Table 2) of the lymphomas were heterozygous in one or both of these loci but they exhibited no difference in SSCP band patterns in comparison to heterozygous control tissue (data not shown). This result indicates that the wild-type allele was retained in these lymphomas.
Functional Analysis of Mutated FAS
To assess the impact of the identified mutations on the function of FAS, the five mutations detected in three DLBCL, which were all located in coding exons (cases 1, 2, and 3; Table 2), were introduced in mammalian FAS expression vectors by site-directed mutagenesis and transfected in T47D breast carcinoma and Jurkat T-cells. Both cell lines express endogenous FAS constitutively and undergo apoptosis in response to FASL.
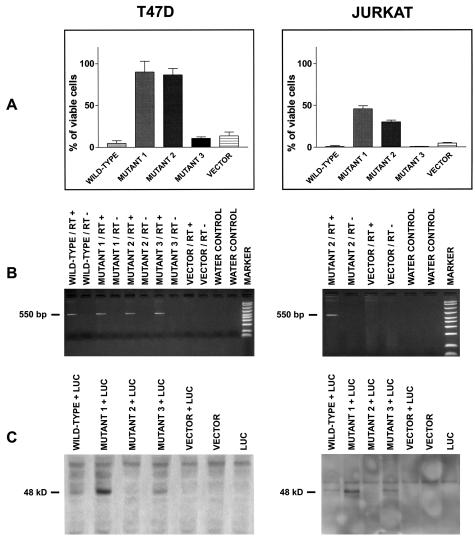
The cells were transfected with either wild-type FAS, mutant FAS or pCDNA3.1/V5-HIS-C vector, together with the luciferase expression vector pGL3. Forty-eight (T47D) or 24 (Jurkat) hours after transfection, aliquots of transfected cells were incubated with human sFASL for 24 hours, and then assayed for luciferase activity. The mean values and standard deviations of three such assays, each performed in triplicate, are shown in Figure 2. The mutations identified in cases 1 and 2 (Table 2), which were located in exon 2 and 9, strongly inhibited FAS-mediated cell death in both cell lines (Figure 2A). The mutation in exon 9 of lymphoma case 2 (Table 2) inhibited sFASL-triggered cell death also without the additional mutation in exon 2. Furthermore, the exon 2 mutation did not enhance the apoptosis inhibitory potential of the exon 9 mutation in transient transfection assays (data not shown).
Figure 2.
Biological activity of CD 95 mutants isolated from three cases of gastric DLBCL (cases 1, 2, and 3 in Table 2). A: T47D breast carcinoma and Jurkat T cells were cotransfected with either FAS wild-type, FAS mutant 1, 2, or 3 (corresponding to cases 1, 2, and 3 in Table 2), or vector control, together with the luciferase reporter vector, pGL3. After transfection cells were cultured with or without sFASL (T47D 30 ng/ml; Jurkat cells 10 ng/ml) for 24 hours. Luciferase expression was assayed thereafter, and percent viability was calculated by the following formula: (luciferase activity with sFASL/luciferase activity without sFASL) × 100. The data shown are the mean results (± SEM) of three independent experiments, each performed in triplicate. The P values for the differences between FAS wild-type and mutant 1, 2, and 3 transfected cells in comparison to vector-only transfectants were: T47D cells; wild-type, P < 0.08, mutant 1, P < 0.0004, mutant 2, P < 0.00004, mutant 3, P < 0.6. Jurkat cells: wild-type, P < 0.0008, mutant 1, P < 0.00001, mutant 2, P < 0.00001, mutant 3, P < 0.0005 (Student’s t-test). B: Detection of transfected FAS wild-type and mutant 1, 2, and 3 mRNA expression by RT-PCR. RT+ = 1. strand cDNA reaction with reverse transcriptase; RT− = negative control without reverse transcriptase in 1. strand cDNA reaction; water control = negative control without addition of cDNA reaction product to the PCR. C: Detection of transfected FAS wild-type and mutant 1, 2, and 3 protein by Western blotting with an antibody against a V5 sequence tag located at the 3′ end of the transfected genes. In the lane “mut 2” no protein band was detected, because this mutant harbors a stop codon in the intracellular death domain, located 5′ to the V5 sequence tag, which is therefore not expressed.
In summary, these transfection results suggest that the FAS mutants of cases 1 and 2 (Table 2) act as dominant-negative molecules, as they have lost their apoptosis inducing activity and interfere with the function of the normal, wild-type FAS. The mutations in case 3 (Table 2), which both affect exon 2 and localize to the signal peptide and the extracellular domain of FAS, did not inhibit sFASL-stimulated cell death (Figure 2A). Instead, the mutant seemed even to enhance cell death similar to transfected wild-type FAS (Figure 2A).
The expression of the transfected wild-type and mutant FAS cDNAs was verified by reverse transcriptase (RT)-PCR and Western blotting (Figure 2). RT-PCR analysis resulted in an amplification product of the expected size and thus confirmed the presence of mRNA transcribed from the expression vectors (Figure 2B). The FAS protein translated from the expression vector encoded mRNA was identified on Western blots by an antibody directed against a V5 epitope tag that was cloned in frame 3′ to the FAS cDNAs. A FAS protein of the expected molecular weight could be demonstrated in protein extracts of wild-type or mutant 1 and mutant 3 (cases 1 and 3 in Table 2) transfected cells (Figure 2C). The mutant 2 (case 2 in Table 2) harbored a stop codon mutation in the intracellular death domain, located 5′ to the V5-tag. Thus verification of the protein expression of this construct was not amenable to Western blotting and, as anticipated, no band corresponding to FAS protein was detected (Figure 2C).
FAS Polymorphisms
SSCP analysis detected known single-nucleotide polymorphisms in exon 3 and 7,32 which do not lead to an amino acid substitution, and in intron 8, with an insertion of 7 nucleotides (forward - 5′-CTATTTT-3′; nucleotides 186–192, corresponding to the numbering of GenBank/EMBL FAS gene sequence accession number X82286). This insertion is absent in two other FAS genomic DNA sequences deposited in GenBank/EMBL (accession numbers X81342 and Z66557).
Discussion
Malignant lymphomas, including gastric lymphomas, are frequently resistant to apoptosis induction via FAS.16,17 Furthermore, mutations of FAS are often present in extragastric MALT- and DLBCL.17,26–28 These findings prompted us to search for FAS gene mutations in primary gastric MALT- and DLBCL and we identified seven mutations in five gastric lymphomas (1 MALT lymphoma and 4 DLBCL; two DLBCL had two mutations) by SSCP analysis. The sensitivity of the SSCP method was 90%. Therefore, statistically we should not have missed more than one mutation, if any at all.
Two mutations involved the intracellular death domain, which is essential for apoptosis signaling.34 Mutations in this domain are associated with the most severe phenotype and increased risk for the development of malignant lymphomas in patients with ALPS Ia.33 Codon 244, which was mutated in one of the DLBCL studied, has been shown previously to have a profound effect on the death domain.35 Substitution of the wild-type aspartic acid with alanine greatly reduced self-association of the death domain and binding to the downstream signaling partner FADD in vitro. The second death domain mutation introduced a stop codon that would result in a truncated FAS protein. Both death domain mutations were introduced in FAS-sensitive T47D and Jurkat cells in transient transfection assays to assess their biological activity. They strongly inhibited the induction of cell death by sFASL, thus demonstrating the functional significance of the mutations. The mutants acted in a dominant-negative mode, because T47D and Jurkat cells also express endogenous wild-type FAS. A dominant-negative mechanism is likely to act also in vivo because the lymphomas were heterozygous for the FAS mutations.
Two mutations were located in exon 2, which encodes part of the extracellular domain of the FAS molecule. They are likely to decrease the binding to FASL.33 One mutation affected the amino-terminal 16 amino acid signal peptide, which is cleaved and absent from the mature FAS.36 This mutation may impair proper cleavage of the signal peptide or disturb the transfer of FAS to the cell membrane; however, introduction of two mutations that were present in one DLBCL and affected the signal peptide and the extracellular domain of FAS, did not inhibit sFASL stimulated cell death in T47D and Jurkat cells. This result, however, does not necessarily mean that these two mutations would not decrease FASL sensitivity in the lymphoma from which they were isolated. FAS signaling requires the formation of receptor trimers20,21 and the transfected mutated FAS may have been recruited with the endogenous FAS to form active receptor trimers, as the mutant molecule possesses an intact transmembrane and intracellular death domain. In the gastric lymphoma cells the wild-type and mutated FAS protein may exist in a stochiometric ratio that differs from the expression pattern of ectopic FAS analyzed in the transfection assay and would result in reduced sensitivity to FASL. This hypothesis is supported by experiments performed on thymocytes from heterozygous FAS-knockout mice, which exhibit a reduced FAS-induced apoptosis.37 Furthermore, a loss of function rather than a dominant-negative mode of action has also been concluded from functional studies with extracellular domain mutations identified in ALPS patients.33
Two mutations were located in introns. They are likely to affect RNA splicing. One of the mutations altered the invariable position + 2 of the donor splice site in intron 8. This mutation was also noted previously in a thyroid lymphoma and caused skipping of exon 8.27 A lack of exon 8 in FAS mRNA was associated with recurrent lymphadenopathy in a kindred with ALPS Ia.38 The effect of the mutation in the donor splice region of intron 3 at position + 17, however, is not yet clear.
FAS mutations seem to be rare in gastric MALT lymphomas as in our study only one of 18 cases (5.6%) was mutated. This low incidence is in accordance with a previous report by Bertoni et al,39 who did not detect any FAS mutation in 27 marginal zone B-cell lymphomas (18 extranodal, five splenic, and four nodal), including 13 gastric MALT lymphomas. However, Seeberger et al17 reported a FAS mutation in one of two gastric MALT lymphomas studied. In summary, the incidence of FAS mutations in gastric MALT lymphomas was 6.1% (2 of 33) in the three studies. This low incidence contrasts with the reported high frequency of FAS mutations in extragastric MALT lymphomas (3 of 3, 3 of 5, 6 of 8).17,26,27 Whether this discrepancy reflects different pathomechanisms is unclear. However, extragastric MALT lymphomas, eg, of the thyroid or salivary glands, are often linked with autoimmune disease, whereas those developing in the stomach are associated with an infectious microorganism (H. pylori).
FAS mutations were reported to be frequent in extragastric DLBCL (9 of 43, 21%; 5 of 10, 50%)26,27 with a remarkable preference for extranodal sites. However, the incidence of FAS gene alterations in primary gastric DLBCL is largely unknown, except that Seeberger et al17 reported on a FAS missense mutation in one of three gastric DLBCL. However, the functional significance of this alteration was not determined. In our study six mutations were identified in 4 of 28 primary gastric DLBCL. The four DLCBL with FAS mutations were lymphomas without a low-grade component. It is thus unclear whether they represent transformed MALT lymphomas or have arisen de novo.
FAS is necessary for the clonal selection of B lymphocytes in germinal centers and the elimination of effector lymphocytes at the end of an immune reaction.20–22 Gastric lymphomas often develop in a background of chronic inflammation triggered by antigenic stimulation exerted by H. pylori infection. The acquisition of a FAS mutation may render gastric mucosal B cells less sensitive to death signals that regulate the immune response to H. pylori and thereby escape their elimination. These regulatory cell death stimuli may be exerted by T cells that use FAS ligand as a cytotoxic effector molecule. Alternatively, an autocrine FAS/FASL loop, which has been demonstrated to participate in the elimination of T-cells,40,41 might be operative also in B cells and would be disrupted by FAS mutation.
It is unclear whether FAS mutations cause a different response to therapy. The mutated lymphomas in our study were archival cases that were treated by gastrectomy. At present the use of antibiotics represents the first-line therapy in gastric MALT lymphomas and more than half of cases regress with antibiotics alone1; however, regression after H. pylori eradication is not restricted to MALT lymphoma and has also been observed in a limited number of patients with early-stage gastric DLBCL,42,43 thus demonstrating that even high-grade lymphomas may still be antigen-dependent. Considering the role of FAS in the termination of an immune response,20,21 it seems possible that FAS mutations may influence the efficacy of antigen withdrawal with antibiotic therapy. Furthermore, although a matter of controversy, FAS may participate in the process of drug-induced cell death and could thus modulate the sensitivity to chemotherapy.44,45 It remains to be determined in further prospective studies whether gastric lymphomas with a FAS mutation are different in their response to current treatment regimens.
Acknowledgments
We thank S. Ender for technical assistance, Dr. G. Oberhuber for sharing data on the lymphoma patients, Dr. P. K. Krammer for the FAS cDNA, M. Eisenbauer for Jurkat cells, and A. Jäger for graphical work.
Footnotes
Address reprint requests to Leonhard Müllauer, M.D., D.M.Sci., Department of Pathology, University of Vienna, General Hospital Vienna, Währinger Gürtel 18–20, A-1090 Vienna, Austria. E-mail: leonhard.muellauer@akh-wien.ac.at.
Supported by the Bürgermeisterfonds (grant number 1688), the Jubiläumsfonds der Österreichischen Nationalbank (grant number 8240), the Anton-Dreher Gedächtnisschenkung (grant number 328), and the Herzfeldersche Familienstiftung.
References
- Zucca E, Bertoni F, Roggero E, Cavalli F. The gastric marginal zone B-cell lymphoma of MALT type. Blood. 2000;96:410–419. [PubMed] [Google Scholar]
- Du MQ, Isaacson PG. Gastric MALT lymphoma: from aetiology to treatment. Lancet Oncol. 2002;3:97–104. doi: 10.1016/s1470-2045(02)00651-4. [DOI] [PubMed] [Google Scholar]
- Chan JK, Ng CS, Isaacson PG. Relationship between high-grade lymphoma and low-grade B-cell mucosa-associated lymphoid tissue lymphoma (MALToma) of the stomach. Am J Pathol. 1990;136:1153–1164. [PMC free article] [PubMed] [Google Scholar]
- Ott G, Katzenberger T, Greiner A, Kalla J, Rosenwald A, Heinrich U, Ott MM, Müller-Hermelink HK. The t(11;18)(q21;q21) chromosome translocation is a frequent and specific aberration in low-grade but not high-grade malignant Non-Hodgkin’s lymphomas of the mucosa-associated lymphoid tissue (MALT-) type. Cancer Res. 1997;57:3944–3948. [PubMed] [Google Scholar]
- Baens M, Maes B, Steyls A, Geboes K, Marynen P, De Wolf-Peeters C. The product of the t(11;18), an API2-MLT fusion, marks nearly half of gastric MALT type lymphomas without large cell proliferation. Am J Pathol. 2000;156:1433–1439. doi: 10.1016/S0002-9440(10)65012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye H, Liu H, Attygalle A, Wotherspoon AC, Nicholson AG, Charlotte F, Leblond V, Speight P, Goodlad J, Lavergne-Slove A, Martin-Subero JI, Siebert R, Dogan A, Isaacson PG, Du MQ. Variable frequencies of t(11;18)(q21;q21) in MALT lymphomas of different sites: significant association with CagA strains of H pylori in gastric MALT lymphoma. Blood. 2003;102:1012–1018. doi: 10.1182/blood-2002-11-3502. [DOI] [PubMed] [Google Scholar]
- Dierlamm J, Baens M, Wlodarska I, Stefanova-Ouzounova M, Hernandez JM, Hossfeld DK, De Wolf-Peeters C, Hagemeijer A, Van den Berghe H, Marynen P. The apoptosis inhibitor gene API2 and a novel 18q gene MLT, are recurrently rearranged in the t(11;18)(q21;q21) associated with mucosa-associated lymphoid tissue lymphomas. Blood. 1999;93:3601–3609. [PubMed] [Google Scholar]
- Uren AG, O’Rourke K, Aravind LA, Pisabarro MT, Seshagiri S, Koonin EV, Dixit VM. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one which plays a key role in MALT lymphoma. Mol Cell. 2000;6:961–970. doi: 10.1016/s1097-2765(00)00094-0. [DOI] [PubMed] [Google Scholar]
- Liu H, Ruskon-Fourmestraux A, Lavergne-Slove A, Ye H, Molina T, Bouhnik Y, Hamoudi RA, Diss TC, Dogan A, Megraud F, Rambaud JC, Du MQ, Isaacson PG. Resistance of t(11;18) positive gastric mucosa-associated lymphoid tissue lymphoma to Helicobacter pylori eradication therapy. Lancet. 2001;357:39–40. doi: 10.1016/S0140-6736(00)03571-6. [DOI] [PubMed] [Google Scholar]
- Streubel B, Lamprecht A, Dierlamm J, Cerroni L, Stolte M, Ott G, Raderer M, Chott A. T(14;18)(q32;q21) involving IGH and MALT1 is a frequent chromosomal aberration in MALT lymphoma. Blood. 2003;101:2335–2339. doi: 10.1182/blood-2002-09-2963. [DOI] [PubMed] [Google Scholar]
- Sanchez-Izquierdo D, Buchonnet G, Siebert R, Gascoyne RD, Climent J, Karran L, Marin M, Blesa D, Horsman D, Rosenwald A, Staudt LM, Albertson DG, Du MQ, Ye H, Marynen P, Garcia-Conde J, Pinkel D, Dyer MJS, Martinez-Climent JA. MALT1 is deregulated by both chromosomal translocation and amplification in B-cell non-Hodgkin lymphoma. Blood. 2003;101:4539–4546. doi: 10.1182/blood-2002-10-3236. [DOI] [PubMed] [Google Scholar]
- Du M, Peng H, Singh N, Isaacson PG, Pan L. The accumulation of p53 abnormalities is associated with progression of mucosa-associated lymphoid tissue lymphoma. Blood. 1995;86:4587–4593. [PubMed] [Google Scholar]
- Wotherspoon AC, Pan LX, Diss TC, Isaacson PG. Cytogenetic study of B-cell lymphoma of mucosa-associated lymphoid tissue. Cancer Genet Cytogenet. 1992;58:35–38. doi: 10.1016/0165-4608(92)90130-z. [DOI] [PubMed] [Google Scholar]
- Du MQ, Peng H, Liu H, Hamoudi RA, Diss TC, Willis TG, Ye H, Dogan A, Wotherspoon AC, Dyer MJS, Isaacson PG. BCL10 gene mutation in lymphoma. Blood. 2000;95:3885–3890. [PubMed] [Google Scholar]
- Willis TG, Jadayel DM, Du MQ, Peng H, Perry AR, Abdul-Rauf M, Price H, Karran L, Majekodunmi O, Wlodarska I, Pan L, Crook T, Hamoudi R, Isaacson PG, Dyer MJ. Bcl10 is involved in t(1;14)(p22;q32) of MALT B cell lymphoma and mutated in multiple tumor types. Cell. 1999;96:35–45. doi: 10.1016/s0092-8674(00)80957-5. [DOI] [PubMed] [Google Scholar]
- Plumas J, Jacob MC, Chaperot L, Molens JP, Sotto JJ, Bensa JC. Tumor B cells from Non-Hodgkin’s lymphoma are resistant to CD95 (Fas/Apo-1)-mediated apoptosis. Blood. 1998;91:2875–2885. [PubMed] [Google Scholar]
- Seeberger H, Starostik P, Schwarz S, Knörr C, Kalla J, Ott G, Müller-Hermelink HK, Greiner A. Loss of Fas (CD95/APO-1) regulatory function is an important step in early MALT-type lymphoma development. Lab Invest. 2001;81:977–986. doi: 10.1038/labinvest.3780310. [DOI] [PubMed] [Google Scholar]
- Trauth BC, Klas C, Peters AM, Matzku S, Moller P, Falk W, Debatin KM, Krammer PH. Monoclonal antibody-mediated tumor regression by induction of apoptosis. Science. 1989;245:301–305. doi: 10.1126/science.2787530. [DOI] [PubMed] [Google Scholar]
- Yonehara S, Ishii A, Yonehara M. A cell-killing monoclonal antibody (anti-Fas) to a cell surface antigen downregulated with the receptor of tumor necrosis factor. J Exp Med. 1989;169:1747–1756. doi: 10.1084/jem.169.5.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer PH. CD95’s deadly mission in the immune system. Nature. 2000;407:789–795. doi: 10.1038/35037728. [DOI] [PubMed] [Google Scholar]
- Siegel RM, Chan FKM, Chun HJ, Lenardo MJ. The multifaceted role of Fas signaling in immune cell homeostasis and autoimmunity. Nature Immunol. 2000;1:469–474. doi: 10.1038/82712. [DOI] [PubMed] [Google Scholar]
- Takahashi Y, Ohta H, Takemori T. Fas is required for clonal selection in germinal centers and the subsequent establishment of the memory B cell repertoire. Immunity. 2001;14:181–192. doi: 10.1016/s1074-7613(01)00100-5. [DOI] [PubMed] [Google Scholar]
- Watanabe-Fukunaga R, Brannan CI, Copeland NG, Jenkins NA, Nagata S. Lymphoproliferation disorder in mice explained by defects in Fas antigen that mediates apoptosis. Nature. 1992;356:314–317. doi: 10.1038/356314a0. [DOI] [PubMed] [Google Scholar]
- Fisher GH, Rosenberg FJ, Straus SE, Dale JK, Middelton LA, Lin AY, Strober W, Lenardo MJ, Puck JM. Dominant interfering Fas gene mutations impair apoptosis in a human autoimmune lymphoproliferative syndrome. Cell. 1995;81:935–946. doi: 10.1016/0092-8674(95)90013-6. [DOI] [PubMed] [Google Scholar]
- Rieux-Laucat F, Le Deist F, Hivroz C, Roberts IAG, Debatin KM, Fischer A, de Villartay JP. Mutations in Fas associated with human lymphoproliferative syndrome and autoimmunity. Science. 1995;268:1347–1349. doi: 10.1126/science.7539157. [DOI] [PubMed] [Google Scholar]
- Gronbaek K, Straten P, Ralfkiaer E, Ahrenkiel V, Andersen MK, Hansen NE, Zeuthen J, Hou-Jensen K, Guldberg P. Somatic Fas mutations in Non-Hodgkin’s lymphoma: association with extranodal disease and autoimmunity. Blood. 1998;92:3018–3024. [PubMed] [Google Scholar]
- Takakuwa T, Dong Z, Takayama H, Matsuzuka F, Nagata S, Aozasa K. Frequent mutations of Fas gene in thyroid lymphoma. Cancer Res. 2001;61:1382–1385. [PubMed] [Google Scholar]
- Müschen M, Rajewsky K, Krönke M, Küppers R. The origin of CD95-gene mutations in B-cell lymphoma. Trends Immunol. 2002;23:75–80. doi: 10.1016/s1471-4906(01)02115-9. [DOI] [PubMed] [Google Scholar]
- Radaszkiewicz T, Dragosics B, Bauer P. Gastrointestinal malignant lymphomas of the mucosa-associated lymphoid tissue: factors relevant to prognosis. Gastroenterology. 1992;102:1628–1638. doi: 10.1016/0016-5085(92)91723-h. [DOI] [PubMed] [Google Scholar]
- Jaffe ES, Harris NL, Stein H, Vardiman JW. Lyon, France: IARC Press,; Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. 2001 [Google Scholar]
- Huang QR, Morris D, Manolios N. Identification and characterisation of polymorphisms in the promoter region of the human Apo-1/Fas (CD95) gene. Mol Immunol. 1997;34:577–582. doi: 10.1016/s0161-5890(97)00081-3. [DOI] [PubMed] [Google Scholar]
- Fiucci G, Ruberti G. Detection of polymorphisms within the Fas cDNA gene sequence by GC-clamp denaturing gradient gel electrophoresis. Immunogenetics. 1994;39:437–439. doi: 10.1007/BF00176163. [DOI] [PubMed] [Google Scholar]
- Vaishnaw AK, Orlinick JR, Chu JL, Krammer PH, Chao MV, Elkon KB. The molecular basis for apoptotic defects in patients with CD95 (Fas/Apo-1) mutations. J Clin Invest. 1999;103:355–363. doi: 10.1172/JCI5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh N, Nagata S. A novel protein domain required for apoptosis. J Biol Chem. 1993;268:10932–10937. [PubMed] [Google Scholar]
- Huang B, Eberstadt M, Olejniczak ET, Meadows RP, Fesik SW. NMR structure and mutagenesis of the Fas (APO-1/CD95) death domain. Nature. 1996;384:638–641. doi: 10.1038/384638a0. [DOI] [PubMed] [Google Scholar]
- Itoh N, Yonehara S, Ishii A, Yonehara M, Mizushima SI, Sameshima M, Hase A, Seto Y, Nagata S. The polypeptide encoded by the cDNA for human cell surface antigen Fas can mediate apoptosis. Cell. 1991;66:233–243. doi: 10.1016/0092-8674(91)90614-5. [DOI] [PubMed] [Google Scholar]
- Adachi M, Suematsu S, Kondo T, Ogasawara J, Tanaka T, Yoshida N, Nagata S. Targeted mutation in the Fas gene causes hyperplasia in peripheral lymphoid organs and liver. Nat Genet. 1995;11:294–300. doi: 10.1038/ng1195-294. [DOI] [PubMed] [Google Scholar]
- Tighe PJ, Stevens SE, Dempsey S, Le Deist F, Rieux-Laucat F, Edgar JD. Inactivation of the Fas gene by Alu insertion: retrotransposition in an intron causing splicing variation and autoimmune lymphoproliferative syndrome. Genes Immun. 2002;3(Suppl 1):S66–S70. doi: 10.1038/sj.gene.6363864. [DOI] [PubMed] [Google Scholar]
- Bertoni F, Conconi A, Luminari S, Realini C, Roggero E, Baldini L, Carobbio S, Cavalli F, Neri A, Zucca E. Lack of CD95/Fas gene somatic mutations in extranodal, nodal and splenic marginal zone B cell lymphomas. Leukemia. 2000;14:446–448. doi: 10.1038/sj.leu.2401708. [DOI] [PubMed] [Google Scholar]
- Dhein J, Walczak H, Bäumler C, Debatin KM, Krammer PH. Autocrine T-cell suicide mediated by APO-1/(Fas/CD95). Nature. 1995;373:438–441. doi: 10.1038/373438a0. [DOI] [PubMed] [Google Scholar]
- Rathmell JC, Cooke MP, Ho WY, Grein J, Townsend SE, Davis MM, Goodnow CC. CD95 (Fas)-dependent elimination of self-reactive B cells upon interaction with CD4+ T cells. Nature. 1995;376:181–184. doi: 10.1038/376181a0. [DOI] [PubMed] [Google Scholar]
- Morgner A, Miehlke S, Fischbach W, Schmitt W, Muller-Hermelink, Greiner A, Thiede C, Schetelig J, Neubauer A, Stolte M, Ehninger G, Bayerdorffer E. Complete remission of primary high-grade B-cell gastric lymphoma after cure of Helicobacter pylori infection. J Clin Oncol. 2001;19:2041–2048. doi: 10.1200/JCO.2001.19.7.2041. [DOI] [PubMed] [Google Scholar]
- Chen LT, Lin JT, Shyu RY, Jan CM, Chen CL, Chiang IP, Liu SM, Su IJ, Cheng AL. Prospective study of Helicobacter pylori eradication therapy in stage I (E) high-grade mucosa-associated lymphoid tissue lymphoma of the stomach. J Clin Oncol. 2001;19:4245–4251. doi: 10.1200/JCO.2001.19.22.4245. [DOI] [PubMed] [Google Scholar]
- Friesen C, Herr I, Krammer PH, Debatin KM. Involvement of the CD95 (APO-1/Fas) receptor/ligand system in drug-induced apoptosis in leukemia cells. Nat Med. 1996;2:574–577. doi: 10.1038/nm0596-574. [DOI] [PubMed] [Google Scholar]
- Debatin KM. Activation of apoptosis pathways by anticancer treatment. Toxicol Lett. 2000;112–113:41–48. doi: 10.1016/s0378-4274(99)00252-0. [DOI] [PubMed] [Google Scholar]