Abstract
Untreated groundwater is responsible for about half of the waterborne disease outbreaks in the United States. Human enteric viruses are thought to be leading etiological agents of many of these outbreaks, but there is relatively little information on the types and levels of viruses found in groundwater. To address this problem, monthly samples from 29 groundwater sites were analyzed for 1 year for enteroviruses, hepatitis A virus, Norwalk virus, reoviruses, and rotaviruses by multiplex reverse transcription-PCR (RT-PCR). A procedure with which to remove environmental RT-PCR inhibitors from groundwater samples was developed. The procedure allowed an average of 71 liters of the original groundwater to be assayed per RT-PCR, with an average virus recovery rate of 74%, based on seeded samples. Human enteric viruses were detected in 16% of the groundwater samples analyzed, with reoviruses being the most frequently detected virus group.
The reported incidence of waterborne disease has declined over the past century as a result of better water treatment technology and the use of water quality indicators. Despite the decline, outbreaks, especially those associated with untreated groundwater, continue to occur (3, 4, 5, 10, 14, 17, 20). About 10% of waterborne outbreaks in the United States are reported to be caused by viruses. However, given the onset and duration of symptoms, it is thought that viruses are the cause of disease in many of the outbreaks associated with agents of unknown etiology. Viruses are also a major agent of concern in groundwater that is not under the influence of surface water (13). Enteric viruses that may cause waterborne outbreaks include the astroviruses, enteroviruses, reoviruses, rotaviruses, and Norwalk and related caliciviruses (6, 18, 21). These viruses result in illnesses such as paralysis, meningitis, encephalitis, infectious hepatitis, respiratory illnesses, gastroenteritis, fever, and skin rashes.
Enteric viruses can move from sources of contamination, such as broken sewage pipes and septic tanks, into groundwater aquifers (24). Viruses are typically detected in contaminated waters by concentrating virus particles from large volumes of water on positively charged filters (12, 29). Viruses normally are eluted from the filters with beef extract, which is further concentrated from the eluate by organic flocculation. Viruses present in the concentrate often are detected with plaque or quantal cell culture assays (2, 11, 12). These cell culture analytical methods are labor intensive, requiring about 1 week to more than 6 weeks. Moreover, these methods detect primarily enteroviruses and reoviruses (12). Environmental isolates of many other virus groups are difficult to culture, and Norwalk-like caliciviruses cannot be propagated in established cell lines.
The reverse transcription-PCR (RT-PCR) method is flexible enough to detect all waterborne human enteric viruses. However, there have been two major obstacles to the use of RT-PCR for the detection of viruses in environmental waters: (i) the presence of inorganic and organic inhibitors of the enzymes that are used to amplify viral genomes in environmental samples (15, 27) and (ii) the very small volume that can be assayed in an RT-PCR, typically representing <1 liter to about 10 liters of the original water sample filtered for virus analysis (1, 2). The small volume assayed may lead to false-negative samples because virus titers in contaminated water are often very low.
The U.S. Environmental Protection Agency (EPA) conducted a national groundwater survey for viruses in response to comments received at an informational meeting held in 1990 on a proposed Groundwater Disinfection Rule. The design of the study, the physicochemical characteristics of the sites, and the results of fecal indicator and virus culture assays have been published previously (7, 19, 30). In the present report, we describe the application of a new procedure for the removal of inhibitors of amplification from groundwater samples and a new multiplex RT-PCR assay for human enteric viruses. The major advantages of these procedures are that they result in excellent virus recovery while providing a large additional concentration of virus in the sample and reduce the number of RT-PCR assays that must be performed on each water sample.
MATERIALS AND METHODS
Virus preparations.
A preparation of [14C]leucine-labeled poliovirus (Chat strain) was prepared in HeLa cells, and virus was released by three freeze-thaw cycles. Following centrifugation at low speed to remove cell debris, the virus in the preparation was concentrated by ultracentrifugation as described by Rueckert and Pallansch (25). Concentrated virus was purified by sedimentation for 75 min into a 7.5 to 45% sucrose gradient with an SW50.1 rotor (Beckman Coulter) at 243,000 × g and 15°C. The sucrose solutions were prepared as weight-per weight solutions in phosphate-buffered saline (PBS) with 0.01% bovine serum albumin (BSA; fraction V). Virus-containing peak fractions were pooled and purified by density gradient centrifugation on a 20 to 45% cesium chloride (CsCl) gradient (25). CsCl was removed by chromatography on a column (0.9 by 21 cm) of Bio-Gel A-5m (Bio-Rad Laboratories catalog no. 151-0740) equilibrated in PBS with 0.01% BSA. Virus-containing peak fractions were pooled and measured for infectivity by plaque assay (11) and for particle counts by optical density determination as described by Rueckert and Pallansch (25). It should be noted that radiolabeled virus primarily was used to identify virus-containing peak fractions during the purification process and that virus was purified to obtain physical virus particle counts for recovery experiments. It is not necessary to use either radioactive or purified virus for routine positive controls for molecular assays. The final poliovirus preparation contained 8.8 × 108 particles/μl and 6.5 × 106 PFU/μl, giving a total particle to infectious particle ratio of 136 to 1. A working stock was prepared by diluting the preparation to 18 PFU (2,500 physical particles) per μl in PBS with 0.1% BSA (crystalline grade was used for all stocks, and reagents was used in molecular assays), aliquoted, and stored at −70°C.
Reovirus type 3 was grown in Buffalo green monkey (BGM) kidney cells, and the virus was released by three freeze-thaw cycles. Following removal of cell debris by centrifugation at low speed, the stock preparation containing 2 × 107 PFU/ml was stored at −70°C. A working stock was prepared by diluting the stock preparation 1:100 in PBS with 0.1% BSA. Aliquots of the working stock were then stored at −70°C.
Rotavirus (Wa strain) was grown on MA104 cells, and the virus was released by three freeze-thaw cycles. Following removal of cell debris by centrifugation at low speed, the stock preparation containing 2,000 PFU/ml was stored at −70°C. A working stock was prepared by diluting the stock preparation 1:100 in PBS with 0.1% BSA. The working stock was stored in aliquots at −70°C.
A stock preparation of hepatitis A virus (HAV) strain HM-175 was obtained from Mark D. Sobsey, University of North Carolina. The preparation had been grown on BS-C-1 cells and determined to contain 1.9 × 106 radioimmunofocus units (RFU)/ml. The virus was diluted to 250 RFU/μl in PBS with 0.1% BSA and stored in aliquots at −70°C.
Norwalk virus was prepared by extracting a 10% suspension of a Norwalk virus-positive stool specimen (in PBS with 0.1% BSA) with an equal volume of trichlorofluoroethane (DuPont; this reagent is no longer available, but current alternatives should be adequate [22]). The stool specimen was from a volunteer study funded by the National Marine Fisheries Service and kindly provided by Gary Richards (Charleston Laboratory, National Marine Fisheries Service). Extracted virus was stored at 4°C and diluted in PBS with 0.1% BSA just prior to assay.
Virus mixtures used in multiplex RT-PCRs.
The molecular titer of each virus stock was measured in terms of RT-PCR units by performing single-virus RT-PCR assays (see below) on 10-fold serial dilutions of each stock. One unit is the amount of virus present in the highest dilution that gives a positive result. A mixture of poliovirus, reovirus, and rotavirus was prepared by mixing 10 μl of working stocks of each virus with 70 μl of PBS with 0.1% BSA. This mixture, designated virus mixture A, was prepared, aliquoted, and stored at −70°C. A mixture of HAV and Norwalk virus was prepared for use in RT-PCRs designed to detect these viruses by mixing 10 μl of HAV working stock and 10 μl of a 10−3 dilution of Norwalk virus with 80 μl of PBS and 0.1% BSA. This mixture, designated virus mixture B, was prepared, aliquoted, and stored at −70°C. Virus mixture A and B preparations were rapidly thawed and used at a 10−1 dilution in RT-PCR assays. This dilution gives a final concentration of 50 to 100 RT-PCR units per reaction for each virus.
Groundwater samples.
Each groundwater sample was collected for virus analyses by passing water through a positively charged 10-in. Zetapor 1MDS cartridge filter (Cuno catalog no. 45144-01-1MDS) placed in a standard filter apparatus. The standard filter apparatus consisted of a backflow regulator, a pressure regulator and gauge, a 10-in. cartridge housing containing the 1MDS filter, a water meter, and a flow control valve. The pressure regulator was set at 207 kPa, and the flow control valve was set at 11.4 liters/min. The standard filter apparatus was cleaned and sterilized prior to each use. Samplers were trained for sample collection through the use of a virus sampling training video. Large water samples were typically collected by allowing water to run through the sampling apparatus overnight and then shipped by overnight courier to the analysis laboratory.
Virus was eluted from each 1MDS filter with a modification of the celite elution procedure of Dahling and Wright (7, 8). Briefly, two elutions were performed with 1,600 ml of 1.5% powdered beef extract (Adams Scientific catalog no. 4900-107), pH 9.5. The first elution was performed immediately on receipt of a sampling apparatus. The second elution was performed by storing the cartridge housing filled with 1.5% beef extract overnight at room temperature prior to elution. Viruses present in each sample eluate were concentrated by the addition of 1.6 g of celite (Ohio Valley Specialty Chemical catalog no. MCAFA), adjustment of the pH to 4.0, stirring for 10 min at room temperature, and collection of the celite onto sterile 75-cm-diameter prefilters (Millipore Corporation catalog no. AP20 075 00) by vacuum filtration. Adsorbed viruses were eluted by allowing 80 ml of 0.15 M sodium phosphate, pH 9.0 to 9.5, to filter through the celite with no vacuum. For each groundwater sample to be analyzed by RT-PCR, 20 ml each of the first and second celite eluates was mixed and stored at −70°C.
Inhibitor removal procedure.
On the day before the day on which samples were to be processed, SW28 ultracentrifuge tubes (Beckman catalog no. 344058) and Microcon-100 units (Millipore catalog no. 42414) were filled with PBS containing 0.2% BSA (crystalline grade) and soaked overnight at 4°C. On the day of processing, the groundwater molecular subsamples were rapidly thawed. Following the addition of 80 μl of 5% BSA, the viruses present in 32 ml of each subsample was pelleted through a 5-ml pad of 30% sucrose in 20 mM Tris-1 M NaCl-1 mM EDTA-5 mM EGTA-0.1% crystalline BSA at 131,000 × g for 4.5 h at 10°C. The supernatants were immediately aspirated at the end of the run, and the pellets were resuspended in 2 × 100 μl of PBS with 0.2% BSA.
Each resuspended ultracentrifuge pellet was extracted with 200 μl of 0.01% dithiozone (diphenyl thiocarbazone; Fisher catalog no. D90)-0.01 M 8-hydroxyquinoline-butanol-methanol-trichloroethane (0.1/0.9/1/0.25/0.25, vol/vol). The mixture was prepared fresh with each use with stock solutions of 0.01% dithiozone and 0.01 M 8-hydroxyquinoline (Fisher catalog no. 0261) in chloroform. Stock solutions were stored for up to 1 month at 4°C. Following the addition of the chemical-solvent mixture, samples were vortexed for 30 s, allowed to sit for 15 s at room temperature, vortexed for another 30 s, allowed to sit for another 30 s, and then centrifuged at 14,000 × g in a microcentrifuge for 5 min at 4°C. The aqueous layer was removed and concentrated in a Microcon-100 unit as described by the manufacturer. Samples were washed with 80 μl of PBS-0.2% BSA, reconcentrated to a total volume of about 40 μl, frozen, and then stored at −70°C until assayed by RT-PCR.
Percent recovery of virus during the inhibitor removal procedure.
Negative celite extracts were prepared by adding celite to sterile beef extract as described above. Following pH adjustment and stirring for 10 min, the celite was collected on prefilters. Sodium phosphate, pH 9.0 to 9.5, was passed through the celite and then adjusted to pH 7.0 to 7.5. Thirty-two-milliliter volumes of negative celite extracts were seeded with 50 PFU of poliovirus and run through the inhibitor removal procedure to determine virus recovery. Percent virus recovery was calculated from the amount of virus recovered in the entire 40-μl concentrated sample and the amount of virus in the seed, as measured by plaque assay (11).
Laboratory-derived celite extracts were also seeded with 400 to 1,400 RT-PCR units of each virus and run through the inhibitor removal process. The concentrated sample and sample diluted 10- and 100-fold were assayed by RT-PCR 2-μl assay volumes as described below.
Oligonucleotide primers and probes.
Oligonucleotide PCR primer and hybridization probe sets were designed to detect reoviruses, HAV, rotaviruses, and Norwalk virus with the DNASTAR and OLIGO 4.0 software packages. The primers and probes of each set were designed on the basis of conserved regions of viral genomes so that all of the human (but not animal) strains present in the available GenBank and EMBL databases could be detected. As much as possible, primer sets were designed to give similar melting temperature characteristics. The enterovirus set has been previously described (9). Table 1 lists the sequences of the primers and probes used in this study.
TABLE 1.
Oligonucleotide primers and probes used in this study
| Name | Sequence (5′-3′) |
|---|---|
| MRD 13 | ACC GGA TGG CCA ATC CAA |
| MRD 14 | CCT CCG GCC CCT GAA TG |
| MRD 32 | ACT ACT TTG GGT GTC CGT GTT TC |
| MRD 154 | GCT GGC GTG TCT ATG GAT TCA |
| MRD 155 | CAA AAC GGG AGT GGG GAG C |
| MRD 156 | GTA ATC ATC GGA ATC AGA CTC TG |
| MRD 157 | GTA ATC TTC ATA GTC AGA ATC TGC TT |
| MRD 158 | CAT TTT CTG TTC TTA GTT TCA TGT TT |
| MRD 185 | CTT CTA ACG TTG CTT CCC ATG TCA G |
| MRD 186 | CCA TTT TCC CTC TGT TAG CTT TTC C |
| MRD 187 | CAT CCA TAG CAT GAT AAA GAG GAG C |
| MRD 188 | ACG TTG TCG CAA TGG AGG TGT |
| MRD 189 | GTG CTG AGA TTG TTT TGT CCC AT |
| MRD 190 | GAC ACT CGT CCT TCA AAT GCG TTA |
| MRD 191 | GCG TTG TTA ATC AAG TCC ACG ACC T |
| MRD 192 | GCG TTG TTA ATC AAG TCC ACG ATC T |
| MRD 193 | AAT GCC TTC TGG GTC TCC TTG C |
| MRD 194 | TCA AAC TCA GCG TTA CTT CTC TGC C |
| MRD 195 | GAA AAG TCA ATT CTG AAA CTG GGT TC |
| MRD 211 | CAA GCC CCC CAA GGT GAA T |
| MRD 212 | GGC GCA TGG TTT GTT GAT TTC |
| MRD 214 | CCA GGG GGT ATG CAG GAA AC |
Primers and probes were synthesized on an Applied Biosystems model 381 DNA synthesizer in accordance with the manufacturer's instructions. They were then purified on oligonucleotide purification cartridges (Applied Biosystems catalog no. 400771) as described by the manufacturer. The oligonucleotide purification cartridges remove noncomplete strands by binding the 5′-trityl group of full-length oligonucleotides.
RT-PCR conditions.
RT reactions were performed by adding 2 μl of a virus stock or groundwater sample to a mixture containing 10 mM Tris (pH 8.3), 50 mM KCl, 1.5 (single-virus and mixture A multiplex reactions) or 1.7 (mixture B multiplex reactions) mM MgCl2, 0.67 mM each deoxyribonucleotide triphosphate (dNTP), and 1.67 μM downstream primers in a final reaction volume of 30 μl. The downstream primers were MRD13 (enteroviruses), MRD155 (rotaviruses), and MRD189 (reoviruses) for mixture A and MRD186 (HAV), MRD194 (HAV), and MRD212 (Norwalk virus) for mixture B. Single-virus reaction mixtures contained only the appropriate primer(s) for the virus group. Each reaction tube was overlaid with 50 μl of sterile mineral oil, and viral RNA was released by heating at 99°C for 5 min. After quenching on ice, 30 U of recombinant RNasin (Promega catalog no. N2515) and 50 U of murine leukemia virus reverse transcriptase (Applied Biosystems catalog no. N8080018) were added. cDNA was prepared by incubation at 43°C for 60 min, followed by a 94°C step for 5 min to inactivate the reverse transcriptase.
PCR was performed by adding 70 μl of a mixture containing 10 mM Tris (pH 8.3), 50 mM KCl, 3.0 or 2.75 mM MgCl2, 0.5 μM each upstream primer, and 5 U of Taq polymerase (Applied Biosystems catalog no. N8080153). The upstream primers were MRD14 (enteroviruses), MRD154 (rotaviruses), and MRD188 (reoviruses) for mixture A and MRD185 (HAV), MRD193 (HAV), and MRD211 (Norwalk virus) for mixture B. Single-virus reaction mixtures contained only the appropriate primer(s) for the virus group. Viral cDNA was amplified with 40 cycles each consisting of 60 s at 95°C, followed by 130 s at 59°C. Following the 40 cycles, samples were incubated at 72°C for 15 min and then kept at 4 or −20°C for long-term storage.
Agarose gel electrophoresis.
Four microliters of RT-PCR product was added to 1 μl of 0.04% bromophenol blue-0.04% xylene cyanol-50% glycerol and run on 1.5% agarose (Amresco catalog no. E776) or 3% NuSieve agarose (FMC catalog no. 50082) gels in 40 mM Tris-5 mM sodium acetate-1 mM EDTA, pH 8.0 (TAE buffer), at 100 V for 45 to 80 min. Gels were stained with TAE buffer containing 1 μg of ethidium bromide per μl. Results were recorded with Polaroid film.
Dot blot hybridization conditions.
Five microliters of PCR product was denatured in 0.4 M NaOH-0.01 M EDTA for at least 10 min at room temperature. Ammonium acetate was added to a final concentration of 2 M, and the sample was immediately spotted onto Magnagraph nylon membranes. The membranes were baked at 80°C for 60 min and hybridized with probes labeled on the 3′ end with digoxigenin-ddUTP in accordance with the manufacturer's instructions (Roche catalog no. 1175033). Membranes were prehybridized for 1 h and hybridized overnight at 51°C. Nonspecifically bound probes were removed by washing for 2 × 10 min at 51°C. Probes and wash conditions are given in Table 2.
TABLE 2.
Hybridization conditions used in this study
| Virus | Probe(s) | Wash solutiona |
|---|---|---|
| Enterovirus | MRD 32 | 0.25× SSC-0.1% SDS |
| Rotavirus | MRD 156, MRD 157, MRD 158 | 0.23× SSC-0.1% SDS |
| HAV | MRD 187, MRD 195 | 0.25× SSC-0.1% SDS |
| Reovirus | MRD 190, MRD 191, MRD 192 | 0.19× SSC-0.1% SDS |
| Norwalk virus | MRD 214 | 0.43× SSC-0.1% SDS |
1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate. SDS; sodium dodecyl sulfate.
Following the stringency wash, membranes were blocked and treated with anti-digoxigenin-alkaline phosphatase conjugate in accordance with the manufacturer's directions (Roche catalog no. 1175041). Hybridized probes were detected with the chemiluminescent substrate CSPD as described by the manufacturer (Applied Biosystems catalog no. CD025).
Quality assurance.
A number of precautions were taken during the study to minimize false-positive results. Separate individuals and rooms were used to perform the following major steps: (i) 1MDS filter elution and celite concentration, (ii) inhibitor removal, (iii) PCR, and (iv) gel electrophoresis and hybridization. Ultracentrifuge buckets and surfaces were cleaned after use with a 10% dilution of commercial bleach. To test for false-positive and false-negative results, negative and positive controls were run with each sample set. The negative controls consisted of an RT-PCR negative control (PBS with 0.2% BSA) and a negative process control (distilled H2O) that was processed in the same way as groundwater samples and run with each set of samples processed for inhibitor removal in the laboratory. Positive controls consisted of an RT-PCR positive control (virus-seeded negative process control) and virus-seeded groundwater samples. All PCR results were confirmed by hybridization. Only samples that were positive by hybridization and not presumptively false positive or false negative were counted as positive.
RESULTS
Inhibitor removal procedure.
Preliminary RT-PCR assays of most celite extracts of groundwater samples (without additional inhibitor removal) demonstrated negative results when seeded with virus-positive controls, indicating that they contained RT-PCR inhibitors (data not shown). The primary published procedures available for inhibitor removal at the start of this study were spin chromatography of concentrated eluates on Sephadex G-100 or Sephadex G-100-Chelex-100 columns and polyethylene glycol (PEG) precipitation (1, 28). Purified, 14C-labeled poliovirus was used to rapidly evaluate virus recovery by these methods. Average virus recoveries from seeded celite concentrates were 15% ± 7% by spin chromatography and 32% ± 14% by PEG (data not shown). The addition of protein and/or glycogen to reduce nonspecific binding of virus to surfaces and to aid in the formation of a precipitate did not significantly improve recovery. Similar recoveries were also obtained with a wide range of different PEG and NaCl concentrations, PEG at a 6,000 to 40,000 average molecular weight, and different times and temperatures of precipitation.
Because simpler methods did not show good levels of virus recovery, it was decided to examine ultracentrifugation through a sucrose pad, a common method for initial purification and concentration of picornaviruses (25). The conditions of ultracentrifugation were designed so that viruses pelleted through the sucrose while less dense ribosomes did not. Thus, it was theorized that the sucrose pad could remove a number of inhibitors. Initial experiments indicated that greater than 80% of poliovirus could be recovered by this method and that it removed the inhibitors present in groundwater from an initial test site. However, as other sites were tested, it was found that ultracentrifugation alone was not adequate.
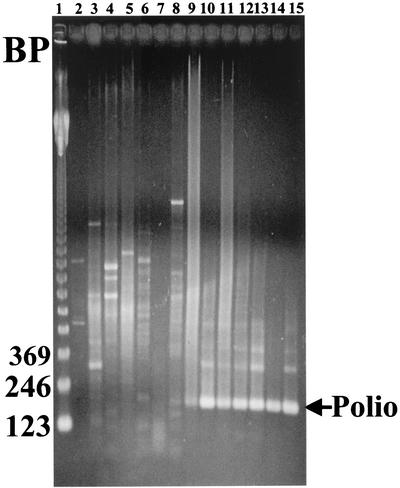
A series of chemical and solvent mixtures were tested for the ability to remove inhibitors. These experiments led to the use of a mixture of 0.01% dithiozone and 0.01 M 8-hydroxyquinoline in chloroform-butanol-methanol-trichloroethane (0.1/0.9/1/0.25/0.25, vol/vol). Figure 1 shows the results obtained with this mixture when it was tested on seven groundwater samples. The positive results seen in lanes 10 to 15 suggest that inhibitors were effectively removed.
FIG. 1.
Inhibitor removal from groundwater samples. Single-enterovirus primer RT-PCRs were performed on seven solvent-treated groundwater samples. Unseeded samples (lanes 3 to 8) or samples seeded with 500 particles of poliovirus (lanes 10 to 15) were analyzed. PBS and virus-seeded PBS were run as a negative (lane 2) and positive controls (lane 9). Products were analyzed on a 1.5% agarose gel. Lane 1 contains a 123-bp ladder. The arrow indicates the location of the 196-bp poliovirus RT-PCR fragment.
The final step of the inhibitor removal procedure was to concentrate the solvent extract with Microcon-100 concentrators. The overall procedure was then tested to determine virus recovery with poliovirus and a plaque assay (Table 3). These tests demonstrated that an overall 74% ± 7% recovery could be achieved when at least 0.1% BSA was used to precoat the ultracentrifuge tubes and Microcon-100 units. It was also necessary to have at least 0.1% BSA in the PBS used to resuspend the ultracentrifuge pellet (data not shown). On the basis of these results, it was decided to use PBS with 0.2% BSA for these steps.
TABLE 3.
Overall virus recovery during inhibitor removal
Treatment is in reference to the amount of BSA that was used to precoat ultracentrifuge tubes and Microcon-100 units and used in the buffer to resuspend the ultracentrifuge pellet.
Percent recovery of poliovirus following the complete inhibitor removal process based on virus plaque assay counts of starting and processed samples. Starting samples were seeded with about 50 PFU of virus.
Starting sample was seeded with about 200 PFU.
Percent recovery based on the average of two replicates.
Percent recovery and standard deviation based on four replicates.
A series of experiments were performed to estimate whether the other viruses used as controls in the study were recovered following the inhibitor removal process. Viruses were seeded into celite extract, processed for inhibitor removal, and assayed by the multiplex RT-PCR procedure described below. Positive results were obtained with all five virus types when celite concentrates were seeded at a level at which 20 to 70 RT-PCR units of each virus was present in the RT-PCR assay.
Multiplex RT-PCR.
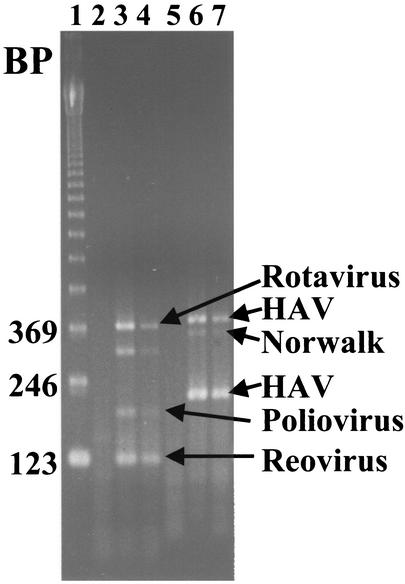
PCR primer and hybridization probe sets were designed or chosen to detect enteroviruses, HAV, Norwalk virus, reoviruses, and rotaviruses in environmental samples. Each set was designed to produce a unique PCR fragment size that could be detected by agarose gel electrophoresis (Fig. 2). The PCR products were 126 bp (lanes 3 and 4) for reovirus, 196 bp for enteroviruses (lanes 3 and 4), 380 bp for rotaviruses (lanes 3 and 4), 222 and 396 bp for HAV (lanes 6 and 7), and 361 bp for Norwalk virus (lanes 6 and 7). In addition to specific bands, a minor rotavirus-related band of about 305 bp (lanes 3 and 4) was often observed.
FIG. 2.
Multiplex RT-PCR. Standard multiplex RT-PCR mixture A (lanes 2 to 4) and B (lanes 5 to 7) reactions were performed and analyzed on a 3% NuSieve agarose gel. Mixture A viruses (poliovirus, reovirus, and rotavirus) were added to the reaction mixture shown in lane 3. A 10-fold dilution of mixture A was added to the reaction mixture shown in lane 4. Mixture B viruses (HAV and Norwalk virus) were added to the reaction mixture shown in lane 6 (RT-PCR mixture B contains two primer sets for HAV, generating two unique PCR fragments). A 10-fold dilution of mixture B was added to the reaction mixture shown in lane 7. Lanes 2 and 5 contained mixture A and B negative control samples, respectively. Lane 1 contains a 123-bp ladder.
A multiplex RT-PCR format was developed to reduce the number of assays needed for each sample. Attempts to combine all five viruses into a single reaction mixture were unsuccessful, as one or more viruses always failed to be amplified under all of the conditions tested. The multiplex RT-PCR procedure required careful optimization of a number of parameters. The optimal levels of magnesium, primers, dNTPs, RNase inhibitor, and reverse transcriptase and the optimal annealing temperature and reverse transcriptase type were determined (data not shown). Changes in the magnesium concentration of as little as ±0.2 mM seriously impacted the ability to detect enteroviruses and Norwalk virus, while different levels of dNTPs and primers had minimal effects (data not shown). After careful optimization, two multiplex reaction mixtures successfully amplified all five viruses (Fig. 2). The first reaction mixture (A) amplifies enteroviruses, reoviruses, and rotaviruses. The second mixture (B) amplifies HAV and Norwalk virus. Even with optimal conditions, enteroviruses and Norwalk virus were not amplified as efficiently as other virus groups.
Although PCR primer sets were designed to generate unique fragment sizes, extraneous bands were always present in groundwater samples (Fig. 1). Thus, gel electrophoresis was a useful tool for optimization studies but could not be used to determine whether viruses were present in groundwater samples. Specific hybridization probes were designed for each primer set, and conditions were optimized so that all hybridization reactions could be performed at the same temperature (data not shown).
Groundwater analyses.
A total of 321 monthly samples from 29 groundwater sites were analyzed by the multiplex RT-PCR method (Table 4). A total of 16% of the samples and 72% of the sites were found to be positive for human enteric viruses. Enteroviruses were present in 5% of the samples, reoviruses were present in 10%, HAV was present in 1%, and Norwalk virus was present in 3% of the samples. Rotaviruses were not detected.
TABLE 4.
Virus analyses of groundwater samples
| Virus | No. of positive samples or sites (% positive)
|
|||
|---|---|---|---|---|
| Initial conditionsa per sample | Final conditionsb per sample | Totalc
|
||
| Per sample | Per sited | |||
| Enterovirus | 8 (8) | 7 (3) | 15 (5) | 11 (38) |
| Reovirus | 6 (6) | 27 (12) | 33 (10) | 18 (62) |
| Rotavirus | 0 (0) | 0 (0) | 0 (0) | 0 (0) |
| HAV | 3 (3) | 1 (0.4) | 4 (1) | 4 (14) |
| Norwalk virus | 3 (3) | 6 (3) | 9 (3) | 6 (21) |
| Totale | 16 (17) | 34 (15) | 50 (16) | 21 (72) |
The percentage is based on 94 samples that were run with 0.01% BSA during the inhibitor removal steps.
The percentage is based on 227 samples that were run with the final inhibitor removal and multiplex RT-PCR conditions.
Total number of samples or sites positive for specific virus types. The percentage is based on 321 samples and 29 sites.
A site was considered positive if one or more samples from the site were positive.
Total samples or sites positive for one or more virus types.
A negative process control and a negative RT-PCR control were added to the inhibitor removal portion of each set of groundwater samples assayed to determine the degree to which false-positive PCRs occur. Virus-seeded groundwater samples were also run to determine the degree to which false-negative PCRs occur. Overall, 6 and 14% of the samples gave presumptively false-positive and false-negative results, respectively.
The percentage of samples that were positive for viruses was the same for both the initial and final conditions. However, the initial samples tended to have larger sample volumes, which may have compensated for the lower recovery (Table 3). The average equivalent amounts of groundwater tested in each RT-PCR assay were 100 liters for the initial samples and 59 liters for the final conditions, with an overall average of 71 liters.
Eleven samples, including 6 of the 20 that were positive by a cell culture assay (see reference 19), were not assayed by the multiplex RT-PCR method. Surprisingly, only 1 of the 14 cell culture-positive samples was positive for a culturable virus type by multiplex RT-PCR. The 14 samples were retested with 10 μl of concentrated sample per assay and a single-virus RT-PCR assay. On retest, 10 samples (71%) were positive, 3 were negative, and 1 gave a potentially false-positive result.
DISCUSSION
An effective inhibitor removal and multiplex RT-PCR method has been developed to test environmental water samples for enteric viruses. This method allows the testing of concentrated samples representing large volumes of the original water sample per reaction. Viruses were concentrated from groundwater with the positively charged 1MDS cartridge filters. Virus particles present on the filters were eluted with a nonflocculating beef extract and concentrated initially by a celite method (8). While this is not the standard method that is typically used by environmental virology laboratories (12), it offers the advantages that virus recovery from seeded water filtered through 1MDS filters and eluted by the celite method is about 90% (7) and that the nonflocculating beef extract is not inhibitory, by itself, to RT-PCR (data not shown).
It was decided that a primary initial criterion for choosing a method for inhibitor removal was to identify one that would give at least 50% virus recovery. Simple spin chromatography and PEG concentration methods did not meet this criterion. An ultracentrifugation step was used for initial removal of inhibitors from celite concentrates. It was necessary to precoat the ultracentrifuge tubes with BSA to obtain reproducible virus recoveries (Table 3). When it was found that ultracentrifugation alone was not adequate for removal of all inhibitors, several solvents were tested for removal efficacy. Solvents were initially selected on the basis of the ability to remove UV and visible light-absorbing material from shellfish tissue over a range of 200 to 640 nm (G. S. Fout and B. C. Martinson, unpublished data). The final solvent combination was derived from those that (i) provided the greatest reduction in absorbing material, (ii) gave a virus recovery following extraction of 14C-labeled poliovirus in celite extract of greater than 85%, and (iii) reduced inhibition. Dithiozone and 8-hydroxyquinoline were added to the solvent mixture because they are chelators of heavy metal ions, which are known to cause inhibition (16, 27). The Microcon-100 concentration step was added to remove residual solvents and other inhibitors of less than 100,000 Da and to provide a greater degree of sample concentration. The overall procedure resulted in effective reduction of inhibitors from groundwater samples (Fig. 1 and Table 4) and an average virus recovery of 74% ± 7% (Table 3).
Groundwater samples were assayed by multiplex RT-PCR for enteroviruses and reoviruses because these are the virus groups that are usually detected by culture assays (12). Rotaviruses were included because they are the leading cause of gastroenteritis in children in the United States and because they caused a waterborne disease outbreak in the United States in 1981 (14). HAV and Norwalk virus were included because they are known to have caused a number of waterborne disease outbreaks (e.g., see reference 4).
Reasons for developing a multiplex RT-PCR procedure were to minimize the number of reactions needed and reduce the cost of water sample screening for enteric viruses. Because an ultracentrifuge rotor holds only six samples, five groundwater samples and a negative process control were processed for inhibitor removal and analyzed by multiplex RT-PCR as sets. The use of the multiplex procedure reduced the number of RT-PCR assays from 70 to 26 per set. With an average reagent cost of about $4.50 per RT-PCR assay, the cost for groundwater sample analysis by the multiplex RT-PCR procedure was about $24.00 per sample, exclusive of labor and equipment costs. It would be desirable to develop a single multiplex reaction with internal controls (26), which could reduce the reagent cost to about $7.00 per water sample.
Overall, 16% of 321 monthly samples and 72% of 29 groundwater sites in the continental United States, the Virgin Islands, and Puerto Rico were positive for human enteric viruses by the multiplex RT-PCR method (Table 4). However, 62% of the samples that were positive for virus by RT-PCR were from seven groundwater sites (24% of the sites). Microscopic particulate analysis was used to rate the risk that these sites are under the influence of surface water (19). Three of the seven sites that showed the most positive samples were rated as being at moderate risk, and four were rated as being at low risk. The three sites that were at moderate risk were in karst formations. The other four sites were in sedimentary, alluvial, or glacial deposits. Reovirus was the most frequently detected virus, with 10% of the samples and 62% of the sites tested being positive. Of the viruses most often associated with waterborne outbreaks, HAV was detected in 1% of the samples and 14% of the sites while Norwalk virus was detected in 3% of the samples and 21% of the sites.
A total of 6% of the samples and 24% of the sites were positive by a culture assay (7, 19). Despite good recoveries with the inhibitor removal procedure (Table 3), there was no good statistical correlation between the culture and molecular assays (data not shown). A main cause for the lack of agreement between the assays was a large difference in assay volumes. The average amount of sample concentrate used in this study for culture and molecular assays was equivalent to 2,181 and 71 liters of groundwater, respectively. When samples that were positive by cell culture were retested with a single primer set assay and a 10-μl assay volume, the two methods were in agreement for 7 of 10 samples tested. The remaining lack of correlation may have resulted from the detection of noninfectious virus or nonculturable coxsackie A viruses.
Because of its importance to public health decisions, the relevance of positive molecular results to public health risk needs to be adequately addressed. It is likely that in certain environmental water types there will be a relevance, but even if there is none, multiplex assays may prove to be useful for rapid screening of water for evidence of virus occurrence. Positive results would then have to be followed up with other tests, such as the integrated cell culture-PCR assay (23), to determine the significance of the findings.
The use of a process negative control in addition to a PCR negative control was important for the identification of presumptively false-positive results, which occurred with 6% of the samples. The level of false-positive results was surprising because of the extent to which all portions of the work were separated. However, most of these false results were associated with the reovirus primer set. This set generated the smallest PCR fragment and appeared to have the highest efficiency of amplification.
False-negative results occurred with 14% of the samples. The inhibitor removal process did not completely remove inhibition from 1 of the 29 groundwater sites, where 47% of the samples gave potentially false-negative values. Most of the other false-negative results were associated with the enterovirus and Norwalk virus primer sets and may have been associated with their poorer efficiency of amplification (Fig. 2). Very low seed levels were used for this study, and this may also have contributed to some of the false-negative results. It is recommended that the seed levels be increased to 100 to 200 RT-PCR units for future studies and that working stocks be carefully aliquoted so that they are frozen and thawed only once.
The U.S. EPA has proposed a Groundwater Rule (www.epa.gov/safewater/gwr.html) that will require public groundwater sites considered to be vulnerable to fecal pollution following sanitary surveys to be monitored monthly for fecal indicators. Sites at which indicators are found must be treated with corrective action that can range from removal of pollution sources to disinfection. The sites assayed for viruses in this study were chosen because they were considered vulnerable and because their water was already being disinfected. Therefore, the positive virus findings of this study do not constitute a public health risk. However, the lack of correlation of virus findings to traditional indicators (19) at highly vulnerable sites puts into question whether indicators will adequately predict public health risk at less vulnerable sites.
Acknowledgments
This study was jointly funded by the U.S. EPA and the American Water Works Association Research Foundation (AWWARF) under a CRADA agreement. This report has been subjected to the Agency’s technical and administrative review and approved for publication as an EPA document. Michael W. N. Moyer was a fellowship recipient of the Oak Ridge Institute for Science and Education Internship Program for the Technical Support Center, Office of Ground Water and Drinking Water, U.S. EPA.
We appreciate the project coordination of Richard J. Lieberman, the EPA Project Manager, and Martin J. Allen, the AWWARF Project Manager. We also acknowledge the technical assistance of Shari R. Crout, Daniel J. Williams, Gretchen T. Sullivan, and Ishrat Abubaker.
REFERENCES
- 1.Abbaszadegan, M., M. S. Huber, C. P. Gerba, and I. L. Pepper. 1993. Detection of enteroviruses in groundwater with the polymerase chain reaction. Appl. Environ. Microbiol. 59:1318-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abbaszadegan, M., P. Stewart, and M. LeChevallier. 1999. A strategy for detection of viruses in groundwater by PCR. Appl. Environ. Microbiol. 65:444-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beller, M., A. Ellis, S. H. Lee, M. A. Drebot, S. A. Jenkerson, E. Funk, M. D. Sobsey, O. D. Simmons III, S. S. Monroe, T. Ando, J. Noel, M. Petric, J. P. Middaugh, and J. S. Spika. 1997. Outbreak of viral gastroenteritis due to a contaminated well: international consequences. JAMA 278:563-568. [PubMed] [Google Scholar]
- 4.Centers for Disease Control. 1983. Water-related disease surveillance annual summary 1982. HHS publication no. (CDC)83-8385. Centers for Disease Control, Atlanta, Ga.
- 5.Craun, G. F. 1990. Review of the causes of waterborne outbreaks, p. 1-22. In G. F. Craun (ed.), Methods for the investigation and prevention of waterborne disease outbreaks. EPA/600/1-90/005a. U.S. Environmental Protection Agency, Cincinnati, Ohio.
- 6.Cukor, G., and N. R. Blacklow. 1984. Human viral gastroenteritis. Microbiol. Rev. 48:157-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dahling, D. R. 2002. An improved filter elution and cell culture assay procedure for evaluating public groundwater systems for cultural enteroviruses. Water Environ. Res. 74:564-568. [DOI] [PubMed] [Google Scholar]
- 8.Dahling, D. R., and B. A. Wright. 1986. Recovery of viruses from water by a modified flocculation procedure for second-step concentration. Appl. Environ. Microbiol. 51:1326-1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Leon, R., Y. S. C. Shieh, R. S. Baric, and M. D. Sobsey. 1990. Detection of enteroviruses and hepatitis A virus in environmental samples by gene probes and polymerase chain reaction. Am. Water Works Assoc. Proc. 1989:833-853. [Google Scholar]
- 10.De Serres, G., T. L. Cromeans, B. Levesque, N. Brassard, C. Barthe, M. Dionne, H. Prud'homme, D. Paradis, C. N. Shapiro, O. V. Nainan, and H. S. Margolis. 1999. Molecular confirmation of hepatitis A virus from well water: epidemiology and public health implications. J. Infect. Dis. 179:37-43. [DOI] [PubMed] [Google Scholar]
- 11.Environmental Protection Agency. 1984. USEPA manual of methods for virology. Publication no. EPA-600/4-84-013. U.S. Environmental Protection Agency, Cincinnati, Ohio. [Online.] www.epa.gov/microbes.
- 12.Fout, G. S., F. W. Schaefer III, J. W. Messer, D. R. Dahling, and R. E. Stetler. 1996. ICR microbial laboratory manual. Publication no. EPA/600/R-95/178. U.S. Environmental Protection Agency, Washington, D.C. [Online.] www.epa.gov/microbes.
- 13.Goyal, S. M. 1984. Viral pollution of the marine environment. CRC Crit. Rev. Environ. Cont. 14:1-32. [Google Scholar]
- 14.Harris, J. R., M. L. Cohen, and E. C. Lippy. 1983. Water-related disease outbreaks in the United States. J. Infect. Dis. 148:759-762. [DOI] [PubMed] [Google Scholar]
- 15.Ijzerman, M. M., D. R. Dahling, and G. S. Fout. 1997. A method to remove environmental inhibitors prior to the detection of waterborne enteric viruses by reverse-transcription polymerase chain reaction. J. Virol. Methods 63:145-153. [DOI] [PubMed] [Google Scholar]
- 16.Kreader, C. A. 1996. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl. Environ. Microbiol. 62:1102-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawson, H. W., M. M. Braun, R. I. Glass, S. E. Stine, S. S. Monroe, H. K. Atrash, L. E. Lee, and S. J. Englender. 1991. Waterborne outbreak of Norwalk virus gastroenteritis at a southwest US resort: role of geological formations in contamination of well water. Lancet 337:1200-1204. [DOI] [PubMed] [Google Scholar]
- 18.LeBaron, C. W., N. P. Furutan, J. F. Lew, J. R. Allen, V. Gouvea, C. Moe, and S. S. Monroe. 1990. Viral agents of gastroenteritis. Public health importance and outbreak management. Morb. Mortal. Wkly. Rep. Recomm. Rep. 39(RR-5):1-24. [PubMed] [Google Scholar]
- 19.Lieberman, R. J., L. C. Shadix, B. S. Newport, S. R. Crout, S. E. Buescher, R. S. Safferman, R. E. Stetler, D. Lye, G. S. Fout, and D. R. Dahling. 1995. Source water microbial quality of some vulnerable public ground water supplies, p. 1425-1436. In Proceedings of the 1994 Water Quality Technology Conference, part II. American Water Works Association, Denver, Colo.
- 20.Maurer, A. M., and D. Sturchler. 2000. A waterborne outbreak of small round structured virus, campylobacter and shigella co-infections in La Neuveville, Switzerland, 1998. Epidemiol. Infect. 125:325-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Melnick, J. L. 1984. Enteric viruses in water. Monogr. Virol. 15:1-16. [Google Scholar]
- 22.Mendez I. I., L. L. Hermann, P. R. Hazelton, and K. M. Coombs. 2000. A comparative analysis of Freon substitutes in the purification of reovirus and calicivirus. J. Virol. Methods 90:59-67. [DOI] [PubMed] [Google Scholar]
- 23.Reynolds, K. A., C. P. Gerba, and I. L. Pepper. 1996. Detection of infectious enteroviruses by an integrated cell culture-PCR procedure. Appl. Environ. Microbiol. 62:1424-1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robertson, J. B., and S. C. Edberg. 1997. Natural protection of spring and well drinking water against surface microbial contamination. I. Hydrogeological parameters. Crit. Rev. Microbiol. 23:143-178. [DOI] [PubMed] [Google Scholar]
- 25.Rueckert, R. R., and M. A. Pallansch. 1981. Preparation and characterization of EMC virus. Methods Enzymol. 78:315-325. [PubMed] [Google Scholar]
- 26.Schwab, K. J., M. K. Estes, F. H. Neill, and R. L. Atmar. 1997. Use of heat release and an internal RNA standard control in reverse transcription-PCR detection of Norwalk virus from stool samples. J. Clin. Microbiol. 35:511-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shieh, Y. S. C., D. Wait, L. Tai, and M. D. Sobsey. 1995. Methods to remove inhibitors in sewage and other fecal wastes for enterovirus detection by the polymerase chain reaction. J. Virol. Methods 54:51-66. [DOI] [PubMed] [Google Scholar]
- 28.Sobsey, M. D. 1994. Molecular methods to detect viruses in environmental samples, p. 387-400. In R. C. Spencer, E. P. Wright, and S. W. B. Newsom (ed.), Rapid methods and automation in microbiology and immunology. Intercept Limited, Hampshire, United Kingdom.
- 29.Sobsey, M. D., and J. S. Glass. 1980. Poliovirus concentration from tap water with electropositive adsorbent filters. Appl. Environ. Microbiol. 40:201-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stetler, R. E., and F. P. Williams, Jr. 1996. Pretreatment to reduce somatic salmonella phage interference with FRNA coliphage assays: successful use in a one-year survey of vulnerable groundwaters. Lett. Appl. Microbiol. 23:49-54. [DOI] [PubMed] [Google Scholar]