Abstract
A chromosomally lux-marked (Tn5 luxCDABE) strain of nontoxigenic Escherichia coli O157:H7 was constructed by transposon mutagenesis and shown to have retained the O157, H7, and intimin phenotypes. The survival characteristics of this strain in the experiments performed (soil at −5, −100, and −1,500 kPa matric potential and artificial groundwater) were indistinguishable from the wild-type strain. Evaluation of potential luminescence was found to be a rapid, cheap, and quantitative measure of viable E. coli O157:H7 Tn5 luxCDABE populations in environmental samples. In the survival studies, bioluminescence of the starved populations of E. coli O157:H7 Tn5 luxCDABE could be reactivated to the original levels of light emission, suggesting that these populations remain viable and potentially infective to humans. The attributes of the construct offer a cheap and low-risk substitute to the use of verocytotoxin-producing E. coli O157:H7 in long-term survival studies.
The incidence of food poisoning outbreaks has increased in recent years, with Escherichia coli O157:H7 emerging as a pathogen of increasing public health concern. The infective dose of E. coli O157:H7 in humans has been estimated to be very low (11), and infection can result in a wide range of clinical manifestations, including life-threatening hemolytic-uremic syndrome (37). E. coli O157:H7 resides harmlessly in the digestive system of cattle and many other animals and is excreted in animal feces, from where it can enter the environment (15, 45). Recent reports have attributed E. coli O157:H7 outbreaks to a diverse range of sources, including swimming pools, water supplies, raw vegetables, and, more commonly, beef products (15, 28).
There is little information regarding the behavior and metabolic status of E. coli O157:H7 in the environment, although some reports suggest the potential for considerable survival in cattle feces, soil, and water (9, 14, 18, 42). Vertical transmission of E. coli O157:H7 from cattle feces through soil has also been demonstrated (10), and this finding highlights a possible transmission route leading to the contamination of private drinking water. All of these studies, however, have utilized culture-based methods, which rely on disruption of cells from environmental material followed by plating on selective media, such as cefixime- and potassium tellurite-containing sorbitol MacConkey agar (6). These approaches will, however, fail to target E. coli O157:H7 populations that may be in a viable but nonculturable state. The occurrence of the viable but nonculturable state in enteric bacteria is highly disputed by some (3), while other reports suggest it does occur in E. coli commensal and O157:H7 populations in water and under saline conditions (21, 31, 42). Additionally, the ability to disrupt bacterial cells which are tightly adhered to soil particles may also be inefficient, leading to an underestimation of target population size. Molecular techniques, such as quantitative PCR, eliminate some of the biases of culture-based methods for estimation of pathogen abundance in soil, although, in most cases, an enrichment step is still required (22). Despite this, PCR-based approaches rely on direct extraction of nucleic acids or cells (followed by nucleic acid extraction) from soil, which may also be biased depending on the extraction method employed (23). The possibility of PCR detection of intact DNA from nonviable pathogens also limits the value of this approach (16).
As an alternative approach, the lux genes, which encode bioluminescence, have been successfully used in studies of the fate of microorganisms in the environment, including soil (33). Use of constructs which have been chromosomally marked with the full luxCDABE cassette offer the same advantages for localization of the target organism as marking with jellyfish green fluorescent protein (GFP) (33) but also offer potential assessment of metabolic activity. Although a GFP-marked E. coli O157:H7 strain has been described recently (14), the fluorescent phenotype of that strain was not used other than for verification of culturable plate counts. In contrast to the GFP fluorescence phenotype, which does not change on starvation or entry into the viable but nonculturable state, the bioluminescence phenotype is dependent on the energy status of the cell (39). Application of lux marker systems has therefore enabled measurement of population activity (26), detection of viable but nonculturable cells (7), and nonextractive estimation of active biomass of target populations in soil (25, 27).
The present study first describes the construction of a chromosomally lux-marked nontoxigenic strain of E. coli O157:H7. Second, the applicability of this stable chromosomally lux-marked strain for survival studies in soil and artificial groundwater samples is demonstrated. A potential luminescence assay has been applied to provide a quick in situ estimation of the size of potentially active bioluminescent E. coli O157:H7 populations.
MATERIALS AND METHODS
Bacterial strain and culture conditions.
E. coli O157:H7 strain 3704 was kindly provided by Fiona Thompson-Carter (E. coli reference laboratory, University of Aberdeen, United Kingdom). The strain was originally isolated from a farm drain and has been proven to be nontoxigenic due to the absence of toxin activity (by Verocell assay) and toxin genes (by PCR) (F. Thomson-Carter, unpublished observation). The strain and subsequent constructs were maintained on LB agar. Stock cultures were kept in 10% (vol/vol) glycerol at −80°C. Cell numbers were quantified on both sorbitol MacConkey agar (SMAC) (Oxoid Ltd., United Kingdom), and tryptone soy agar (TSA) (Oxoid Ltd., United Kingdom), at 37°C for 24 h to estimate differences due to sublethal injury (40).
Chromosomal lux marking of E. coli O157:H7.
E. coli O157:H7 strain 3704 Tn5 luxCDABE was constructed by biparental mating of a spontaneous rifampin-resistant mutant of E. coli O157:H7 strain 3704 with a donor strain, followed by suicide plasmid delivery and transposon mutagenesis. The donor strain, E. coli S17 λpir luxCDABE Km2, was a kind gift from P. Hill (University of Nottingham, United Kingdom) and contains the luxCDABE cassette from Photorhabdus luminescens and the antibiotic resistance genes for ampicillin and kanamycin (43). The rifampin-resistant mutant of E. coli O157:H7 strain 3704 was made by plating serial dilutions of an overnight culture onto Luria-Bertani (LB) plates containing rifampin (100 μg ml−1). Transconjugants in which Tn5 luxCDABE had inserted into the chromosome were initially selected on the basis of growth on LB containing rifampin (100 μg ml−1) and kanamycin (50 μg ml−1) and then by visible bioluminescence in the dark. The absence of the plasmid (which conferred ampicillin resistance) was confirmed by small-scale plasmid DNA preparations and by the lack of growth of the transconjugants on LB plates containing ampicillin (50 μg ml−1).
The stability of the lux phenotype was examined by successive subculturing of the selected E. coli O157:H7 strain 3704 Tn5 luxCDABE in LB broth without addition of kanamycin and subsequent confirmation of colony growth on LB agar with versus without kanamycin (50 μg ml−1). To confirm that mutagenesis had not disrupted the O157, H7, or intimin phenotype of the strain, multiplex PCR as described by Campbell et al. (4) was performed.
Southern blot conditions.
Genomic DNA from the donor, host, and chromosomally lux-marked transconjugants was isolated according to standard procedures (34) and digested to completion with NdeI. The digested genomic DNA was probed with a 1,273-bp fragment of the luxCDABE cassette (containing a single NdeI site) which had been amplified with the primer pair JR42 (5′-CGC TGT CGG AAA TTA TAC GG-3′) and JR43 (5′-GTT ACG GTA AAT GTC GTA GG-3′). The specific PCR conditions used to generate the lux fragment were 95°C for 1 min 30 s, then 29 cycles of 94°C for 30 s, 55°C for 1 min, and 72°C for 1 min, followed by a final extension at 72°C for 10 min. The probe was purified from a 1% agarose gel and labeled according to the manufacturer's instructions (ECL direct nucleic acid labeling and detection system; Amersham Pharmacia, United Kingdom). Southern blotting conditions were as recommended by the manufacturer of the labeling kit.
Growth experiments comparing chromosomally lux-marked and wild-type E. coli O157:H7.
Batch culture growth experiments were carried out in triplicate in tryptone soy broth (TSB) (Oxoid Ltd., United Kingdom). The cultures were incubated at 37°C at 200 rpm, and 1-ml samples were removed at regular intervals for analysis of optical density at 600 nm and bioluminescence (relative light units [RLU]). Bioluminescence was measured with a Jade luminometer (Labtech International Ltd., United Kingdom).
Potential bioluminescence assay.
A slight modification of the method by Duncan et al. (7) was used. Briefly, 9 ml of warm TSB (37°C) was inoculated with either pure culture or inoculated groundwater (1 ml) or inoculated soil (1 g) from the survival studies described below. The mixture was incubated at 37°C with continuous shaking at 200 rpm for 30 min. After this activation step, luminescence was measured from 1-ml undiluted aliquots of the mixture as described above. When potential luminescence was measured from soil, aliquots of the TSB-sample mixture (1.5 ml) were first centrifuged at 4,000 × g for 5 s, and 1 ml of supernatant was used for luminescence measurements. Cell numbers were quantified with the remainder of the TSB-sample mixture.
Potential luminescence as an indicator of potentially active population size of E. coli O157:H7 in soil.
To investigate potential luminescence as an indicator of the size of potentially active E. coli O157:H7 populations, stationary-phase (15-h) cultures (washed and resuspended in one-quarter-strength Ringer's solution) of chromosomally lux-marked E. coli O157:H7 were subjected to 10-fold dilutions in one-quarter-strength Ringer's solution. Aliquots (1 ml) of this dilution series were used for the potential luminescence assay described above. To investigate the response in soil, 0.8 ml of the dilution series was added to 10 g (fresh weight) of Insch soil (n = 3) to achieve a soil matric potential of −100 kPa, and the potential luminescence assay was performed with the inoculated soil. The response of the potential luminescence to starved populations of E. coli O157:H7 was also tested by incubating the washed stationary-phase culture for up to 15 days at 15°C. The potential luminescence assay was performed with and without the inclusion of nalidixic acid (10 μg ml−1) in order to investigate whether growth could take place during the course of the assay. The remainder of the dilution series was used for estimation of cell numbers on SMAC and TSA.
Validation of substitute strain for survival studies in natural water samples.
The survival responses of the wild-type and the lux-marked construct were also compared in sterile artificial groundwater. Both strains were grown to late exponential phase in TSB, washed twice with 1 volume of sterile artificial groundwater (Table 1), and incubated at 15°C in the dark for 70 days. Viable cell numbers were established at set time points by plate counts on SMAC and TSA following dilution in one-quarter-strength Ringer's solution. Potential luminescence of the chromosomally lux-marked strain was measured as described above.
TABLE 1.
Composition of groundwatera
| Constituent | Concn (mM) |
|---|---|
| NaCl | 0.07 |
| MgCl2 | 0.07 |
| MgSO4 | 0.07 |
| CaSO4 | 0.045 |
| K2SO4 | 0.02 |
| (NH4)2NO3 | 0.02 |
| KH2PO4 | 0.01 |
The pH was 7.0.
Validation of marked strain as a substitute in a soil survival study.
Topsoil (a sandy loam) was collected from Insch, northeastern Scotland, sieved to 3 mm, oven dried (105°C), and stored at 4°C until required. A stationary-phase (15-h) culture of chromosomally lux-marked E. coli O157:H7 strain 3704 or wild-type strain 3704 was centrifuged for 10 min at 11,000 × g, after which cell pellets were resuspended in one-quarter-strength Ringer's solution and left to starve for 24 h. A volume of resuspended cells was then added to sieved and dried soil to achieve soil matric potentials, after equilibration, of −5, −100, and −1,500 kPa. Matric potential was assessed by a combination of pressure plate and tension table measurements (30). The final matric potentials were prepared in accordance to the moisture release characteristics of the soil. The inoculated soil (30 g) was weighed into glass jars, which were sealed with parafilm and incubated at 15°C. Uninoculated control soil, which was equilibrated to the same matric potentials, was also included. The microcosms were sampled in triplicate after 1, 5, 10, 20, 35, 60, 90, and 158 days of incubation at 15°C. At each sampling point, potential luminescence of samples was measured with the methods described above. Numbers of chromosomally lux-marked E. coli O157:H7 were determined by serial dilution of soil in one-quarter-strength Ringer's solution and plating appropriate dilutions on SMAC and TSA.
Statistical analysis.
One-way analysis of variance (with Excel 97) was performed to compare differences in specific growth rates between the wild-type and chromosomally lux-marked E. coli O157:H7. One-way analysis of variance was also performed to test differences in survival rates in the soil and water survival experiments at each time point. Differences were considered significant at the P ≤ 0.05 level.
RESULTS
Characterization of chromosomally lux-marked against wild-type E. coli O157:H7.
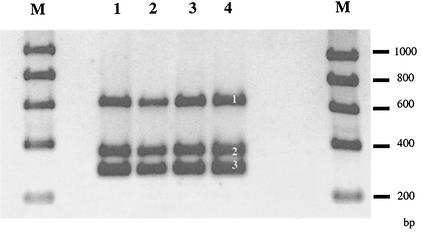
Batch culture experiments comparing the wild-type and the chromosomally lux-marked E. coli O157:H7 strain 3704 showed that lux marking slightly affected the specific growth rate at 37°C. The wild-type strain displayed a specific growth rate of 1.13 ± 0.03 h−1, whereas the lux-marked strain grew slightly faster at 1.24 ± 0.01 h−1. The difference was statistically significant (P < 0.05). Stability experiments involving successive subculturing of the conjugant in the absence of antibiotics found that the lux phenotype was stable for over 100 generations (data not shown). The insertion of the lux cassette had not disrupted the genes for production of the O157, H7, or intimin phenotype, as shown by the identical sizes of the PCR products for these markers (Fig. 1). Southern analysis confirmed a single chromosomal insertion of the luxCDABE cassette in E. coli O157:H7 strain 3704. Two bands were detected in the transconjugants due to the presence of the NdeI site in the probe fragment. However, only one band was present in the donor strain as the plasmid containing the luxCDABE cassette includes only one restriction site for NdeI. No bands were detected from the wild-type E. coli O157:H7 strain 3704 (data not shown).
FIG. 1.
Mutagenesis of wild-type strain (lanes 1 and 2) with the chromosomal insertion of a lux cassette did not interrupt the products for the H7 (band 1), intimin (band 2), or O157 (band 3) markers in the lux-marked mutant (lanes 3 and 4). Lanes M, Bioline Hyperladder I molecular size markers.
Potential luminescence as an indicator of potentially active population size of E. coli O157:H7.
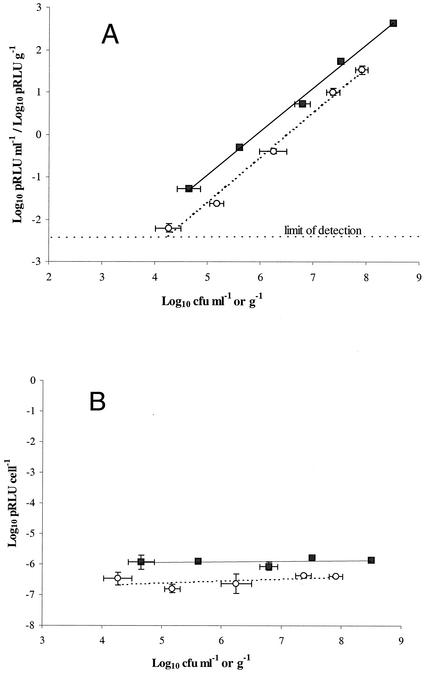
Potential luminescence was proportional to the number of chromosomally lux-marked E. coli O157:H7, either as pure culture washed in one-quarter-strength Ringer's solution or as a soil inoculum (Fig. 2A). In these experiments, increasing numbers of cells resulted in a proportional increase in luminescence within the 30-min assay period, i.e., potential luminescence per cell was the same over a range of cell densities between 104 and 108 CFU per ml or per g of sample (Fig. 2B). The results suggested that the potential luminescence assay was sensitive enough to detect >4.0 × 103 CFU ml−1 when cells were presented in a clear matrix, in this case, one-quarter-strength Ringer's solution. Similarly, although the presence of soil had a significant masking effect, the assay could detect >1.5 × 104 CFU per g of Insch soil. Starvation of the cells over 15 days did not result in a significant loss of potential luminescence per cell (Fig. 3). There was no increase in cell numbers during the course of potential luminescence assays (P > 0.05). In addition, inclusion of nalidixic acid (10 μg ml−1) to inhibit cell division had no statistically significant effect on potential luminescence (data not shown).
FIG. 2.
(A) Potential luminescence response of a dilution series of a pure culture of chromosomally lux-marked E. coli O157:H7 in one-quarter-strength Ringer's solution (▪) and when inoculated into Insch soil (○); (B) their corresponding potential luminescence values per cell. Data represent the mean of three replicates ± standard deviation.
FIG. 3.
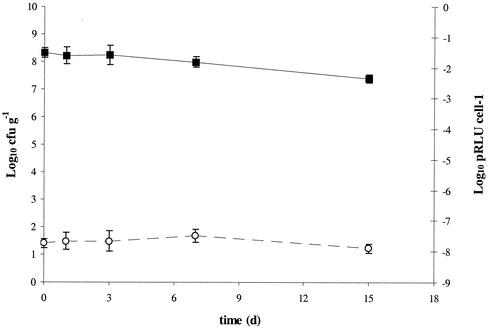
Survival of chromosomally lux-marked strain to starvation in one-quarter-strength Ringer's solution for 15 days, as assessed by culturable cell counts (▪) and potential luminescence (pRLU) per cell (○). Data represent the mean of three replicates ± standard deviation.
Survival of wild-type and lux-marked E. coli O157:H7 in artificial groundwater.
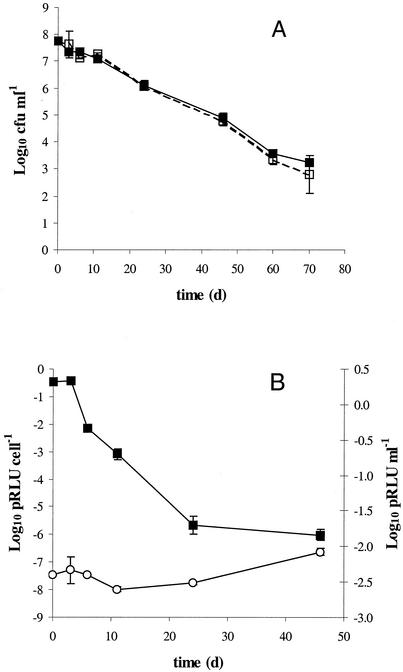
The population sizes of both the wild-type and the chromosomally lux-marked strain declined by approximately 5 log units over the course of the 70 days of incubation in artificial groundwater at 15°C (Fig. 4A). There was no statistical difference between the population sizes of the strains at any of the time points (P > 0.05) or of the cell numbers retrieved on either TSA and SMAC.
FIG. 4.
(A) Survival of wild-type (▪) and chromosomally lux-marked (□) E. coli O157:H7 in artificial groundwater at 15°C. (B) The potential luminescence (pRLU) of the lux-marked strain (▪) dropped at an equivalent rate to the cell numbers, as seen from the potential luminescence per cell (○) response. Only data until day 45 are shown, as potential luminescence values dropped below the detection level after this time point. Data represent the mean of three replicates ± standard deviation.
Survival of E. coli O157:H7 and the lux-marked strain in soil at different matric potentials.
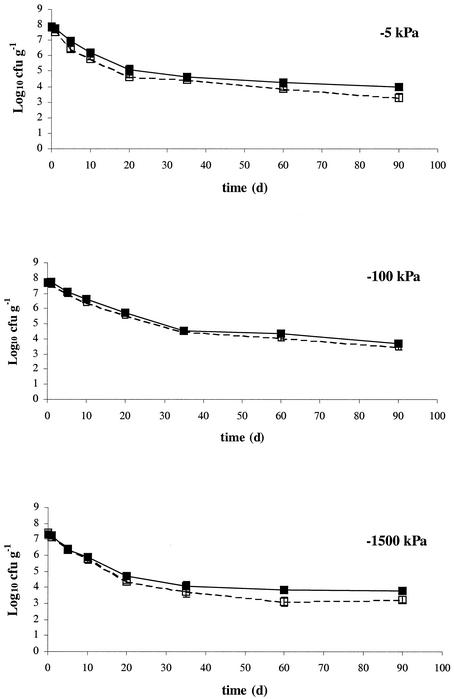
The effects of soil matric potential on survival of wild-type and chromosomally lux-marked E. coli O157:H7 strain 3704 was investigated in Insch soil at 15°C equilibrated to matric potentials of −5 kPa, −100 kPa, and −1,500 kPa (Fig. 5). Both wild-type and lux-marked E. coli O157:H7 populations declined relatively quickly (by approximately 3.5 log units) during the first 35 days of the experiment. Population decline was less rapid for the remainder of the 90-day experiment. There was no statistically conclusive evidence with regards to different soil matric potentials influencing the survival characteristics of E. coli O157:H7 populations (Table 2). Additionally, although suggestive at some sampling points, there was no conclusive statistical evidence for a difference between the wild-type and lux-marked strains.
FIG. 5.
Survival of wild-type (▪) and chromosomally lux-marked (□) E. coli O157:H7 in Insch soil at 15°C and matric potentials of −5 kPa, −100 kPa, and −1,500 kPa. Data represent the mean of three replicates ± standard deviation.
TABLE 2.
Recovery of E. coli strains from spiked Insch soil at 15°C and different matric potentialsa
| Day | Recovery (% of inoculum) at potential (kpa):
|
|||||
|---|---|---|---|---|---|---|
| −5
|
−100
|
−1500
|
||||
| WT | clux | WT | clux | WT | clux | |
| 0 | 100 | 100 | 100 | 100 | 100 | 100 |
| 1 | 74.7 ab | 46.7 a | 103.3 b | 77.6 ab | 86.3 b | 49.9 a |
| 5 | 12.1 ab | 3.9 a | 24.2 c | 20.3 bc | 11.6 a | 9.4 a |
| 10 | 1.94 ab | 0.91 a | 8.74 c | 5.00 d | 4.06 bd | 2.61 ab |
| 20 | 0.190 ab | 0.061 a | 0.958 c | 0.724 d | 0.263 b | 0.097 a |
| 35 | 0.059 a | 0.041 a | 0.067 a | 0.061 a | 0.064 a | 0.024 b |
| 60 | 0.0271 ab | 0.0105 c | 0.0437 d | 0.0220 a | 0.0335 bd | 0.0058 c |
| 90 | 0.0121 a | 0.0031 a | 0.0094 a | 0.0063 a | 0.0357 b | 0.0071 a |
Values with different letters within a row were significantly different. WT, wild type; clux, chromosomally lux marked.
Potential luminescence as an indicator of metabolic activity in survival studies.
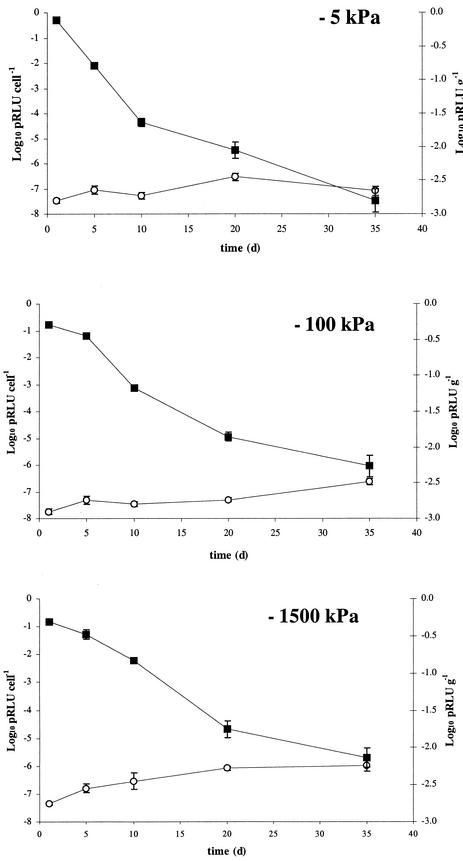
In both the artificial groundwater experiment (Fig. 4B) and at all matric potentials used in Insch soil (Fig. 6), values of potential luminescence per cell remained relatively constant. This indicated that there was no decrease of potential metabolic activity in the cells over time. Potential luminescence values in the assays dropped below the detection limit after 45 days in artificial groundwater and after 35 days in the soil matric potential experiments due to the decreases in total cell number, and potential luminescence per cell could thus not be calculated for these time points. In addition, the slight increases in potential luminescence at the last time points shown in each of these figures are most likely an artifact of the combination of very low potential luminescence values and small cell numbers in these samples.
FIG. 6.
Corresponding values (to the cell numbers in Fig. 5) of potential luminescence (▪) and luminescence per cell (○) in microcosms of Insch soil at 15°C and matric potentials of −5, −100, and −1,500 kPa, spiked with a culture of chromosomally lux-marked E. coli O157:H7. Data shown are for the first 35 days, as potential luminescence dropped below the detection limit after this time point. Data represent the mean of three replicates ± standard deviation.
DISCUSSION
In the European Community, verocytotoxin-producing E. coli O157:H7 has recently been reclassified from Hazard Group 2 to a Hazard Group 3 pathogen in response to a number of laboratory-acquired infections (5, 8). This level of classification limits the ability of most European research laboratories to conduct studies with the toxigenic organism. Nontoxigenic isolates of E. coli O157:H7 provide an alternative approach to study pathogen behavior and movement. Such nontoxigenic strains appear to possess the same characteristics as toxigenic isolates with the exception of the genes coding for the cytotoxins stx1 and stx2 (35). Whether the presence of the cytotoxins constitutes an advantage in survival and transport of E. coli O157:H7 in the environment is debatable. The toxicity of Shiga toxin and similar toxins to eukaryotic cells has been well documented, and it has been speculated that this cytotoxicity may aid the survival of E. coli O157 in eukaryotic cells (17, 19). In a recent, comprehensive study of the survival of various O157:H7 isolates in manures and manure slurries, however, identical or very similar survival patterns were obtained for toxigenic and nontoxigenic strains (18). The use of nontoxigenic isolates can thus be viewed as a valid alternative approach to study pathogen behavior and movement in environmental samples.
Use of a lux-marked E. coli O157:H7 to monitor adherence to food has been reported previously (36). The use of this construct enabled real-time visualization of plasmid-marked E. coli O157:H7 adherence to animal carcass tissue and clearly highlighted the potential values of a lux marker system in E. coli O157:H7 survival studies. Due to potentially high copy numbers, plasmid lux-marked constructs may exhibit higher luminescence than chromosomally lux-marked constructs (1). The disadvantage of such constructs, however, is that a selective pressure (such as addition of antibiotics) must be applied to maintain the plasmid. Such constructs are, therefore, only suitable for short-term studies where plasmid loss is unlikely to occur. In contrast, use of stable chromosomally lux-marked constructs, such as that reported in this study, eliminates the requirement of applying a selective pressure and is thus desirable for long-term survival experiments. Although the construction of a chromosomally lux-marked E. coli O157:H7 strain has been reported previously (41), this was a luxAB mutant, which is dependent on addition of n-decanol for display of the luminescent phenotype. The construct described in this study alleviates this need for additions prior to analysis.
Although there was a slight difference in growth rate between the chromosomally lux-marked and the wild-type E. coli O157:H7 strain at 37°C, this difference was not noted in the long-term survival experiments. In experiments with soil and artificial groundwater, there was no difference in survival behavior between the wild-type and chromosomally lux-marked strain. These findings support the validity of the construct as a substitute for the wild-type strain for long-term experiments. In addition, the observed survival rates correspond well with published data of survival of toxigenic O157:H7 strains in similar environmental samples (14, 24, 29, 42). Although the experiments presented here have been conducted in essentially sterile environments in order to provide preliminary validation for the use of this lux-marked construct, recent findings also suggested the validity of application in nonsterile environmental samples (2).
The data obtained here suggest that contrasting matric potentials did not influence the survival of E. coli O157:H7 in Insch soil. Although matric potential is thought to affect bacterial movement below −20 kPa and, similarly, decrease bacterial activity below −50 kPa (44), no difference in survival of E. coli O157:H7 at matric potentials between field capacity (−5 kPa) and wilting point (−1,500 kPa) was noted. Similar observations were made by Meikle et al. (25) and Turnbull et al. (38) when survival of Pseudomonas fluorescens in soil was studied. In the study by Meikle et al. (25), some indication that increased matric stress reduces bacterial survival was found, but, as in this study, these indications were not confirmed by statistical differences. Although survival of E. coli O157:H7 may be similar in soil at most realistic matric potentials, the differences in matric potential will most certainly have an effect on the localization of bacteria within the soil in the event of a sudden rainfall. In the drier soils, the cells will be translocated into the smaller pores within the soil matrix (32), which will limit their potential transport by leaching. Thus, although there may be an equal risk of infection by direct ingestion of soils of different matric potentials, the infection risk through leaching should be investigated further.
Potential luminescence of lux-marked E. coli O157:H7 introduced into artificial groundwater and soil was used to assess the viability and metabolic activity of the strain in situ. Although the cell densities needed for adequate detection of potential luminescence may appear high for environmental samples, such cell densities have been shown to occur in fecal material of actively shedding cattle (45) and may thus be within a similar range in top layers of soil and/or soil leachates. In both the artificial groundwater and the soil survival experiments, final values of potential luminescence per cell were similar, irrespective of the incubation time of the chromosomally lux-marked E. coli O157:H7 populations in soil. This suggested that E. coli O157:H7 populations were capable of reactivation to similar metabolic levels as when they had been introduced into soil or water.
Despite the possibility of experiencing starvation conditions, our results suggest there was no noticeable effect of starvation or matric stress on the potential metabolic activity of the cells. These findings may bear important considerations with regard to ingestion of environmental material contaminated with E. coli O157:H7. The capacity for an ingested starved E. coli O157:H7 population to reactivate may have implications regarding expression of virulence factors (e.g., intimin binding gene, Shiga toxin genes) which may influence the likelihood or severity of infection.
Currently, there are no data that document the effects of starvation in soil on the virulence traits of E. coli O157:H7. Starvation of E. coli O157:H7 in water has, however, been shown to influence the expression of the O157 antigen (12) and the development of a chlorine-resistant phenotype (20). Other studies also point to the importance of the physiological condition of E. coli O157:H7 with regard to the expression of cellular adhesion factors (13). The potential luminescence assay described here does not rely on cultivation of cells and requires only an activation step and could therefore be used for the fast quantification of nonoptimal metabolism of starved cells to further investigate the likelihood of infection arising from E. coli O157:H7 in environmental material. In addition, although not observed within the experimental framework of this validation study, trends of potential luminescence over the course of survival studies could be also potentially be used to quantify metabolic states such as sublethal injury and the viable but nonculturable response.
In this study, we successfully engineered a chromosomally lux-marked E. coli O157:H7 construct. With this construct, an assay was developed which enabled predictions of the size of potentially active populations of chromosomally lux-marked E. coli O157:H7 to be made without the limitations of the established detection methods. The in situ, metabolically linked nature of the luminescence-based assay, eliminated the requirement of cell extraction and provided the ability to discriminate viable populations of chromosomally lux-marked E. coli O157:H7 from nonviable cells. This assay provides estimates of chromosomally lux-marked E. coli O157:H7 relative population sizes within 30 min, which is advantageous over alternative, time-consuming techniques such as quantitative PCR and culture-based techniques. The attributes of this construct may thus provide a potentially useful tool for future studies of survival and transfer of E. coli O157:H7 in the environment.
Acknowledgments
This work was supported by a grant from the Scottish Executive Rural Affairs Department and the Soil Health Initiative (a joint research program involving the University of Aberdeen and the Macaulay Institute, Aberdeen, United Kingdom).
Fiona Thomson-Carter (E. coli Reference Laboratory, University of Aberdeen) and David Fenlon (Scottish Agricultural College, Veterinary Science Division, Aberdeen, United Kingdom) are gratefully acknowledged for providing strains and cattle slurry, respectively. We thank Hedda Weitz, University of Aberdeen, for valuable comments on the manuscript.
REFERENCES
- 1.Amin-Hanjani, S., A. Meikle, L. A. Glover, J. I. Prosser, and K. Killham. 1993. Plasmid and chromosomally encoded luminescence marker systems for detection of Pseudomonas fluorescens in soil. Mol. Ecol. 2:47-54. [Google Scholar]
- 2.Artz, R. R. E., and K. Killham. 2002. Survival of Escherichia coli O157:H7 in private drinking water wells: influences of protozoan grazing and elevated copper concentrations. FEMS Microbiol. Lett. 216:117-122. [DOI] [PubMed] [Google Scholar]
- 3.Bogosian, G., P. J. Morris, and J. P. O'Neil. 1998. A mixed culture recovery method indicates that enteric bacteria do not enter the viable but nonculturable state. Appl. Environ. Microbiol. 64:1736-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Campbell, G. R., J. I. Prosser, L. A. Glover, and K. Killham. 2001. Detection of Escherichia coli O157:H7 in soil and water with multiplex PCR. J. Appl. Microbiol. 91:1-7. [DOI] [PubMed] [Google Scholar]
- 5.Coia, J. E. 1998. Clinical, microbiological and epidemiological aspects of Escherichia coli O157 infection. FEMS Immunol. Med. Microbiol. 20:1-9. [DOI] [PubMed] [Google Scholar]
- 6.de Boer, E., and A. E. Heuvelink. 2000. Methods for the detection and isolation of Shigatoxin-producing Escherichia coli. J. Appl. Microbiol. Symp. Suppl. 88:133S-143S. [DOI] [PubMed] [Google Scholar]
- 7.Duncan, S., L. A. Glover, K. Killham, and J. I. Prosser. 1994. Luminescence-based detection of activity of starved and viable but nonculturable bacteria. Appl. Environ. Microbiol. 60:1308-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.European Parliament and the Council of the European Union. 2000. Directive 2000/54/EC on the protection of workers from risks related to exposure to biological agents at work. Official Journal of the European Communities L262/21. Council of the European Union, Brussels, Belgium.
- 9.Fukushima, H., K. Hoshina, and M. Gomyoda. 1999. Long-term survival of Shigatoxin-producing Escherichia coli O26, O111, and O157 in bovine feces. Appl. Environ. Microbiol. 65:5177-5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gagliardi, J. V., and J. S. Karns. 2000. Leaching of Escherichia coli O157:H7 in diverse soils under various agricultural management practices. Appl. Environ. Microbiol. 66:877-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Griffin, P. M., and R. V. Tauxe. 1991. The epidemiology of infection caused by E. coli O157 and other enterohaemorrhagic E. coli and the associated haemolytic uraemic syndrome. Epidemiol. Rev. 13:60-98. [DOI] [PubMed] [Google Scholar]
- 12.Hara-Kudo, Y., M. Miyahara, and S. Kumagai. 2000. Loss of O157 O antigenicity of verotoxin-producing Escherichia coli O157:H7 surviving under starvation conditions. Appl. Environ. Microbiol. 66:5540-5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.James, B. W., and C. W. Keevil. 1999. Influence of oxygen availability on physiology, verocytotoxin expression and adherence of Escherichia coli O157. J. Appl. Microbiol. 86:117-124. [DOI] [PubMed] [Google Scholar]
- 14.Jiang, X., J. A. W. Morgan, and M. P. Doyle. 2002. Fate of Escherichia coli O157:H7 in manure-amended soil. Appl. Environ. Microbiol. 68:2605-2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones, D. L. 1999. Potential health risks associated with the persistence of Escherichia coli O157:H7 in agricultural environments. Soil Use Manag. 15:76-83. [Google Scholar]
- 16.Josephson, K. L., C. P. Gerba, and I. L. Pepper. 1993. Polymerase chain reaction detection of nonviable bacterial pathogens. Appl. Environ. Microbiol. 59:3513-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Konowalchuk, J., N. Dickie, S. Stavric, and J. I. Speirs. 1978. Properties of an Escherichia coli cytotoxin. Infect. Immun. 20:575-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kudva, I. T., K. Blanch, and C. J. Hovde. 1998. Analysis of Escherichia coli O157:H7 survival in ovine or bovine manure and manure slurry. Appl. Environ. Microbiol. 64:3166-3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levine, M. M. 1987. Escherichia coli that cause diarrhea: enterotoxigenic, enteropathogenic, enteroinvasive and enteroadherent. J. Infect. Dis. 155:377-389. [DOI] [PubMed] [Google Scholar]
- 20.Lisle, J. T., S. C. Broadaway, A. M. Prescott, B. H. Pyle, C. Fricker, and G. A. McFeters. 1998. Effects of starvation on physiological activity and chlorine disinfection resistance in Escherichia coli O157:H7. Appl. Environ. Microbiol. 64:4658-4662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Makino, S.-I., T. Kii, H. Asakura, T. Shirahata, T. Ikeda, K. Takeshi, and K. Itoh. 2000. Does enterohemorrhagic Escherichia coli O157:H7 enter the viable but nonculturable state in salted salmon roe. Appl. Environ. Microbiol. 66:5536-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marsh, P., N. Z. Morris, and E. M. H. Wellington. 1998. Quantitative molecular detection of Salmonella typhimurium in soil and demonstration of persistance of an active but non-culturable population. FEMS Microbiol. Ecol. 27:351-363. [Google Scholar]
- 23.Martin-Laurent, F., L. Philippot, S Hallet, R. Chaussod, J. C. Germon, G. Soulas, and G. Catroux. 2001. DNA Extraction from soils: old bias for new microbial diversity analysis methods. Appl. Environ. Microbiol. 67:2354-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maule, A. 2000. Survival of enterocytotoxigenic Escherichia coli O157 in soil, water and on surfaces. J. Appl. Microbiol. Symp. Suppl. 88:71S-78S. [DOI] [PubMed] [Google Scholar]
- 25.Meikle, A., Amin- A. Hanjani, L. A. Glover, K. Killham, and J. I. Prosser. 1995. The effect of matric potential on survival and activity of a Pseudomonas fluorescens inoculum in soil. Soil Biol. Biochem. 27:881-892. [Google Scholar]
- 26.Meikle, A., K. Killham, J. I. Prosser, and L. A. Glover. 1992. Luminometric measurement of population activity of genetically modified Pseudomonas fluorescens in soil. FEMS Microbiol. Lett. 99:217-220. [DOI] [PubMed] [Google Scholar]
- 27.Meikle, A., L. A. Glover, K. Killham, and J. I. Prosser. 1994. Potential luminescence as an indicator of activation of genetically modified Pseudomonas fluorescens in liquid culture and in soil. Soil Biol. Biochem. 26:747-755. [Google Scholar]
- 28.Meng, J., and M. P. Doyle. 1998. Microbiology of Shiga toxin-producing Escherichia coli in foods, p. 92-108. In J. B. Kaper and A. D. O'Brien (ed.), Escherichia coli O157:H7 and other Shiga toxin-producing E. coli strains. American Society for Microbiology, Washington, D.C.
- 29.Mubiru, D. N., M. S. Coyne, and J. H. Grove. 2000. Mortality of Escherichia coli O157:H7 in two soils with different physical and chemical properties. J. Environ. Qual. 29:1821-1825. [Google Scholar]
- 30.Mullins, C. E. 1991. Matric potential, p. 65-93. In K. A. Smith (ed.), Soil analysis: physical methods. Marcel Decker, New York, N.Y.
- 31.Pommepuy, M., M. Butin, A. Derrien, M. Gourmelon, R. R. Colwell, and M. Cormier. 1996. Retention of enteropathogenicity by viable but nonculturable Escherichia coli exposed to seawater and sunlight. Appl. Environ. Microbiol. 62:4621-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Postma, J., and J. A. van Veen. 1990. Habitable pore space and survival of Rhizobium leguminosarum biovar trifolii introduced into soil. Microb. Ecol. 21:149-161. [DOI] [PubMed] [Google Scholar]
- 33.Prosser, J. I., K. Killham, L. A. Glover, and E. A. S. Rattray. 1996. Luminescence-based systems for detection of bacteria in the environment. Crit. Rev. Biotechnol. 16:157-183. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 35.Schmidt, H., J. Scheef, H. I. Huppertz, M. Frosch, and H. Karch. 1999. Escherichia coli O157:H7 and O157:H− strains that do not produce Shiga toxin: phenotypic and genetic characterization of isolates associated with diarrhea and hemolytic-uremic syndrome. J. Clin. Microbiol. 37:3491-3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Siragusa, G. R., K. Nawotka, S. D. Spilman, P. R. Contag, and C. H. Contag. 1999. Real-time monitoring of Escherichia coli O157:H7 adherence to beef carcass surface tissues with a bioluminescent reporter. Appl. Environ. Microbiol. 65:1738-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Su, C. Y., and L. J. Brandt. 1995. Escherichia coli O157:H7 infections in humans. Ann. Intern. Med. 123:698-714. [DOI] [PubMed] [Google Scholar]
- 38.Turnbull, G. A., J. A. Morgan, W. J. M. Whipps, and J. R. Saunders. 2001. The role of bacterial motility in the survival and spread of Pseudomonas fluorescens in soil and in the attachment and colonisation of wheat roots. FEMS Microbiol. Ecol. 36:21-31. [DOI] [PubMed] [Google Scholar]
- 39.Unge, A., R. Tombolini, L. Molbak, and J. K. Jansson. 1999. Simultaneous monitoring of cell number and metabolic activity of specific bacterial populations with a dual gfp-luxAB marker system. Appl. Environ. Microbiol. 65:813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uyttendale, M., I. Taverniers, and J. Debevere. 2001. Effect of stress induced by subobtimal growth factors on survival of Escherichia coli O157:H7. Int. J. Food Microbiol. 66:31-37. [DOI] [PubMed] [Google Scholar]
- 41.Waddell, T. E., and C. Poppe. 2000. Construction of mini-Tn10luxABcam/Ptac-ATS and its use for developing a bacteriophage that transduces bioluminescence to Escherichia coli O157:H7. FEMS Microbiol. Lett. 182:285-289. [DOI] [PubMed] [Google Scholar]
- 42.Wang, G., and M. P. Doyle. 1998. Survival of enterohemorrhagic Escherichia coli O157:H7 in water. J. Food Prot. 61:662-667. [DOI] [PubMed] [Google Scholar]
- 43.Winson, M. M., S. Swift, P. J. Hill, C. M. Sims, G. Griesmayr, B. Bycroft, P. Williams, and G. S. A. B. Stewart. 1998. Engineering the luxCDABE genes from Photorhabdus luminescens to provide a bioluminescent reporter for constitutive and promoter probe plasmids and mini-Tn5 constructs. FEMS Microbiol. Lett. 163:193-202. [DOI] [PubMed] [Google Scholar]
- 44.Wong, P. T. W., and D. M. Griffin. 1976. Bacterial movement at high matric potentials. Soil Biol. Biochem. 8:215-218. [Google Scholar]
- 45.Zhao, T., M. P. Doyle, J. Shere, and L. Garber. 1995. Prevalence of enterohemorrhagic Escherichia coli O157:H7 in a survey of dairy herds. Appl. Environ. Microbiol. 61:1290-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]